Abstract
We define the epidemiology of predominant and sporadic methicillin-resistant Staphylococcus aureus (MRSA) strains in a central teaching and referral hospital in Kuala Lumpur, Malaysia. This is done on the basis of spa sequencing, multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCCmec) typing, and virulence gene profiling. During the period of study, the MRSA prevalence was 44.1%, and 389 MRSA strains were included. The prevalence of MRSA was found to be significantly higher in the patients of Indian ethnicity (P < 0.001). The majority (92.5%) of the isolates belonged to ST-239, spa type t037, and possessed the type III or IIIA SCCmec. The arginine catabolic mobile element (ACME) arcA gene was detected in three (1.05%) ST-239 isolates. We report the first identification of ACME arcA gene-positive ST-239. Apart from this predominant clone, six (1.5%) isolates of ST-22, with two related spa types (t032 and t4184) and a singleton (t3213), carrying type IVh SCCmec, were detected for the first time in Asia. A limited number of community-acquired (CA) MRSA strains were also detected. These included ST-188/t189 (2.1%), ST-1/t127 (2.3%), and ST-7/t091 (1%). Panton-Valentin leukocidin (PVL) was detected in all ST-1 and ST-188 strains and in 0.7% of the ST-239 isolates. The majority of the isolates carried agr I, except that ST-1 strains were agr III positive. Virulence genes seg and sei were seen only among ST-22 isolates. In conclusion, current results revealed the predominance of ST-239-SCCmec III/IIIA and the penetration of ST-22 with different virulence gene profiles. The emergence in Malaysia of novel clones of known epidemic and pathogenic potential should be taken seriously.
Methicillin-resistant Staphylococcus aureus (MRSA) is an established human pathogen that causes both health care-associated (HA) and community-acquired (CA) infections. Modern MRSA has evolved from several successful clonal lineages of methicillin-susceptible S. aureus strains via acquisition of a mobile genetic element called staphylococcal cassette chromosome mec (SCCmec). This element contains the mecA gene, which encodes penicillin-binding protein 2′ (PBP2′) with significantly reduced affinity for β-lactams (36).
Increased emergence of multidrug resistance among MRSA strains has become a major concern in the hospital environment, as it invokes a tremendous financial burden and enhanced morbidity and mortality due to hard-to-treat systemic infections (7). Genotyping data from large international studies have shown that a limited number of major clones of MRSA display an enhanced propensity to spread and cause opportunistic human infections in various parts of the world (5, 13, 33). MRSA was introduced into Malaysia in the early 1970s (26), and a review of the records of the microbiology laboratories of all state hospitals in Malaysia showed that the proportion of MRSA isolates from S. aureus-infected individuals had been approximately 21% throughout the last few years.
Phenotypic MRSA typing methods are not suited to detailed epidemiological surveillance (20, 40). Several superior molecular typing methods can be used for supporting infection control and for tracing the nosocomial sources and transmission routes of bacterial pathogens. Among these methods, multilocus sequence typing (MLST) has recently been proven to be the best for long-term global epidemiological and bacterial population genetics studies. It is a discriminatory genotypic method where isolates are typed by sequencing variable regions of housekeeping genes. A combination of alleles from the seven loci forms a sequence type (ST). Similar STs are grouped into clonal complexes (13). spa typing has been shown to be as discriminatory as the current gold standard method, pulsed-field gel electrophoresis (PFGE) (10). spa types can be determined via amplification and sequencing of the X region of the protein A gene (spa), which contains polymorphic direct repeats. StaphType (version 1.4; Ridom GmbH, Würzburg, Germany) provides a software tool enabling straightforward sequence analysis and designation of spa types by synchronization via a central server (42). spa typing is useful in analyzing hospital outbreaks and in identifying genetic changes that occur over a relatively short time span (14, 43). Molecular typing data on HA MRSA isolates in Malaysia are sparse in comparison with those on strains deriving from Europe, the United States, or Japan. This is notwithstanding the steadily increasing but already high rates of MRSA infections in Malaysia (38). Pilot data hinted at the predominance of a single MRSA genotype in Malaysia. This is similar to the situation in many other Asian countries (1, 23, 31).
For the present study, clinical MRSA isolates associated with various infections were collected from the largest public tertiary referral hospital in Kuala Lumpur, the capital of Malaysia. The epidemiology of MRSA in this hospital (HKL) will most likely reflect the nation's epidemiology as it is the major government referral hospital for patients from all states in Malaysia. Each year at least 1 million people (∼3.8% of the total Malaysian population) are treated here. HKL records an annual MRSA prevalence over 40%. Many of these MRSA strains are multidrug resistant. Additional epidemiological studies are clearly warranted in order to increase the insight into the dynamics of MRSA epidemiology in Malaysia.
The aims of the present study were to characterize the current Malaysian MRSA isolates and determine their molecular epidemiology by MLST, spa typing, SCCmec typing, and virulence gene profiling.
MATERIALS AND METHODS
Clinical setting and bacterial strains.
Hospital Kuala Lumpur (HKL) is the largest government tertiary referral hospital, with 81 wards and 2,502 beds. It is located in Kuala Lumpur. An average of 40% MRSA prevalence is recorded annually in this hospital. For conducting an adequate statistical analyses, taking into account the prevalence of MRSA in this hospital, the sample size (SS) was determined using the equation SS = Z2 × p × (1 − p)/c2, where Z is the statistic for the level of confidence (for a level of confidence of 95% or a 5% type I error, which is conventional, Z is 1.96), p is the estimated proportion of an attribute that is present in the population (in the present study, p = 0.468), and c is the desired level of precision, which is 0.05 (8, 25).
MRSA samples used in the current study were collected from October 2007 to September 2008. Out of a total number of 1,887 MRSA strains, a random selection of 389 strains was investigated in molecular detail. Thirty-two nonrepetitive MRSA samples per month (approximately eight per week) were collected from clinical samples obtained from hospitalized patients. The first 8 isolates (irrespective of the ward) per week were collected. Demographic and culture data such as age, gender, ethnicity, ward, site of isolation, and type of sample were all recorded. All isolates previously identified to the species level by the hospital laboratory were reconfirmed in the microbiology laboratory at Universiti Putra Malaysia by standard methods such as Gram staining, catalase testing, tube coagulation, mannitol testing, and MRSA screen latex agglutination testing (Denka Seiken Co., Ltd., Tokyo, Japan). Isolates were confirmed as MRSA by oxacillin and cefoxitin susceptibility testing according to the CLSI guidelines (6). All isolates were also reevaluated for the presence of the mecA gene by PCR (32). Confirmed MRSA isolates were then stored at −80°C in Luria-Bertani broth supplemented with 20% glycerol awaiting further investigation.
SCCmec typing, spa typing, and MLST.
Chromosomal DNA was extracted using the DNeasy kit (Qiagen Inc.) according to the manufacturer's instructions. SCCmec typing was carried out according to the protocol described by Zhang et al. (50). Confirmation of SCCmec type V and subtyping of type IV were done based on the variation in the ccrC and ccrA2B2 genes, as recently described by Kondo et al. (24) and Milheiriço et al. (29).
All MRSA isolates, irrespective of their SCCmec types, were subjected to spa typing according to the method of Shopsin et al. (41) using the primers SpaF (AGACGATCCTTCGGTGA) and SpaR (CAGCAGTAGTGCCGTTTG). The amplified spa gene fragments were purified and sequenced. spa types were assigned by using StaphType software (version 1.4; Ridom GmbH, Würzburg, Germany), as described by Harmsen et al. (16). By application of the BURP (based upon repeat pattern) algorithm, spa types with more than five repeats were clustered into distinct groups.
MLST was conducted for representatives of each spa lineage, including the new and uncommon ones, as described previously (13). Twenty-six selected strains were processed for MLST. Sequence types (STs) and clonal clusters (CCs) were assigned using the S. aureus MLST database (www.mlst.net) hosted by Imperial College in London, United Kingdom.
PCR-based assays for virulence factors of S. aureus.
Four different categories of virulence factors were screened. These included cytolytic toxin genes such as pvl (28); superantigen genes including sea, seb, sec, sed, see, seg, seh, sei, and tsst (21); exotoxin genes such as eta and etb (26); genes encoding adhesions such as cna (2), fnbA (51), ica, and icaD (37); and the newly identified arginine catabolic mobile element (ACME) gene cluster, which is involved in bacterial growth and development (4). Multiplex PCR-based protocols for allotyping agr classes I to IV were performed as described elsewhere (27).
Statistical analysis.
Statistical analyses were made with SPSS version 14 software (SSPS, Inc., Chicago, IL). Categorical variables between two groups were compared by means of chi-square or Fisher exact tests. The latter was done if 20% of the expected values were smaller than 5. A P value of <0.05 indicated statistical significance. A goodness-of-fit test was used to test the prevalence of MRSA strains among different ethnic groups in Malaysia. The model used for the test is the Malaysian population statistics (http://www.statistics.gov.my), which shows that 65%, 26%, 7.7%, and 1.3% of the Malaysian population are Malay (bumiputras), Chinese, Indians, and others, respectively.
RESULTS
Demographical characterization of MRSA strains.
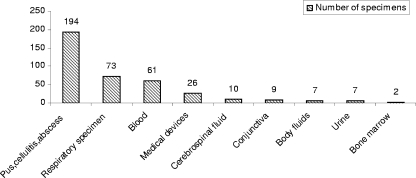
From October 2007 to September 2008, 44.1% (n = 1,887) of all S. aureus strains (n = 4,280) causing infections in the hospital were identified as MRSA and 389 of them were collected for molecular investigation. The frequencies of MRSA isolates obtained from various wards and different type of specimens are summarized in Table 1 and Fig. 1.
TABLE 1.
Distribution of MRSA isolates based on the ward of isolation
| Ward | No. (%) of MRSA isolates characterized in the present study | Total no. (%) of MRSA isolates during the study period |
|---|---|---|
| General medicine | 97 (24.9) | 378 (20.1) |
| Pediatrics | 30 (7.7) | 174 (9.2) |
| General surgery | 54 (13.9) | 212 (11.2) |
| Urology/nephrology | 60 (15.4) | 297 (15.7) |
| Neurosurgery | 51 (13.1) | 286 (15.2) |
| Orthopedic surgery | 68 (17.5) | 243 (12.8) |
| Maternity | 2 (0.5) | 15 (0.8) |
| ICUa | 27 (6.9) | 94 (5.1) |
| Unknown | 188 (9.9) | |
| Total | 389 (100) | 1,887 (100) |
ICU, intensive care unit.
FIG. 1.
Number of MRSA strains isolated from different types of specimens: pus, cellulites, and abscess, referred as wound and soft-tissue infections; respiratory specimens (pulmonary secretion, sputum, and tracheal aspirate); medical devices (central nervous system tip and catheter); cerebrospinal fluid (CSF); conjunctiva; body fluids (peritonea and synovial); and bone marrow.
All patients were divided into four groups according to age. The infant group ages ranged from 1 to 11 months (9 subjects, 2.3%), the child group ages ranged from 1 to 15 years (31 subjects, 8%), the adult group ages ranged from 16 to 65 years (271 subjects, 69.7%), and the elderly were more than 65 years old (78 subjects, 20.1%). The median age of patients was 47.8 years, and the age range was 4 days to 88 years (standard deviation [SD], ±21.1 years). The male-to-female patient ratio was 61.2:38.8, which showed no significance (χ2 = 0.282) for gender relatedness of MRSA infection. The highest prevalence of MRSA was seen in the adult category. When data were stratified according to the three major ethnic groups in Malaysia, it was found that MRSA was isolated from 207 (53.2%) Malays, 88 (22.6%) Chinese, and 94 (24.2%) Indians. The prevalence of MRSA infection among Indians was found to be significantly elevated (goodness χ2 = 88.09, P < 0.001).
SCCmec typing, spa typing, and MLST.
SCCmec typing identified that the vast majority (92.8%) of isolates carried either SCCmec type III (n = 78 or 20%) or its variant IIIA (n = 283 or 72.8%), while 22 (5.7%) and 6 (1.5%) isolates harbored SCCmec types V and IVh, respectively.
spa typing of the 389 isolates revealed 13 different spa types. Repeats among the spa types varied from 4 (t4150) to 17 (t4184). The majority (73.5%) of the isolates belonged to t037. The BURP algorithm was used to group all isolates into definite clonal lineages on the basis of sequence relatedness. Since clustering parameters excluded spa types shorter than five repeats, one spa type (t4150) was excluded from BURP grouping. The phylogenetic tree produced two different groups, and 4 other spa types were singletons. Group 1 contained 6 spa types, with t037 as the founder, while group 2 contained 2 spa types (t032 and t4184), with t032 as the founder. Among the 13 spa types 3 new spa types were identified and deposited in the spa database. Two of these new types clustered with t037, while one resembled t032.
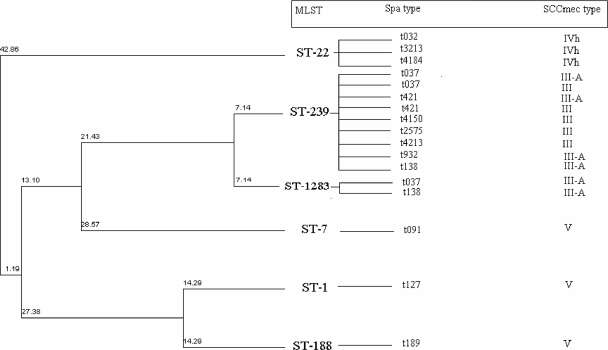
MLST (60 isolates) was performed on all isolates belonging to the different spa types except for the two predominant ones (t037 and t421), for which 10 and 6 isolates, respectively, were MLST typed. This identified six STs, ST-239 (CC8) (n = 360; 92.5%), ST-1 (CC1) (n = 9; 2.3%), ST-188 (CC1) (n = 8; 2.1%), ST-22 (CC22) (n = 6; 1.5%), ST-7 (CC7) (n = 4; 1%), and ST-1283 (CC8), confirming the existence of six distinct clones among the 389 MRSA isolates. Application of the eBURST algorithm to the six STs clustered both ST-239 and ST-1283 in CC8. ST-1283 was described for the first time in this study and deposited at the MLST database. This ST was recovered from a cerebrospinal fluid sample and is a single-locus variant (SLV) of ST-239, differing from this genotype by a single synonymous point mutation in the gene encoding guanylate kinase (gmk). The other four STs and CCs were ST-1 (CC1), ST-22 (CC22), ST-188 (CC1), and ST-7 (CC7). ST-188 is closely related to ST-1, with differences only in the two loci arcC (carbamate kinase gene) and gmk (http://saureus.mlst.net/). Figure 2 illustrates the molecular characterization of strains representing the different SCCmec types, spa types, and STs.
FIG. 2.
Phylogenic tree of MRSA strains included in the present study showing the combination of data from spa typing, MLST, and SCCmec typing. The tree is a dendrogram based on the unweighted pair group method using average linkages (UPGMA) and constructed based on the pairwise differences in the MLST allelic profiles. The numbers above the row lines demonstrate the genetic distances between the strains: two isolates that differ at one of seven loci differ at a distance of 14.3% (1/7). The diagram shows distances along each branch as 7.1%, half that of the single-locus difference. In this way the distance between ST-239 and ST-1283, defined by a single locus, is 7.1% whereas for ST-1 and ST-188, with two different loci, the distance is again 14.3%.
Toxin gene profile.
All MRSA strains irrespective of STs and spa types were examined for the presence of several virulence factors (Table 2). All strains were commonly positive for adhesin genes such as the fibronectin binding protein gene (fnbA) and the biofilm-related genes icaA and icaD, while the collagen adhesin gene (cna) was amplified in 384 (98.7%) strains. Among the superantigen genes, sea was the most prevalent one and was detected in 337 (86.6%) strains (ST-239, -1, -188, and -7), followed by seh, detected in 9 strains (ST-1), while sec was found in 4 strains (ST-22). seg and sei genes were demonstrated only in ST-22 isolates. None of the other superantigenic toxin genes such as seb, sed, and tsst were observed in the strains investigated. The pvl gene was amplified in 19 (4.9%) strains (0.7% of ST-239 strains and 100% of ST-1 and ST-188 strains). Three ST-239 strains (1.1%) harbored the ACME-associated arcA gene, two from wounds and one from a respiratory infection. Analyzing the relationship between the toxin genes and the STs by χ2 test revealed that there is an association between MLST and certain virulence genes: sea and ST-239 are correlated (χ2 = 54.84, P < 0.001) and ST-22 and sec, seg, and sei are also correlated (χ2 = 389, P < 0.001), whereas the association between ST-188, ST-1, and pvl (χ2 = 389, P < 0.001) is clearly significant. agr typing grouped the majority of the isolates in the agr I category, except that ST-1 strains were of the agr III class.
TABLE 2.
Distribution of different virulence factors among MRSA isolates
| No. of isolates | MLST | spa typea | No. (%) of isolates carrying: |
agr type | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACME arcA | sea | seh | sec, seg, and sei | pvl | icaA | icaD | fnbA | cna | ||||
| 284 | ST-239 | t037 (10) | 3 (1.05) | 243 (85.5) | 2 (0.7) | 284 (100) | 284 (100) | 284 (100) | 284 (100) | I | ||
| 1 | ST-1283 | t037 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 61 | ST-239 | t421 (6) | 59 (96.7) | 61 (100) | 61 (100) | 61 (100) | 61 (100) | I | ||||
| 5 | ST-239 | t4213 (5) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | I | ||||
| 4 | ST-239 | t4150 (4) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | I | ||||
| 1 | ST-239 | t138 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 1 | ST-1283 | t138 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 1 | ST-239 | t932 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 1 | ST-239 | t2575 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 4 | ST-22 | t032 (4) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | I | ||||
| 1 | ST-22 | t4184 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | I | ||||
| 1 | ST-22 | t3213 (1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ||||
| 9 | ST-1 | t127 (9) | 9 (100) | 9 (100) | 9 (10) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | III | ||
| 8 | ST-188 | t189 (8) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | 8 (100) | I | |||
| 4 | ST-7 | t091 (4) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | I | |||||
| 3 | ST-239 | NTb (3) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | 3 (100) | I | ||||
The numbers in parentheses indicate the numbers of isolates that were typed by MLST for that particular spa type.
NT, nontypeable.
DISCUSSION
In agreement with earlier reports, the prevalence of MRSA in the study hospital did not vary by gender (22). However, the MRSA infection rates are significantly different between ethnic groups and the rate is clearly elevated in the Indian population. A similar observation was reported for CA MRSA in African-American children (39).
The highest rate of MRSA, as reported from our general medicine ward, is in the dermatology ward, with a high rate of soft-tissue infection. The short hospital stay and adequate antibiotic therapy in otherwise healthy patients in the maternity ward explain the low prevalence in that ward (31, 46).
Several international epidemiological investigations demonstrated that most hospital-acquired MRSA infections are caused by a relatively small number of epidemic MRSA clones spread worldwide. The most frequently encountered clones are the Iberian (ST-247-IA), Brazilian/Hungarian (ST-239-III), Berlin (ST-45-IV), New York/Japan (ST-5-II), pediatric (ST-5-IV), EMRSA-15 (ST-22-IV), and EMRSA-16 (ST-36-II) clones (33). We here show the high endemicity of a limited number of clones including ST-239 (CC8), which is the predominant Malaysian clone. This clone usually is SCCmec type III/IIIA and spa type t037, which is dominant in most other Asian countries except South Korea and Japan (23). Six spa types were obtained for ST-239, which shows that there are at least 6 subtypes circulating in a single hospital. Surprisingly, ST-22 (CC22) of SCCmec type IVh, spa t032, seems to have recently penetrated Malaysia.
We present the first cases of infection by ST-22, SCCmec type IVh, with three different spa types (t932, t3213, and t4184) in Malaysia. These strains were mainly isolated from blood (66.6%) or skin and soft-tissue infection (33.3%). SCCmec IVh, present in a recently described specific subtype of EMRSA-15 (ST-22), was reported for the first time in Portugal (29) but shortly after from places as distant as Sweden (3) and lately from Spain (48). The SCCmec IV element is small in comparison to elements such as SCCmec type III; the small size enables easier spread among S. aureus strains. As a possible consequence of its enhanced mobility, it is also more variable than other SCCmec types. The detection of ST-22 (which is one of the predominant MRSA clones in Europe, Australia, and New Zealand [40]) in Singapore (18) and later in Malaysia strengthens the case for the epidemic nature of this clone and also suggests that it may easily spread to other Southeast Asian countries.
S. aureus produces a wide variety of exoproteins that contribute to its ability to colonize and cause disease in mammalian hosts. The majority of the virulence factors produced in S. aureus are under the control of the agr system. Although agr II and III are found in some well known clones (pediatric, New York/Japan, and EMRSA-16) (15, 21), the most predominant agr group among worldwide MRSA clones is agr group I (47). A similar situation was observed in our isolates, where 96.7% of the isolates were agr I while only 3.3% carried agr III. Our results illustrate that the pvl locus is more common (80.9%) among CA MRSA strains that belonged to ST-1- SCCmec V-agr III and ST-188-SCCmec V-agr I, while a small percentage (0.7%) were seen among the ST-239-SCCmec III and III-A-agr I isolates. pvl-positive ST-1-SCCmec IV-agr III isolates were first reported in the United States, but strains recently detected from Europe and Asia are of agr type I or II (44). Recently, ST-1 and ST-188 MRSA strains with type V SCCmec were reported from Singapore (17, 19). Furthermore, pvl-positive HA MRSA isolates harboring SCCmec type III were found in Algeria (34) and China (49). In general, pvl is carried in less than 5% of the MRSA strains that harbor SCCmec types I to III (9).
The current study for the first time reports the presence of the ACME arcA gene in ST-239 SCCmec type III MRSA isolates. ACME arcA, which contributes to growth and survival by encoding resistance and virulence determinants that enhance fitness and pathogenicity, was first detected in USA300 (ST-8) (4, 11).
Exotoxins with superantigenic properties are responsible for strong inflammatory responses that contribute to the severity of staphylococcal infections. Regardless of the STs, most (86.6%) of the isolates studied carried the sea gene. Our results are in accordance with those obtained for Russian isolates, where 88% of the MRSA strains from clinical samples were sea positive (12). Although in previous studies sea was not detected in the Brazilian clone in general (45), it was found in our isolates which matched the Brazilian and Hungarian clones in MLST and SCCmec type. Enterotoxin H was found to be present only among the Malaysian ST1 isolates. Enterotoxin H has a particularly high affinity for binding to the major histocompatibility complex type II molecules and is usually reported in CA MRSA strains (35). Among all isolates, only the ST-22 clone shared the enterotoxin gene cluster locus (egc), comprising five superantigenic enterotoxin genes (seg, sei, sem, sen, and seo). The combination of classical (enterotoxin C-positive) (21) and variant (enterotoxin C-negative) strains (30) among our isolates reconfirms that diverse ST-22 strains have penetrated our hospital. Despite of high percentages of wound and soft-tissue infections, exfoliative toxin genes (eta and etb) were not detected in any of our strains, indicating the role of other virulence factors involved in establishing the infections.
In conclusion, this study identified three main types of MRSA in HKL. The majority of strains are ST-239, which is predominant in several other Asian countries as well. ST-22, first described in the United Kingdom and later in other European countries, Australia, New Zealand, and Singapore, has now penetrated Malaysia, and this emphasizes the epidemic nature of strains with this ST. In addition, minor groups of CA MRSA strains detected in the current study indicate the ability of such strains to settle in the hospital environment. The combination of ST-1-SCCmec V-agr III and ST-188-SCCmec V-agr I was commonly detected among CA MRSA strains. This is the first study to report the ACME arcA gene in ST-239-SCCmec type III MRSA. The presence of a series of virulence determinants in the local MRSA strains suggests the ability of these strains to cause a broad spectrum of infectious diseases under favorable conditions.
Acknowledgments
We are grateful to Teruyo Ito (Juntendo University) for providing reference SCCmec type strains and confirming the SCCmec type IVh MRSA isolates. We express our special thanks to Dag Harmsen (University Hospital Munster) and Alexander Friedrich for helping us to implement BURP analysis for spa types.
We take this opportunity to thank Hospital Kuala Lumpur for kindly providing all of the MRSA isolates. This work was supported by the Ministry of Science, Technology and Innovation Malaysia (MOSTI) Science Fund with grant numbers 06-01-04-SF0885 and 02-01-04-SF0853.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Aires de Sousa, M., M. I. Crisóstomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciola, C. R., D. Campoccia, S. Gamberini, L. Baldassarri, and L. Montanaro. 2005. Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopedic infections associated to different types of implant. FEMS Microbiol. Lett. 246:81-86. [DOI] [PubMed] [Google Scholar]
- 3.Berglund, C., T. Ito, X. X. Ma, M. Ikeda, S. Watanabe, B. Söderquist, and K. Hiramatsu. 2009. Genetic diversity of methicillin-resistant Staphylococcus aureus carrying type IV SCCmec in Orebro County and the western region of Sweden. J. Antimicrob. Chemother. 63:32-41. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, S., H. Deshmukh, C. Nelson, I. Bae, M. Stryjewski, J. Federspiel, G. Tonthat, T. Rude, S. Barriere, R. Corey, and V. J. Fowler. 2008. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J. Clin. Microbiol. 46:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, M., G. Dickinson, H. De Lencastre, and A. Tomasz. 2004. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J. Clin. Microbiol. 42:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, W. 1999. Biostatistics: a foundation for analysis in the health sciences. John Wiley & Sons, New York, NY.
- 9.Deresinski, S. 2005. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 40:562-573. [DOI] [PubMed] [Google Scholar]
- 10.Deurenberg, R. H., and E. E. Stobberingh. 2008. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 8:747-763. [DOI] [PubMed] [Google Scholar]
- 11.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 12.Dmitrenko, O. A., V. Prokhorov, F. S. Fluer, T. N. Suborova, I. I. Volkov, V. I. Karabak, and A. L. Gintsburg. 2006. Detection of the genes of pyrogenic toxins of superantigens in clinical isolates of methicillin resistant Staphylococcus aureus. Zh. Mikrobiol. Epidemiol. Immunobiol. 2:36-42. [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faria, N. A., J. A. Carrico, D. C. Oliveira, M. Ramirez, and H. de Lencastre. 2008. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Clin. Microbiol. 46:136-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes, A. R., S. Vinga, M. Zavolan, and H. de Lencastre. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu, L. Y., Y. L. Koh, N. L. Chlebicka, T. Y. Tan, P. Krishnan, R. T. Lin, N. Tee, T. Barkham, and T. H. Koh. 2006. Establishment of ST30 as the predominant clonal type among community-associated methicillin-resistant Staphylococcus aureus isolates in Singapore. J. Clin. Microbiol. 44:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, L. Y., T. H. Koh, K. Singh, M. L. Kang, A. Kurup, and B. H. Tan. 2005. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 43:2923-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, L. Y., T. H. Koh, T. Y. Tan, T. Ito, X. X. Ma, R. T. Lin, and B. H. Tan. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus in Singapore: a further six cases. Singapore Med. J. 47:20-26. [PubMed] [Google Scholar]
- 20.Ip, M., D. J. Lyon, F. Chio, M. C. Enright, and A. F. Cheng. 2003. Characterization of isolates of methicillin-resistant Staphylococcus aureus from Hong Kong by phage typing, pulsed-field gel electrophoresis, and fluorescent amplified-fragment length polymorphism analysis. J. Clin. Microbiol. 41:4980-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kairam, N., M. E. Silverman, D. F. Salo, E. Baorto, B. Lee, and C. S. Amato. 8 July 2009, posting date. Cutaneous methicillin-resistant Staphylococcus aureus in a suburban community hospital pediatric emergency department. J. Emerg. Med. [Epub ahead of print.] doi: 10.1016/j.jemermed.2009.05.015. [DOI] [PubMed]
- 23.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawanga, S. K., and S. Lemeshow. 1991. Sample size determination in health studies: a practical manual. WHO, Geneva, Switzerland.
- 26.Lim, V. K., and H. I. Zulkifli. 1987. Methicillin resistant Staphylococcus aureus in a Malaysian neonatal unit. Singapore Med. J. 28:176-179. [PubMed] [Google Scholar]
- 27.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 29.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: 'SCCmec IV multiplex.' J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 30.Moore, P. C. L., and J. A. Lindsay. 2002. Molecular characterization of the dominant UK methicillin-sensitive Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 151:516-521. [DOI] [PubMed] [Google Scholar]
- 31.Nor Shamsudin, M., Z. Sekawi, A. van Belkum, and V. Neela. 2008. First community-acquired meticillin-resistant Staphylococcus aureus in Malaysia. J. Med. Microbiol. 57:1180-1181. [DOI] [PubMed] [Google Scholar]
- 32.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 34.Ramdani-Bouguessa, N., M. Bes, H. Meugnier, F. Forey, M. E. Reverdy, G. Lina, F. Vandenesch, M. Tazir, and J. Etienne. 2006. Detection of methicillin-resistant Staphylococcus aureus strains resistant to multiple antibiotics and carrying the Panton-Valentine leukocidin genes in an Algiers hospital. Antimicrob. Agents Chemother. 50:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren, K., J. D. Bannan, and V. Pancholi. 1994. Characterization and biological properties of a new staphylococcal exotoxin. J. Exp. Med. 180:1675-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohde, H., J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2001. Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J. Clin. Microbiol. 39:4595-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sam, I. C., M. Kahar-Bador, Y. F. Chan, S. K. Loong, and G. Mohd Nor. 2008. Multisensitive community-acquired methicillin-resistant Staphylococcus aureus infections in Malaysia. Diagn. Microbiol. Infect. Dis. 62:437-439. [DOI] [PubMed] [Google Scholar]
- 39.Sattler, C. A., E. J. Mason, and S. L. Kaplan. 2002. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. Pediatr. Infect. Dis. J. 21:910-916. [DOI] [PubMed] [Google Scholar]
- 40.Schlichting, C., C. Branger, J. M. Fournier, W. Witte, A. Boutonnier, C. Wolz, P. Goullet, and G. Döring. 1993. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J. Clin. Microbiol. 31:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tristan, A., M. Bes, H. Meugnier, G. Lina, B. Bozdogan, P. Courvalin, M. E. Reverdy, M. C. Enright, F. Vandenesch, and J. Etienne. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg. Infect. Dis. 13:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tristan, A., T. Ferry, G. Durand, O. Dauwalder, M. Bes, G. Lina, F. Vandenesch, and J. Etienne. 2007. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J. Hosp. Infect. 65:105-109. [DOI] [PubMed] [Google Scholar]
- 46.Udo, E. E., N. Al-Sweih, S. Mohanakrishnan, and P. W. West. 2006. Antibacterial resistance and molecular typing of methicillin-resistant Staphylococcus aureus in a Kuwaiti general hospital. Med. Princ. Pract. 15:39-45. [DOI] [PubMed] [Google Scholar]
- 47.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. Van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vindel, A., O. Cuevas, E. Cercenado, C. Marcos, V. Bautista, C. Castellares, P. Trincado, T. Boquete, M. Pérez-Vázquez, M. Marín, and E. Bouza. 2009. Methicillin-resistant Staphylococcus aureus in Spain: molecular epidemiology and utility of different typing methods. J. Clin. Microbiol. 47:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, F., Z. Chen, C. Liu, X. Zhang, X. Lin, S. Chi, T. Zhou, Z. Chen, and X. Chen. 2008. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalized patients in China. Clin. Microbiol. Infect. 14:381-384. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, H., Z. Y. Xiong, H. P. Li, Y. L. Zheng, and Y. Q. Jiang. 2006. An immunogenicity study of a newly fusion protein Cna-FnBP vaccinated against Staphylococcus aureus infections in a mice model. Vaccine 24:4830-4837. [DOI] [PubMed] [Google Scholar]