Abstract
The sodC gene has been reported to be a useful marker for differentiating nontypeable (NT) Haemophilus influenzae from Haemophilus haemolyticus in respiratory-tract samples, but discrepancies exist as to the prevalence of sodC in NT H. influenzae. Therefore, we used a microarray-based, “library-on-a-slide” method to differentiate the species and found that 21 of 169 (12.4%) NT H. influenzae strains and all 110 (100%) H. haemolyticus strains possessed the sodC gene. Multilocus sequence analysis confirmed that the 21 NT H. influenzae strains were H. influenzae and not H. haemolyticus. An inactive sodC gene has been reported in encapsulated H. influenzae strains belonging to phylogenetic division II. Capsule-specific Southern hybridization and PCR and a lack of copper/zinc-cofactored superoxide dismutase (CuZnSOD) expression indicated that 6 of the 21 sodC-containing NT H. influenzae strains in our study were likely capsule-deficient mutants belonging to phylogenetic division II. DNA sequence comparisons of the 21 H. influenzae sodC genes with sodC from H. haemolyticus or encapsulated H. influenzae demonstrated that the sodC genes of the six H. influenzae capsule-deficient mutants were, on average, 99% identical to sodC from encapsulated H. influenzae but only 85% identical to sodC from H. haemolyticus. The sodC genes from 2/15 NT H. influenzae strains were similarly more closely related to sodC from encapsulated strains, while sodC genes from 13 NT H. influenzae strains were almost 95% identical to sodC genes from H. haemolyticus, suggesting the possibility of interspecies recombination in these strains. In summary, this study demonstrates that sodC is not completely absent (9.2%) in true NT H. influenzae strains.
Haemophilus influenzae asymptomatically colonizes the pharyngeal cavity of humans but may cause either systemic or respiratory-tract infections. Those strains that possess a polysaccharide capsule, serotypes a through f, cause most H. influenzae invasive infections (11), whereas nonencapsulated or nonserotypeable (NT) H. influenzae strains are associated with localized respiratory-tract diseases, such as otitis media and exacerbation of chronic obstructive pulmonary disease (COPD) (19). In addition, a cryptic genospecies of NT H. influenzae is also associated with neonatal invasive infections and adult urogenital infections (36). Phylogenetically, however, the cryptic genospecies is more closely related to Haemophilus haemolyticus than to H. influenzae of the respiratory tract (26, 27).
H. haemolyticus is a commensal of the pharyngeal cavity, and H. haemolyticus and NT H. influenzae isolated from the respiratory tract overlap taxonomically and phylogenetically, likely through the exchange of DNA by natural transformation (1, 20, 21, 32). The species have been traditionally differentiated by the ability of H. haemolyticus to hemolyze horse blood (11, 25), but recent studies have identified significant numbers of nonhemolytic H. haemolyticus in NT H. influenzae collections isolated from throat or sputum samples (21, 37). Since sputum samples are used to monitor COPD exacerbation, accurate differentiation of NT H. influenzae from H. haemolyticus is clinically important (5, 21).
Fung et al. (9) recently suggested that the presence of the sodC gene or activity of its cognate protein, copper/zinc-cofactored superoxide dismutase (CuZnSOD), can differentiate H. haemolyticus from NT H. influenzae, as sodC was present in 20 H. haemolyticus isolates and absent in 20 NT H. influenzae isolates. In addition, a previous study failed to find the sodC gene in 45 NT H. influenzae disease isolates (18). Prior to these studies, however, 12 of 26 NT H. influenzae strains were found to hybridize weakly with a sodC gene probe, and these strains also displayed CuZnSOD activity (13). The discrepancy between these studies has never been explained. Aside from the questionable absence of sodC in NT H. influenzae of the respiratory tract, the gene and its associated CuZnSOD activity are present in the H. influenzae cryptic genospecies associated with neonatal invasive infections and adult urogenital infections (18). In addition, the gene is present among serotype e encapsulated strains and in strains that belong to the capsular phylogenetic division II, a multilocus enzyme electrophoresis (MLEE)-based division that contains some serotype a and b (Hib) strains and all serotype f (Hif) strains (13, 18, 22). When present, the sodC gene in encapsulated strains is adjacent to the bexA to -D genes of the capsule locus. The sodC gene of encapsulated sodC-containing strains shares significant sequence homology with the gene from Haemophilus parainfluenzae, but unlike H. parainfluenzae, encapsulated H. influenzae lacks CuZnSOD activity, presumably due to a substitution mutation that replaces an active-site histidine with tyrosine (13).
In the present study, we further evaluated the potential of the sodC gene to differentiate a large collection of known NT H. influenzae and H. haemolyticus respiratory tract isolates using a whole-genome microarray hybridization technology called library on a slide (39).
MATERIALS AND METHODS
Bacterial growth and strains.
Bacteria were grown on chocolate agar plates (BD, Franklin Lakes, NJ) at 37°C with 5% CO2. Bacteria were also grown on Levinthal agar prior to assessment of CuZnSOD activity (34).
Species designation of 169 NT H. influenzae and 110 H. haemolyticus strains was made previously by phenotypic, genotypic, or phylogenetic assays, and lack of encapsulation of the NT H. influenzae strains was determined by serotyping (8, 20, 28, 31). Microarray positive-control sodC-containing strains included H. parainfluenzae ATCC 33392 and H. haemolyticus ATCC 33390, and negative-control sodC-lacking strains included genome-sequenced H. influenzae strains Rd, 86-028NP, and R2846.
Microarray printing and analysis.
Microarray printing of crude genomic DNA and the generation of labeled sodC and DNA quantification probes have been described elsewhere (12, 14, 28, 38). The sodC gene probe was obtained from H. haemolyticus strain 65P28H9 with 5′ and 3′ UNIVSOD oligonucleotides (14).
A relative measure of probe hybridization to the DNA concentration for each spot was determined as follows. Duplicate slides were hybridized at 65°C in PerfectHyb Plus hybridization buffer (Sigma-Aldrich) with a digoxigenin-labeled control probe, serially washed with low- and high-stringency buffers, and analyzed (12, 28). The slides were then stripped with 4 M NaOH, washed, and rehybridized with a fluorescein-labeled sodC probe. As described previously, Spotfinder v.3.1.1 and MIDAS v.2.19 were used for determining probe intensity and spot standardization, respectively, and programs written in the statistical software “R” were used to graph the frequency of log-transformed sodC hybridization signal to a concentration-control signal ratio (28).
Southern hybridization.
Southern hybridization was performed on the sodC-positive strains using sodC or bexA gene probes. Genomic DNA was digested with EcoRI, electrophoresed on 1% agarose gels, and transferred to nylon membranes (Amersham). Probe labeling and hybridization have been described previously (20).
CuZnSOD activity.
CuZnSOD activity was determined by electrophoresis of whole-cell sonicated extracts on isoelectric focusing (IEF) pH 3 to 10 gels (Invitrogen) and visualized using previously described staining protocols (4, 30). CuZnSOD activity was specifically inhibited with 10 mM diethyl dithiocarbamic acid (DEDC), a copper chelator (17).
DNA isolation, amplification, and sequencing.
Isolation of template DNA and PCR amplification have been described previously (20). Oligonucleotides to amplify capsule-specific genes and to create a bexA gene probe have been described (7, 35). Intragene oligonucleotides, relative to the 5′ and 3′ ends of the published H. influenzae type b and f and H. parainfluenzae sodC gene sequences, were used to amplify partial sodC genes from each of the sodC-containing H. influenzae strains. The primer sequences were F, 5′-ATGATGAAAATGAAAACTCTCKTAGCATTAGC, and R, 5′-TTATTAATCAYRCCACATGCCATACG.
DNA sequence analysis and multilocus sequence analysis (MLSA) using partial DNA sequences of the adk, pgi, recA, infB, and 16S rRNA gene have been described previously (20). MLSA was performed on all sodC-positive strains (excluding strains Mr27, 32324, and H08-25, which had been done previously [20]). Sequence concatenations were done with Lasergene version 7.0 (DNAStar, Madison, WI), and phylogenetic analysis was done with Mega 3.1 (15). Bootstrap consensus minimum-evolution dendrograms for concatenated or individual sequences were made with 1,000 replicates.
Nucleotide sequence accession numbers.
Partial gene sequences for new H. influenzae MLSA sequences are available from GenBank under accession numbers GQ358536 to GQ358553 (for recA), GQ358554 to GQ358571 (for 16S rRNA genes), GQ358572 to GQ358589 (for adk), GQ358590 to GQ358607 (for infB), and GQ358608 to GQ358625 (for pgi). Partial sodC gene sequences from the H. haemolyticus and H. influenzae strains described in this study are under accession numbers GU269205 to GU269227.
RESULTS
Microarray hybridization of the sodC gene.
In whole-genome microarray hybridization, the H. parainfluenzae and H. haemolyticus positive-control strains hybridized with the sodC gene probe while the NT H. influenzae negative-control strains did not hybridize with the probe. In addition, 110/110 (100%) H. haemolyticus strains and 21/169 (12.4%) NT H. influenzae strains hybridized with the sodC gene probe. sodC hybridization to the 21 NT H. influenzae strains was confirmed by Southern blot hybridization. Eight representative NT H. influenzae strains, together with an H. influenzae negative-control strain, Rd, and an H. haemolyticus positive-control strain, 65P28H9, are shown in Fig. 1.
FIG. 1.
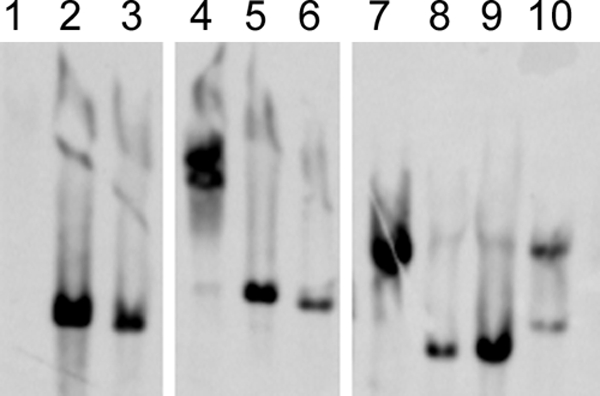
Southern hybridization of a sodC gene probe to microarray-positive NT H. influenzae strains. Lane 1, negative-control H. influenzae strain Rd; lane 2, positive-control H. haemolyticus strain 65P28H9; lanes 3 to 10, eight representative H. influenzae strains, P18-25, P05-24, O05-21, I8809, C21-21, O14-21, C14-21, and C09-23, respectively.
MLSA species confirmation of sodC-containing NT H. influenzae.
Since previous studies have suggested that the sodC gene is present in H. haemolyticus but not in NT H. influenzae, we used MLSA to rule out the possibility that the 21 sodC-containing NT H. influenzae strains were misidentified in their original species designations (21, 37). DNA sequences (partial gene sequences of adk, pgi, recA, infB, and 16S rRNA genes) obtained from the strains were analyzed according to an MLSA system that was previously developed in our laboratory and designed to differentiate NT H. influenzae from hemolytic and nonhemolytic H. haemolyticus (20). Generation of a minimum-evolution bootstrap consensus tree using concatenated sequences from the 21 sodC-containing NT H. influenzae strains, and from the 88 NT H. influenzae and 109 H. haemolyticus strains used in the original MLSA, demonstrated that all 21 sodC-containing strains clustered with known NT H. influenzae isolates (a similar tree including all NT H. influenzae strains and relevant type strains of other species is shown in Fig. 2). These results confirmed the species identification of the sodC-containing NT H. influenzae strains.
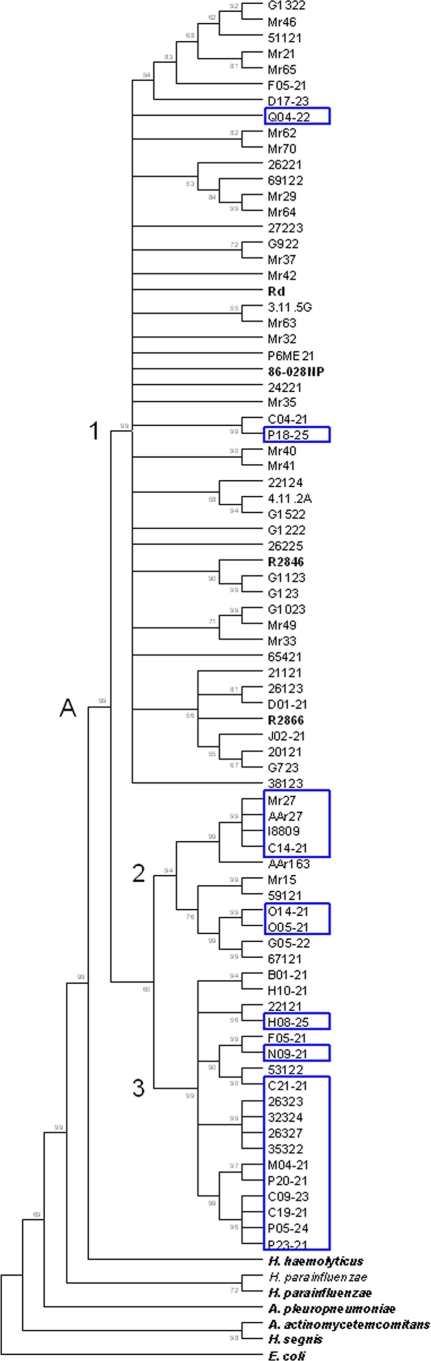
FIG. 2.
Minimum-evolution dendrogram of the Haemophilus sensu stricto cluster containing sodC-positive H. influenzae strains. The tree is rooted by E. coli (strain K-12) and is based on concatenated adk, pgi, recA, infB, and 16S rRNA gene sequences, with ≥50% of 1,000 bootstraps indicated. Node A contains all 88 NT H. influenzae strains used in a previous MLSA (15), together with the 21 H. influenzae strains that contain a sodC gene (blue boxes). The remaining type strains of species bordering H. influenzae are shown in boldface and were used in the original dendrogram (15). Clusters 1, 2, and 3 of node A have been previously shown to vary in their distributions of H. haemolyticus-like traits (15).
Presence of capsule DNA among sodC-containing NT H. influenzae strains.
Previous studies suggested that sodC is not present in NT H. influenzae but is present in encapsulated H. influenzae of phylogenetic division II (9, 13, 18). To investigate the possibility that some or all of the 21 sodC-containing NT H. influenzae strains are capsule-deficient mutants of encapsulated strains, we performed Southern hybridization using a bexA gene probe on the strains. The probe hybridized to 6/21 sodC-containing NT H. influenzae strains (O14-21, O05-21, C14-21, AAr27, I8809, and Mr27). Furthermore, capsule-specific PCR (7) revealed that, of the 6 strains hybridizing to a bexA probe, 2 contained type e (O14-21 and O05-21) and 4 contained type f (C14-21, AAr27, I8809, and Mr27) capsular DNA (Table 1). These results suggest that 6 of the original 21 sodC-containing H. influenzae strains are capsule-deficient mutants of encapsulated strains and that the remaining 15 strains are true NT H. influenzae strains. The results also allow the population prevalence of true sodC-containing NT H. influenzae strains to be readjusted from 21/169 (12.4%) to 15/163 (9.2%).
TABLE 1.
Characterization of the 21 sodC-hybridizing, nonencapsulated H. influenzae strains
| Strain | MLSA clustera | bexA hybridizationb | Cap genotypeb | CuZn-SOD activityb | % Identity to Hh sodCc | % Identity to Hib-like sodCc | sodC gene truncation | Isolation sourced |
|---|---|---|---|---|---|---|---|---|
| P18-25 | A1 | − | − | + | 96.7 | 84.1 | No | Throat |
| Q04-22 | A1 | − | − | + | 95.5 | 84.5 | No | Throat |
| C19-21 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| C21-21 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| H08-25 | A3 | − | − | + | 93.8 | 84.9 | No | Throat |
| M04-21 | A3 | − | − | + | 94.4 | 83.7 | No | Throat |
| N09-21 | A3 | − | − | + | 93.8 | 84.9 | No | Throat |
| P05-24 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| P20-21 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| 26323 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| 26327 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| 32324 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| 35322 | A3 | − | − | + | 94.6 | 83.9 | No | Throat |
| C09-23 | A3 | − | − | + | 86.1 | 95.6 | No | Throat |
| P23-21 | A3 | − | − | + | 84.9 | 99.6 | Yes | Throat |
| O14-21 | A2 | + | e | ± | 84.9 | 99.6 | Yes | Throat |
| O05-21 | A2 | + | e | − | 84.9 | 99.6 | Yes | Throat |
| C14-21 | A2 | + | f | − | 84.1 | 98.8 | Yes | Throat |
| AAr27 | A2 | + | f | − | 84.1 | 98.8 | Yes | ME |
| I8809 | A2 | + | f | − | 84.1 | 98.8 | Yes | ME |
| Mr27 | A2 | + | f | − | 84.1 | 98.8 | Yes | ME |
Subclade from dendrogram (Fig. 2) containing the strain.
−, negative; +, positive; ±, positive or negative.
Comparison is to the Hh (H. haemolyticus) or Hib-like (Hib, Hif, and H. parainfluenzae) consensus sequence listed.
Throat, isolate obtained from the throat of a healthy child attending day care; ME, middle-ear isolate of a child with otitis media.
CuZnSOD activity among the sodC-containing H. influenzae strains.
The CuZnSOD enzyme has been previously shown to be active in H. haemolyticus and H. parainfluenzae but inactive in encapsulated H. influenzae of phylogenetic division II (9, 18). Therefore, we determined the CuZnSOD activities of the 21 sodC-containing H. influenzae strains in this study to assess their relationship with the previously described haemophili. Native protein electrophoresis using whole-cell lysates of bacteria run on 3 to 10 IEF gels with subsequent staining for CuZnSOD activity (17) demonstrated that the positive-control H. haemolyticus strain, ATCC 33390, and all 15 true NT H. influenzae strains containing a sodC gene displayed consistent CuZnSOD activity, as evidenced by two achromatic bands, a feature previously observed for H. haemolyticus (Fig. 3, left, and Table 1) (9). CuZnSOD activity in the NT H. influenzae strains could be specifically inhibited by 10 mM DEDC, a copper-chelating agent (Fig. 3, right). One of the capsule-deficient strains (O14-21) displayed CuZnSOD activity in only one of several experiments. Inhibition with DEDC could not be done with this strain. None of the five remaining capsule-deficient strains had CuZnSOD activity, suggesting that their sodC genes are related to the sodC gene from encapsulated H. influenzae of phylogenetic division II. In addition, the results suggest that sodC of NT H. influenzae strains differs from the sodC of encapsulated strains.
FIG. 3.
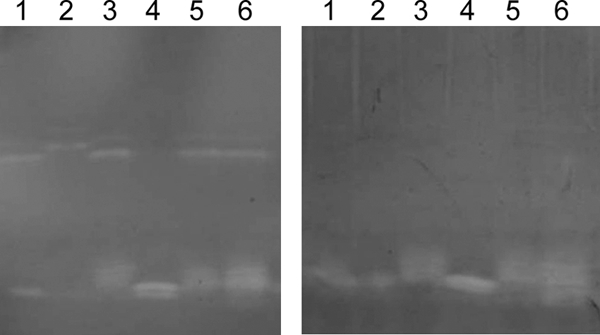
SOD activities of representative nonencapsulated H. influenzae strains. Achromatic zones of SOD activity are shown in the left panel as bands in the upper and lower regions of the gel. Only the two upper bands are absent in the right panel, where the copper chelator DEDC was used to inhibit CuZnSOD activity. The lanes containing sonicated cellular extracts are as follows: lane 1, H. influenzae N09-21; lane 2, H. haemolyticus ATCC 33390; and lanes 3 to 6, H. influenzae strains 35322, Mr27, 26323, and C19-21, respectively.
Relationships of the H. influenzae and H. haemolyticus sodC genes.
The relationships between the sodC gene from NT H. influenzae and the sodC gene from encapsulated H. influenzae or from other Haemophilus species are not known. Phylogenetic relationships between the strains and species in the MLSA described above, however, may provide an indication. In our previously published MLSA (20), all 88 NT H. influenzae strains were present in a single clade (A) that contained three subclades: A1, A2, and A3. Although 72/88 NT H. influenzae strains were found in subclade A1, the remaining 16 strains were divided between subclades A2 and A3. In addition, strains in subclades A2 and A3 were found to possess higher proportions of H. haemolyticus-like phenotypic and genotypic traits than NT H. influenzae strains present in subclade A1, suggesting the possibility of interspecies recombination for strains found in clades A2 and A3 (20). Interestingly, 19 of the 21 sodC-containing H. influenzae strains in our current study clustered with the NT H. influenzae strains originally present in subclades A2 and A3 (Fig. 2 and Table 1). These data suggest the possibility that some H. influenzae strains possess a sodC gene through interspecies recombination with H. haemolyticus.
The sodC genes from 6 of the 19 sodC-containing strains in subclades A2 and A3, however, would be predicted to be related to encapsulated H. influenzae of phylogenetic division II, rather than to the sodC gene from H. haemolyticus. To investigate these potential differences, we compared the sodC gene sequences of our 21 H. influenzae strains with sodC from H. haemolyticus and encapsulated H. influenzae. sodC genes were PCR amplified from two H. haemolyticus strains (ATCC 33390 and 65P28H9) and all 21 sodC-containing H. influenzae strains using oligonucleotides representing the 5′ and 3′ ends of sodC nucleic acid sequences previously published from encapsulated Hib and Hif and H. parainfluenzae (13, 29). DNA sequences obtained from the 21 sodC PCR products of the H. influenzae strains in this study revealed an internal 536- to 548-bp fragment, depending on the strain. Sequence comparisons revealed that the two H. haemolyticus sodC genes were 95.2% identical to each other, while the sodC genes from Hib, Hif, and H. parainfluenzae have been previously shown to be 96 to 98.4% identical to each other (13, 29). The H. haemolyticus sodC genes were 84.1 to 87.3% identical to a Hib, Hif, and H. parainfluenzae sodC consensus sequence (representing an internal 536-bp fragment) (data not shown). Further DNA sequence comparisons revealed that the sodC genes from 13 of the 21 H. influenzae strains were, on average, 94.7% identical to a consensus sequence generated from the two H. haemolyticus sodC gene sequences and 84.1% identical to the Hib, Hif, and H. parainfluenzae sodC consensus sequence (P < 0.05) (Table 1). All 13 strains were bexA-negative true NT H. influenzae strains. In contrast, the sodC sequences from the remaining 8 strains were on average 84.7% identical to the H. haemolyticus consensus sequence but 98.7% identical to the Hib, Hif, and H. parainfluenzae consensus sequence (henceforth referred to as Hib-like sodC) (P < 0.05) (Table 1). Of the eight strains with Hib-like sodC genes, six were capsule-deficient H. influenzae strains and two (C09-23 and P23-21) were bexA-negative NT H. influenzae strains. Therefore, sodC sequence analysis substantiates the existence of capsule mutant strains in this collection and the possibility that some NT H. influenzae strains may have acquired sodC through recombination with H. haemolyticus.
Interestingly, translation of the partial sodC gene sequences from all 21 strains revealed that the sodC genes of 7/8 strains with Hib-like sodC genes would be inactivated due to various frameshift insertions. This differs from the previously described inactive sodC genes of encapsulated strains, where substitution mutations were thought to affect active-site residues (13). Four of the seven strains (AAr27, C14-21, I8809, and Mr27) had a 7-bp tandem duplication at position 169, and three strains (P23-21, O14-21, and O05-21) had a 1-bp insertion at position 302 (both positions are based on the full-length sodC gene from Hib, Hif, and H. parainfluenzae) (13). In contrast, all 13 strains with H. haemolyticus-like sodC genes and one Hib-like sodC gene possessed open reading frames.
One NT H. influenzae strain, C09-23, was atypical in that it possessed a Hib-like sodC gene with CuZnSOD activity (data not shown). The sodC gene of strain C09-23 differed from other Hib-like sodC genes in that it did not contain a truncated reading frame. One explanation is that the sodC gene of this strain may have originated from H. parainfluenzae. sodC in H. parainfluenzae is very similar to sodC of encapsulated H. influenzae and displays CuZnSOD activity (13). Alternatively, NT H. influenzae strain C09-23 was also found to possess two EcoRI fragments that hybridized with a sodC gene probe (Fig. 1, lane 10), and it is possible that CuZnSOD activity may have come from a second, non-PCR-amplified sodC gene. One other strain (P05-24) in this study also contained doublet bands hybridizing to a sodC gene probe (Fig. 1, lane 4), suggesting that multiple copies of sodC are possible in some H. influenzae strains.
In addition to C09-23, two other sodC-containing strains were atypical. One NT H. influenzae strain, P23-21, and one unencapsulated type e H. influenzae strain, O14-21, both displayed CuZnSOD activity despite having Hib-like sodC genes that contained frameshift mutations. In repeated experiments, CuZnSOD activity was always present in strain P23-21 but was observed only one time in strain O14-21. A second, unidentified copy of sodC would provide an explanation for the discrepancy between CuZnSOD activity and truncated sodC genes in these strains. Further studies, however, to confirm and characterize sodC copy numbers and expression would be outside the scope of our original objective, which was to identify sodC-containing NT H. influenzae strains and to estimate the population prevalence of the gene.
DISCUSSION
In this study, microarray hybridization analysis of a large population of respiratory-tract NT H. influenzae strains identified two groups of sodC-containing, unencapsulated H. influenzae strains (Table 1): (i) true NT strains (nonserotypeable strains lacking any capsule-associated genes) that possessed CuZnSOD activity and (ii) capsule-deficient mutants of encapsulated strains (containing bexA and capsule-specific DNA) that lacked CuZnSOD activity (with the exception of one type e strain, O14-21, which had inconsistent activity). Preliminary work in our laboratory indicates that the six bexA-positive strains represent all of the capsule-deficient mutants among the 169 nonserotypeable H. influenzae strains used in this study (unpublished results). Therefore, sodC was present in 15/163 (9.2%) true NT H. influenzae strains. sodC in capsule-deficient type e and f mutant strains are reflective of earlier studies that identified the sodC gene with no CuZnSOD activity in phylogenetic division II encapsulated strains (13, 18). The lack of CuZnSOD activity in sodC-containing encapsulated strains was proposed to occur due to a substitution mutation that changed an active-site histidine to a tyrosine. Although all of the sodC genes from the capsule derivatives in the current study possessed a tyrosine at the same position (unpublished results), the genes also contained various frameshift insertions that resulted in truncated reading frames.
As mentioned above, previous studies have reported discrepancies in the prevalence of sodC among NT H. influenzae strains where sodC and CuZnSOD activity were found in nearly half of the NT H. influenzae strains of one study (13) but were not found among NT H. influenzae strains in two later studies (9, 18). Based on our current results, NT H. influenzae possessing sodC and CuZnSOD activity in the initial report may have been correct. Alternatively, the sodC-containing strains may have been nonhemolytic H. haemolyticus or capsule-deficient mutants of encapsulated H. influenzae that were misclassified as NT H. influenzae (21, 40).
Another possible explanation for the discrepancy in the literature, however, may be found in the selection of NT H. influenzae strains examined. In our study, 3/56 (5.4%) H. influenzae isolates (Mr27, AAr27, and I8809) that contained a sodC gene were obtained from the middle ears of children with otitis media, while 18/113 (15.9%) H. influenzae isolates that contained a sodC gene were obtained from the throats of healthy children or adults (unpublished results) (Table 1). Similarly, Norskov-Lauritsen (23) recently found that only 6/480 (1.25%) clinical H. influenzae isolates possessed a sodC gene and that all six strains were related to encapsulated phylogenetic division II H. influenzae strains. Commensal isolates were not examined in that study. Therefore, the presence or absence of sodC and CuZnSOD activity among NT H. influenzae strains used in earlier studies may have been due to the virulence potential of the isolates examined (18). These observations also highlight recent studies that document a number of genetic differences between disease and commensal NT H. influenzae strains (10, 24, 37).
The role of sodC and related CuZnSOD activity in the virulence of Gram-positive and Gram-negative pathogenic bacteria has been the subject of considerable study (2, 6). In pathogenic bacteria that have a functional sodC gene, the translated protein is a periplasmic or membrane-bound enzyme thought to inactivate superoxide released during the respiratory burst of phagocytes, thereby contributing to the virulence of the pathogen (2). The role of sodC and corresponding CuZnSOD activity in the pathogenesis of NT H. influenzae would be expected to be negligible, since only a small proportion of strains carry the gene, and those strains are predominantly commensal isolates. The role of sodC in the host interactions of commensal NT H. influenzae or in commensal H. haemolyticus and H. parainfluenzae is not known (14, 16). Aside from superoxide dismutase activity, some bacterial sodC genes are known to encode structural variants that contain species-specific insertions, deletions, and substitutions that provide non-cofactor-related, metal-binding properties via their N termini or subunit-subunit interactions (3, 33). Such diverse functional properties of CuZnSOD in H. haemolyticus, H. parainfluenzae, or H. influenzae, however, remain to be determined. The presence of sodC in commensal species, such as H. haemolyticus and H. parainfluenzae, suggests an important role for the gene in the commensal microbiology of humans. Indeed, a better understanding of commensal microbiology is necessary to differentiate “normal” from disease-related growth of opportunistic pathogens, such as NT H. influenzae, a species that resides in humans predominantly as a commensal bacterium but infrequently as a pathogen.
Acknowledgments
This research was supported by grants from the National Institutes of Health (DC05840 and HL08393) awarded to J.R.G.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Albritton, W. L. 1982. Infections due to Haemophilus species other than H. influenzae. Annu. Rev. Microbiol. 36:199-216. [DOI] [PubMed] [Google Scholar]
- 2.Battistoni, A. 2003. Role of prokaryotic Cu,Zn superoxide dismutase in pathogenesis. Biochem. Soc. Trans. 31:1326-1329. [DOI] [PubMed] [Google Scholar]
- 3.Battistoni, A., F. Pacello, A. P. Mazzetti, C. Capo, J. S. Kroll, P. R. Langford, A. Sansone, G. Donnarumma, P. Valenti, and G. Rotilio. 2001. A histidine-rich metal binding domain at the N terminus of Cu,Zn-superoxide dismutases from pathogenic bacteria: a novel strategy for metal chaperoning. J. Biol. Chem. 276:30315-30325. [DOI] [PubMed] [Google Scholar]
- 4.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 5.Chin, C. L., L. J. Manzel, E. E. Lehman, A. L. Humlicek, L. Shi, T. D. Starner, G. M. Denning, T. F. Murphy, S. Sethi, and D. C. Look. 2005. Haemophilus influenzae from patients with chronic obstructive pulmonary disease exacerbation induce more inflammation than colonizers. Am. J. Respir. Crit. Care Med. 172:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desideri, A., and M. Falconi. 2003. Prokaryotic Cu, Zn superoxide dismutases. Biochem. Soc. Trans. 31:1322-1325. [DOI] [PubMed] [Google Scholar]
- 7.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon. 1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farjo, R. S., B. Foxman, M. J. Patel, L. Zhang, M. M. Pettigrew, S. I. McCoy, C. F. Marrs, and J. R. Gilsdorf. 2004. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr. Infect. Dis. J. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 9.Fung, W. W., C. A. O'Dwyer, S. Sinha, A. L. Brauer, T. F. Murphy, J. S. Kroll, and P. R. Langford. 2006. Presence of copper- and zinc-containing superoxide dismutase in commensal Haemophilus haemolyticus isolates can be used as a marker to discriminate them from nontypeable H. influenzae isolates. J. Clin. Microbiol. 44:4222-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juliao, P. C., C. F. Marrs, J. Xie, and J. R. Gilsdorf. 2007. Histidine auxotrophy in commensal and disease-causing nontypeable Haemophilus influenzae. J. Bacteriol. 189:4994-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilian, M. 2003. Haemophilus, p. 623-635. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 12.Kong, Y., M. D. Cave, L. Zhang, B. Foxman, C. F. Marrs, J. H. Bates, and Z. H. Yang. 2006. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. J. Clin. Microbiol. 44:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroll, J. S., P. R. Langford, and B. M. Loynds. 1991. Copper-zinc superoxide dismutase of Haemophilus influenzae and H. parainfluenzae. J. Bacteriol. 173:7449-7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroll, J. S., P. R. Langford, K. E. Wilks, and A. D. Keil. 1995. Bacterial [Cu,Zn]-superoxide dismutase: phylogenetically distinct from the eukaryotic enzyme, and not so rare after all! Microbiology 141:2271-2279. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignments. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 16.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1992. Copper-zinc superoxide dismutase in Haemophilus species. J. Gen. Microbiol. 138:517-522. [DOI] [PubMed] [Google Scholar]
- 17.Langford, P. R., A. Sansone, P. Valenti, A. Battistoni, and J. S. Kroll. 2002. Bacterial superoxide dismutase and virulence. Methods Enzymol. 349:155-166. [DOI] [PubMed] [Google Scholar]
- 18.Langford, P. R., B. J. Sheehan, T. Shaikh, and J. S. Kroll. 2002. Active copper- and zinc-containing superoxide dismutase in the cryptic genospecies of Haemophilus causing urogenital and neonatal infections discriminates them from Haemophilus influenzae sensu stricto. J. Clin. Microbiol. 40:268-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrs, C. F., G. P. Krasan, K. W. McCrea, D. L. Clemans, and J. R. Gilsdorf. 2001. Haemophilus influenzae—human specific bacteria. Front. Biosci. 6:E41-E60. [DOI] [PubMed] [Google Scholar]
- 20.McCrea, K. W., J. Xie, N. Lacross, M. Patel, D. Mukundan, T. F. Murphy, C. F. Marrs, and J. R. Gilsdorf. 2008. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J. Clin. Microbiol. 46:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: A human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 22.Musser, J. M., D. M. Granoff, P. E. Pattison, and R. K. Selander. 1985. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 82:5078-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norskov-Lauritsen, N. 2009. Detection of cryptic genospecies misidentified as Haemophilus influenzae in routine clinical samples by assessment of marker genes fucK, hap, and sodC. J. Clin. Microbiol. 47:2590-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 70:3551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pittman, M. 1953. A classification of the hemolytic bacteria of the genus Haemophilus: Haemophilus haemolyticus Bergey et al. and Haemophilus parahaemolyticus nov. spec. J. Bacteriol. 65:750-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quentin, R., C. Martin, J. M. Musser, N. Pasquier-Picard, and A. Goudeau. 1993. Genetic characterization of a cryptic genospecies of Haemophilus causing urogenital and neonatal infections. J. Clin. Microbiol. 31:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quentin, R., R. Ruimy, A. Rosenau, J. M. Musser, and R. Christen. 1996. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J. Clin. Microbiol. 34:1380-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandstedt, S. A., L. Zhang, M. Patel, K. W. McCrea, Z. Qin, C. F. Marrs, and J. R. Gilsdorf. 2008. Comparison of laboratory-based and phylogenetic methods to distinguish between Haemophilus influenzae and H. haemolyticus. J. Microbiol. Methods 75:369-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satola, S. W., P. L. Schirmer, and M. M. Farley. 2003. Genetic analysis of the capsule locus of Haemophilus influenzae serotype f. Infect. Immun. 71:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinman, H. M. 1985. Bacteriocuprein superoxide dismutases in pseudomonads. J. Bacteriol. 162:1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahata, S., T. Ida, N. Senju, Y. Sanbongi, A. Miyata, K. Maebashi, and S. Hoshiko. 2007. Horizontal gene transfer of ftsI, encoding penicillin-binding protein 3, in Haemophilus influenzae. Antimicrob. Agents Chemother. 51:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toro, I., C. Petrutz, F. Pacello, M. D'Orazio, A. Battistoni, and K. Djinovic-Carugo. 2009. Structural basis of heme binding in the Cu,Zn superoxide dismutase from Haemophilus ducreyi. J. Mol. Biol. 386:406-418. [DOI] [PubMed] [Google Scholar]
- 34.Turk, D. C., and J. R. May. 1967. Haemophilus influenzae; its clinical importance. English University Press, London, United Kingdom.
- 35.van Ketel, R. J., B. de Wever, and L. van Alphen. 1990. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J. Med. Microbiol. 33:271-276. [DOI] [PubMed] [Google Scholar]
- 36.Wallace, R. J., Jr., C. J. Baker, F. J. Quinones, D. G. Hollis, R. E. Weaver, and K. Wiss. 1983. Nontypable Haemophilus influenzae (biotype 4) as a neonatal, maternal, and genital pathogen. Rev. Infect. Dis. 5:123-136. [DOI] [PubMed] [Google Scholar]
- 37.Xie, J., P. C. Juliao, J. R. Gilsdorf, D. Ghosh, M. Patel, and C. F. Marrs. 2006. Identification of new genetic regions more prevalent in nontypeable Haemophilus influenzae otitis media strains than in throat strains. J. Clin. Microbiol. 44:4316-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, L., B. Foxman, J. R. Gilsdorf, and C. F. Marrs. 2005. Bacterial genomic DNA isolation using sonication for microarray analysis. Biotechniques 39:640, 642-644. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, L., U. Srinivasan, C. F. Marrs, D. Ghosh, J. R. Gilsdorf, and B. Foxman. 2004. Library on a slide for bacterial comparative genomics. BMC Microbiol. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, J., D. K. Law, M. L. Sill, and R. S. Tsang. 2007. Nucleotide sequence diversity of the bexA gene in serotypeable Haemophilus influenzae strains recovered from invasive disease patients in Canada. J. Clin. Microbiol. 45:1996-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]