Abstract
Introduction of a new influenza virus in humans urges quick analysis of its virological and immunological characteristics to determine the impact on public health and to develop protective measures for the human population. At present, however, the necessity of executing pandemic influenza virus research under biosafety level 3 (BSL-3) high-containment conditions severely hampers timely characterization of such viruses. We tested heat, formalin, Triton X-100, and β-propiolactone treatments for their potencies in inactivating human influenza A(H3N2) and avian A(H7N3) viruses, as well as seasonal and pandemic A(H1N1) virus isolates, while allowing the specimens to retain their virological and immunological properties. Successful heat inactivation coincided with the loss of hemagglutinin (HA) and neuraminidase (NA) characteristics, and β-propiolactone inactivation reduced the hemagglutination titer and NA activity of the human influenza virus 10-fold or more. Although Triton X-100 treatment resulted in inconsistent HA activity, the NA activities in culture supernatants were enhanced consistently. Nonetheless, formalin treatment permitted the best retention of HA and NA properties. Triton X-100 treatment proved to be the easiest-to-use influenza virus inactivation protocol for application in combination with phenotypic NA inhibitor susceptibility assays, while formalin treatment preserved B-cell and T-cell epitope antigenicity, allowing the detection of both humoral and cellular immune responses. In conclusion, we demonstrated successful influenza virus characterization using formalin- and Triton X-100-inactivated virus samples. Application of these inactivation protocols limits work under BSL-3 conditions to virus culture, thus enabling more timely determination of public health impact and development of protective measures when a new influenza virus, e.g., pandemic A(H1N1)v virus, is introduced in humans.
Host switching of viruses from animals to humans may result in an epidemic among humans and can be particularly dangerous for the new, immunologically naïve host. Examples are the introduction of human immunodeficiency virus, severe acute respiratory syndrome coronavirus, and pandemic influenza A viruses in humans. In particular, avian influenza A virus subtypes H5N1, H9N2, and H7N7 have been transmitted directly to humans in the past decade, exhibiting the zoonotic potential of influenza viruses (4, 11, 19, 25). Moreover, the recent introduction of swine origin influenza A(H1N1)v virus in humans initiated the first influenza pandemic of the 21st century (16, 35). Introduction of a new influenza virus in humans urges quick analysis of its virological and immunological characteristics to assist in the determination of the impact on public health and the development of protective measures. At present, however, the necessity of executing pandemic influenza virus research under biosafety level 3 (BSL-3) high-containment conditions hampers timely characterization of such viruses.
Several virological and immunological assays are used for the characterization of a virus and the immune response induced. For antigenic characterization of influenza viruses, hemagglutination assays and hemagglutination inhibition (HI) assays are the “gold standard” tests. In addition, since the global emergence of antiviral-resistant influenza viruses is becoming an increasing problem, the characterization of influenza virus susceptibilities to the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir is a clinical necessity (2, 9, 13, 17, 23). For investigating the immune response against influenza viruses, the HI assay determines protective humoral responses (8). Finally, in addition to HI assay results, assessment of the human T-cell responses against influenza virus infection has been reported previously to provide an important marker of protection (3, 10, 22). Until now, these assays have been performed mostly by applying live virus, hence necessitating the use of BSL-3 conditions for studying (potential) pandemic influenza virus. Although numerous studies of virus inactivation, e.g., by means of virucidal compounds, UV light, or gamma irradiation treatment, have been performed, these studies have not comprehensively documented the preservation of influenza virus protein function and antigenic characteristics following inactivation (5-7, 14, 18). Specifically, these studies have not addressed whether inactivated virus can be used for phenotypic determination of susceptibilities to NAIs and for characterization of T-cell responses.
In this study, we evaluated the inactivation of influenza viruses of human, avian, and swine origins by heat, formalin, Triton X-100, or β-propiolactone (β-PL) and the retention of hemagglutinin (HA) and NA glycoprotein functions and antigenic integrity. The optimal procedures have been used to demonstrate the proof of principle in antiviral susceptibility assays, antigenic characterization, and T-cell response assays with both seasonal and pandemic influenza A(H1N1) viruses.
MATERIALS AND METHODS
Inclusion of donors and isolation of PBMC.
Buffy coats from healthy individuals were retrieved from the Sanquin Blood Bank North West Region in accordance with human experimental guidelines (project number S03.0015-X). In addition, peripheral blood mononuclear cells (PBMC) were retrieved from two previously healthy individuals (a 51-year-old female and a 55-year-old male) with laboratory-confirmed influenza A(H1N1)v virus infection 13 and 19 days after the start of symptoms, respectively. Both participants provided written informed consent before the start of the study. The study was approved by the Medical Ethical Committee of the Utrecht University Medical Center. Human PBMC were isolated by density centrifugation and were cryopreserved at −135°C in a solution of 90% fetal calf serum (FCS; HyClone, UT) and 10% dimethyl sulfoxide (Sigma-Aldrich, MO) until analysis.
Influenza antiviral drugs.
Oseltamivir carboxylate Ro64-0802 (GS4071) and zanamivir (GG167) were kindly provided by Roche Diagnostics (Germany) and GlaxoSmithKline (The Netherlands), respectively.
Virus expansion.
Human influenza virus isolate A/Wisconsin/67/2005 (H3N2) and low-pathogenicity avian influenza virus isolate A/Mallard/NL/12/2000 (H7N3) were used for initial evaluation of virus inactivation protocols. Both virus isolates were cultured on Madin-Darby canine kidney (MDCK) cells until cytopathic effects (CPE) were observed. The culture flasks were subjected to one freeze-thaw cycle, and the culture supernatant was centrifuged (5 min at 1,500 rpm) to clear it of cell debris. The resulting supernatant was stored at −80°C until further analysis.
Final inactivation experiments were performed using supernatants from cultures of pandemic influenza A(H1N1)v viruses A/California/04/2009, A/Paris/2590/2009, and A/Netherlands/602/2009 and seasonal influenza A(H1N1) viruses A/New Caledonia/20/99 and A/Netherlands/268/2008 on MDCK cells, prepared as described above.
TCID50 determination.
The determination of the 50% tissue culture infectious dose (TCID50) was carried out using 96-well plates containing confluent MDCK cell monolayers. The MDCK cells were incubated with serial 10-fold dilutions of influenza virus culture supernatant in infection medium (Dulbecco's modified Eagle medium [DMEM; Gibco] with antibiotics, nonessential amino acids [Gibco], and 2.5 μg/ml trypsin and without FCS) at 37°C for 60 min. After 1 h, the monolayer was rinsed with phosphate-buffered saline (PBS), overlaid with infection medium, and incubated at 37°C for 5 days. To identify influenza virus-positive wells, the hemagglutination assay was performed. The log TCID50 per milliliter was calculated using the Reed-Muench method as described previously (29).
Heat inactivation.
Influenza virus culture supernatants (400 μl) were incubated for 30 min at room temperature (22°C) and at 35.0, 38.3, 43.7, 49.6, 55.6, 61.3, 66.7, and 70°C in 0.5-ml vials in a T-Gradient thermal cycler (Biometra, Göttingen, Germany) and subsequently stored at −80°C until analysis.
Formalin inactivation.
Influenza virus culture supernatants were incubated for 18 and 72 h at 37°C with a 0.02% final concentration of formalin (Merck, Darmstadt, Germany) in PBS. Immediately after inactivation, formalin was removed by dialysis using Dispodialyzers according to the instructions of the manufacturer (Spectrum Laboratories, CA). The samples were dialyzed against 50 ml DMEM at room temperature on a roller bank two times for 2 h each and then overnight with fresh DMEM each time. The dialyzed samples were stored at −80°C until analysis.
Triton X-100.
For initial experiments, 450-μl influenza virus culture supernatants cleared of cell debris were incubated for 1 h at room temperature after being subjected to a thorough vortex step for 30 s with 50 μl of Triton X-100 (BDH Chemicals, Poole, United Kingdom) to yield final concentrations of 0, 0.1, 0.2, 0.5, and 1.0% (vol/vol) Triton X-100. In the final experiments with a final concentration of 1% Triton X-100, 450 μl of influenza virus culture supernatant cleared of cell debris was mixed in a thorough vortex step for 30 s with 50 μl of freshly prepared 10% (vol/vol) Triton X-100 in infection medium prior to incubation for 1 h at room temperature. Subsequently, for hemagglutination titer determination and virus culture, Triton X-100 was removed from the supernatants by using the column-based absorption Detergent-OUT kit according to the instructions of the manufacturer (Calbiochem, CA), and the flowthrough was stored at −80°C until analysis.
β-PL.
Influenza culture supernatants were incubated for 16 h at 4°C with and without a final concentration of 0.094% β-PL (ACROS Organics, Geel, Belgium). The supernatants were subsequently incubated for 2 h at 37°C to facilitate hydrolysis of β-PL and were stored at −80°C until analysis.
Validation of inactivation by culturing on MDCK cells.
Both control and treated influenza virus culture supernatants were subjected to a maximum of three blind passages on MDCK cells. Confluent MDCK cells were incubated with 50 μl culture supernatant and 5 ml infection medium in 25-cm2 tissue culture flasks at 37°C. After a 10-day incubation period, the flasks were subjected to one freeze-thaw cycle. In the absence of CPE, the harvested culture material was used as an inoculum (50 μl) for a subsequent passage on MDCK cells. All harvested culture supernatants were stored at −80°C until further analysis.
Validation of inactivation by matrix gene PCR analysis.
Total nucleic acid (50 μl) was extracted from 200 μl influenza virus culture supernatant with the MagNA Pure LC total nucleic acid isolation kit on a MagNA Pure LC (version 2.0) extraction robot (Roche, Mannheim, Germany). For semiquantitative analysis of influenza virus negative-sense genomic RNA in the culture material, reverse transcriptase PCR (RT-PCR) was performed using avian myeloblastosis virus RT (Promega, WI) and the sense matrix gene primer 5′ AAG ACC AAT CCT GTC ACC TCT GA 3′ (M-Fw) (40) to generate cDNA. To detect positive-sense viral RNA transcripts in the culture material, total RNA was transcribed into cDNA by using recombinant Tth DNA polymerase (Applied Biosystems, CA), matrix gene antisense primer 5′ CAA AGC GTC TAC GCT GCA GTC C 3′ (M-Rv) (40), and treatment with RNase H (Roche) and RNase A (Sigma-Aldrich) to remove all RNA. Subsequently, a 94-bp matrix gene fragment was amplified using the LightCycler 480 real-time PCR system (Roche), the LightCycler TaqMan master (Roche) with primer pair M-Fw and M-Rv, and amplicon-specific probe 5′ TTT GTG TTC ACG CTC ACC GTG CC 3′ labeled with 6-carboxyfluorescein/Black Hole Quencher 1.
Hemagglutination assay.
Serial twofold dilutions of live and inactivated influenza virus culture supernatants in PBS were prepared and incubated in quadruplet with 0.25% turkey erythrocytes in PBS at 4°C. After 60 min, the hemagglutination titer, expressed as the reciprocal of the highest dilution producing complete hemagglutination, was read.
HI assay.
The inhibition of hemagglutination was assessed with ferret antisera raised against seasonal influenza A(H1N1) viruses A/Texas/36/91, A/New Caledonia/20/99, A/Solomon Islands/03/06, and A/Brisbane/59/07 (kindly provided by the WHO Collaborating Centre, London, United Kingdom). To remove nonspecific hemagglutination activity from the sera, 1 volume of ferret antiserum was mixed with 5 volumes of cholera filtrate, incubated at 37°C for 16 h, and then incubated for 1 h at 56°C to inactivate the receptor-destroying activity of the cholera filtrate. Subsequently, 25-μl twofold serial dilutions of ferret antisera in PBS were prepared and incubated with 25 μl of influenza A virus (4 hemagglutinating units) for 30 min at 37°C, after which 50 μl of 0.5% turkey erythrocytes in PBS was added. The inhibition of hemagglutination was read after 1 h of incubation at 4°C and expressed as the reciprocal of the highest serum dilution producing 100% inhibition of hemagglutination.
NA assay.
The NA activities and NAI (oseltamivir and zanamivir) susceptibilities of the culture supernatants were determined using a fluorescence-based NA inhibition assay as described previously (17, 27). The assay was based on the detection of the fluorescent product 4-methylumbelliferone, released after hydrolysis of the substrate 2-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA; Sigma-Aldrich) by NA. Final NA inhibition data were supplemented with data generated using the NA-Star influenza NAI resistance detection kit according to the instructions of the manufacturer (Applied Biosystems). The NAI susceptibility was expressed as the concentration of NAI needed to inhibit the NA enzyme activity by 50% (the 50% inhibitory concentration [IC50]).
PBMC stimulation and flow cytometric analysis.
PBMC were infected at a multiplicity of infection of 2 with live influenza virus or pulsed with an equal amount of formalin-inactivated virus (as confirmed by three blind passages in tissue culture) and cultured in RPMI medium containing 10% FCS (HyClone), penicillin, streptomycin, and l-glutamine (Gibco BRL, NY) at 106 cells per well in a 48-well plate. Occasionally, where indicated, twofold-larger amounts of inactivated virus were used. Culture supernatant from uninfected cells was formalin inactivated, dialyzed, and used as a negative control. As a positive control, Staphylococcus aureus enterotoxin B (Sigma-Aldrich) was used. The cells were incubated for various times at 37°C in 5% CO2. Phycoerythrin-conjugated antibodies specific for CD107a (BD Biosciences, CA) were added 16 h before the end of culture. For the detection of intracellular cytokine production, monensin (GolgiStop; BD Biosciences) was added during the last 16 h of culture. After incubation, PBMC were harvested and stained with anti-CD4 Pacific Blue antibody (BioLegend, San Diego, CA), anti-CD8-peridinin chlorophyll protein-Cy5.5 (BD Biosciences), anti-gamma interferon-allophycocyanin (BD Biosciences), anti-interleukin-2 (anti-IL-2)-fluorescein isothiocyanate (eBioscience, CA), anti-tumor necrosis factor alpha-phycoerythrin-Cy7 (BD Biosciences), and LIVE/DEAD fixable dead cell stain (Invitrogen, Paisley, United Kingdom) according to the manufacturers' procedures and were acquired using a FACSCanto II system (BD Biosciences). At least 105 viable lymphocytes were acquired based on forward-side scatter characteristics and analysis of viability staining. The results were analyzed using FACSDiva software (BD Biosciences).
RESULTS
Seasonal influenza virus A(H3N2) and avian influenza virus A(H7N3) inactivation.
Since we wished to determine the effect of virus inactivation on viral infectivity and viral protein resilience, the procedures were first executed using human and avian influenza virus strains with completely different HA and NA subtypes.
Influenza virus stocks A/Wisconsin/67/2005 subtype H3N2 (104.8 TCID50/ml) and A/Mallard/NL/12/00 subtype H7N3 (105.3 TCID50/ml) were treated separately with heat, formalin, Triton X-100, and β-PL. Virus infectivity was completely absent after inactivation by temperatures of ≥55.6°C, formalin, Triton X-100 concentrations of ≥0.2%, and β-PL, as confirmed by the absence of CPE, the absence of sense viral mRNA, and the reduction of antisense viral genomic RNA in three subsequent cell culture passages (Table 1). Influenza virus mRNA is not present in the virus itself but is produced only in the infected cell and is therefore a clear measure of virus replication. In contrast to residual virus genomic RNA that is protected by the viral nucleoprotein, posttreatment residual virus mRNA is rapidly degraded in tissue culture. Specimens from cell culture passages showing incomplete virus inactivation, based on phenotypic (CPE) and molecular data, were not passaged further. The combined viral culture and matrix RNA detection data demonstrate that these procedures result in complete inactivation of 104.8 and 105.3 TCID50/ml human and avian influenza virus strains, respectively.
TABLE 1.
Confirmation of A(H3N2) and A(H7N3) influenza virus inactivation by phenotypic and molecular testsa
| Influenza virus | Treatment | Posttreatment |
Passage 1 |
Passage 2 |
Passage 3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vRNA | mRNA | CPE | vRNA | mRNA | CPE | vRNA | mRNA | CPE | vRNA | mRNA | ||
| A(H3N2) | Heat (22°C; control) | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| Heat (35°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (38.3°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (43.7°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (49.6°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (55.6°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (61.3°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (66.7°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (70°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Formalin (18 h) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Formalin (72 h) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (0.0%; control) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Triton X-100 (0.1%) | + | + | − | − | − | − | − | − | − | − | − | |
| Triton X-100 (0.2%) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (0.5%) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (1.0%) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| β-PL (overnight) | + | + | − | + | − | − | − | − | − | − | − | |
| A(H7N3) | Heat (22°C; control) | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| Heat (35°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (38.3°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (43.7°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (49.6°C) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Heat (55.6°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (61.3°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (66.7°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Heat (70°C) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Formalin (18 h) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Formalin (72 h) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (0.0%; control) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Triton X-100 (0.1%) | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| Triton X-100 (0.2%) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (0.5%) | + | + | − | + | − | − | − | − | − | − | − | |
| Triton X-100 (1.0%) | + | + | − | + | − | − | − | − | − | − | − | |
| Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND | |
| β-PL (overnight) | + | + | − | + | − | − | − | − | − | − | − | |
Virus specimens are noted as being positive or negative for a CPE and influenza virus matrix gene RNA and mRNA. Control samples were subjected to the same protocol as the treatment specimens except for the treatment itself: control treatments were heat at 22°C and the absence of formalin, Triton X-100, or β-PL. vRNA, viral genomic RNA; ND, not determined.
HA and NA characteristics of inactivated influenza A(H3N2) and A(H7N3) viruses.
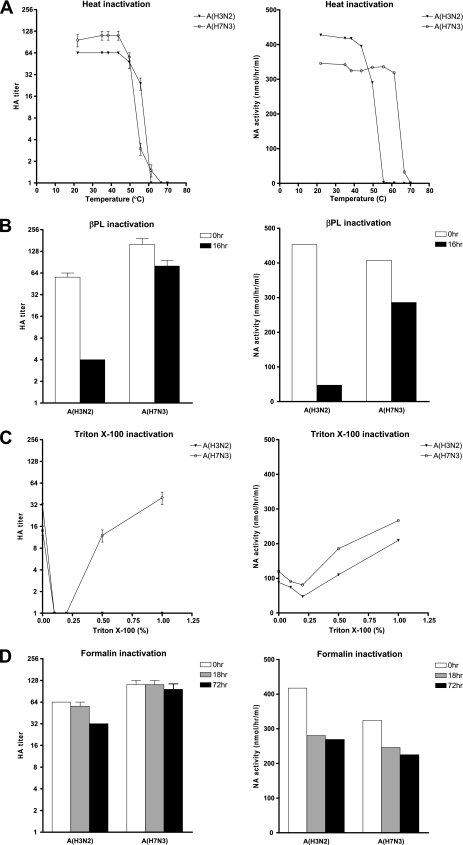
As reported above, virus inactivation may be attained by each of the four described inactivation techniques. To address whether the methods of virus inactivation affected the virological properties, we determined the HA and NA characteristics (Fig. 1) after virus inactivation by each treatment.
FIG. 1.
Hemagglutination titers (left) and NA activities (right) of human influenza A(H3N2) virus and avian influenza A(H7N3) virus after treatment with heat (A), β-PL (B), Triton X-100 (C), or formalin (D). Controls were subjected to the same procedures as the treatment specimens except for the treatment itself: they were heated at 22°C and had no β-PL, Triton X-100, or formalin added. Hemagglutination titers and NA activities after Triton X-100 treatment were determined after the removal of the detergent. Hemagglutination titers of <2 are expressed as 1.
Successful heat inactivation coincided with the loss of HA and NA characteristics for both viruses; however, the human A(H3N2) virus lost HA activity at a slightly higher temperature than the avian A(H7N3) virus (difference in temperature [ΔT], 4°C). In contrast, avian A(H7N3) virus lost NA activity at a higher temperature than the human A(H3N2) virus (ΔT, 14°C). β-PL inactivation did not have a major effect on the avian A(H7N3) virus HA and NA characteristics but reduced the hemagglutination titer and NA activity of the human A(H3N2) virus by 10-fold or more.
Treatments with Triton X-100 and formalin resulted in less significant reduction of HA and/or NA activity than heat and β-PL treatments. Although the log TCID50 of infectious virus was not adversely affected by passing of supernatants over the detergent removal column [log TCID50s of A(H7N3) virus (means ± standard deviations), 5.0 ± 0.25/ml precolumn treatment and 4.75 ± 0.35/ml postcolumn treatment], both the hemagglutination titers and NA activities of the viruses were reduced by subjection to the column step without the addition of Triton X-100 (mock treatment) compared to those of the stock viruses (values were similar to those for the 1-h heat treatment at 22°C) (Fig. 1). Triton X-100 treatment resulted in inconsistent HA activity compared with that of the control, depending on the virus subtype. However, compared to those of the controls, the NA activities of both viruses were enhanced consistently by the addition of Triton X-100 (Fig. 1C). In addition, without the removal of Triton X-100, the NA activities of these viruses were enhanced even further beyond those of the stock viruses (see Fig. 3A). Therefore, we further investigated the use of Triton X-100 treatment in NAI susceptibility assays.
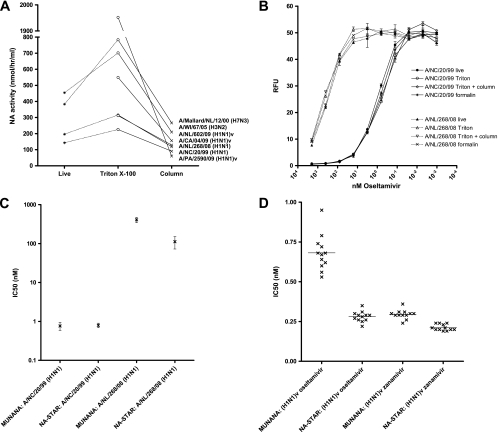
FIG. 3.
Results obtained using inactivated influenza viruses in NA inhibition assays. (A) The NA activity increases after 1% Triton X-100 treatment but is subsequently reduced by column-based detergent removal. Lines connect results from experiments with the same virus strain. (B) Effects of 1% Triton X-100 and formalin treatments on the performance of the MUNANA NA inhibition assay with oseltamivir-sensitive and -resistant influenza viruses. Results obtained after column-based detergent removal (Triton + column) are indicated. RFU, relative fluorescence units. (C) IC50 values obtained with the MUNANA assay (for live and Triton X-100-treated virus samples, virus samples treated with Triton X-100 and subjected to detergent removal, and formalin-treated virus samples) and the NA-Star assay (for live and Triton X-100-treated virus samples and virus samples treated with Triton X-100 and subjected to detergent removal) for NA inhibition with oseltamivir-sensitive and -resistant seasonal influenza A(H1N1) viruses. (D) NAI susceptibilities of 12 1% Triton X-100-treated influenza A(H1N1)v virus isolates as measured with the MUNANA and NA-Star NA inhibition assays.
Nevertheless, formalin treatment best retained both HA and NA activities (Fig. 1) but was more complex to perform than Triton X-100 treatment without detergent removal.
Seasonal influenza A(H1N1) and pandemic influenza A(H1N1)v virus inactivation and HA and NA characteristics.
Based on the results obtained from human A(H3N2) and avian A(H7N3) virus inactivation using four different protocols, Triton X-100 and formalin inactivation protocols were chosen to demonstrate the proof of principle using seasonal and pandemic A(H1N1) viruses. To this end, we validated virus inactivation by Triton X-100 and formalin by using two seasonal A(H1N1) viruses, A/Netherlands/268/2008 (103.8 TCID50/ml) and A/New Caledonia/20/99 (105.0 TCID50/ml), and three pandemic A(H1N1)v virus strains of porcine origin, A/Netherlands/602/2009 (104.1 TCID50/ml), A/California/04/2009 (104.9 TCID50/ml), and A/Paris/2590/2009 (108.0 TCID50/ml). Both 1% Triton X-100 and 18-h 0.02% formalin inactivation protocols effectively inactivated the viruses, as demonstrated by the absence of CPE in three consecutive passages of the treated viruses (Table 2). The results of the RT-PCRs for virus matrix gene RNA and mRNA confirmed that virus replication was absent (Table 2). These data demonstrate that 1% Triton X-100 and 18-h 0.02% formalin inactivation protocols result in complete inactivation of both seasonal A(H1N1) and pandemic A(H1N1)v virus strains at concentrations up to 108.0 TCID50/ml.
TABLE 2.
Confirmation of seasonal and pandemic A(H1N1) influenza virus inactivation by phenotypic and molecular testsa
| Virus | Treatment or sample status | Posttreatment |
Passage 1 |
Passage 2 |
Passage 3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| vRNA | mRNA | CPE | vRNA | mRNA | CPE | vRNA | mRNA | CPE | vRNA | mRNA | ||
| Influenza A/Netherlands/ | Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| 268/2008 (H1N1) virus | 0.02% formalin for 18 h | + | + | − | + | − | − | − | − | − | − | − |
| 1% Triton X-100 | + | + | − | − | − | − | − | − | − | − | − | |
| Influenza A/New Caledonia/ | Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| 20/99 (H1N1) virus | 0.02% formalin for 18 h | + | + | − | + | − | − | − | − | − | − | − |
| 1% Triton X-100 | + | + | − | + | − | − | − | − | − | − | − | |
| Influenza A/Netherlands/ | Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| 602/2009 (H1N1)v virus | 0.02% formalin for 18 h | + | + | − | + | − | − | − | − | − | − | − |
| 1% Triton X-100 | + | + | − | − | − | − | − | − | − | − | − | |
| Influenza A/California/04/ | Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| 2009 (H1N1)v virus | 0.02% formalin for 18 h | + | + | − | + | − | − | − | − | − | − | − |
| 1% Triton X-100 | + | + | − | − | − | − | − | − | − | − | − | |
| Influenza A/Paris/2590/2009 | Control | + | + | + | + | + | ND | ND | ND | ND | ND | ND |
| (H1N1)v virus | 0.02% formalin for 18 h | + | + | − | + | − | − | − | − | − | − | − |
| 1% Triton X-100 | + | + | − | − | − | − | − | − | − | − | − | |
Virus specimens are noted as being positive or negative for a CPE and influenza virus matrix gene RNA and mRNA. Controls were the stock viruses prior to treatment. vRNA, viral genomic RNA; ND, not determined.
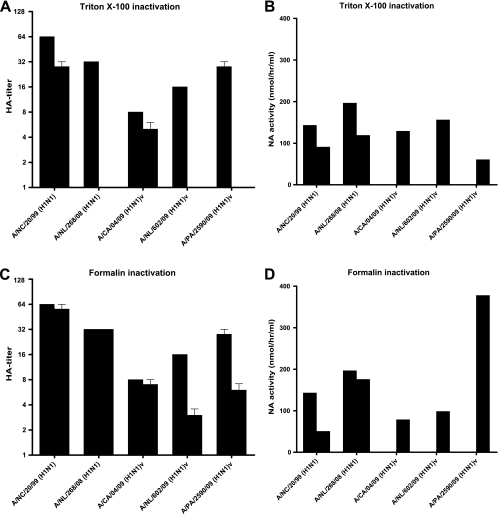
Characterization of the inactivated viruses showed that Triton X-100 treatment again resulted in inconsistent hemagglutination titers (Fig. 2A). After formalin treatment, seasonal influenza A(H1N1) virus hemagglutination titers were retained but A(H1N1)v hemagglutination titers were reduced twofold (Fig. 2C). The NA activities of seasonal influenza A(H1N1) viruses were reduced less than twofold by both inactivation protocols (after Triton X-100 removal) (Fig. 2B and D). Although NA activities of live influenza A(H1N1)v viruses could not be evaluated for technical reasons (no equipment was available in the BSL-3 laboratory), the other viruses showed consistently increased NA activities after treatment with Triton X-100 without removal of the detergent compared to the NA activities of the virus stocks (Fig. 3A). Based on the combined HA and NA characteristics, we concluded that formalin treatment best retained HA and NA activity. In addition, the results confirmed the usefulness of Triton X-100 inactivation without removal of the detergent for the determination of NAI susceptibility.
FIG. 2.
Hemagglutination titers (left) and NA activities (right) of seasonal A(H1N1) and pandemic A(H1N1)v viruses after inactivation using Triton X-100 (A and B) or formalin (C and D). The left bars show the results for the original stock viruses, and the right bars show the results for the viruses after inactivation. Hemagglutination titers and NA activities after Triton X-100 treatment were determined after the removal of the detergent. Hemagglutination titers of <2 are expressed as 1. NA activities of live A(H1N1)v viruses were not determined and are therefore not shown for these viruses in panels B and D.
Analysis of NA inhibition after influenza A(H1N1) virus inactivation.
Before the implementation of routine NAI susceptibility testing using inactivated virus culture supernatants, preservation of NA enzyme characteristics after 1% Triton X-100 or 0.02% formalin treatment needed further evaluation. Therefore, oseltamivir-sensitive and -resistant seasonal influenza A(H1N1) virus isolates were inactivated prior to NAI susceptibility testing with the MUNANA assay. The IC50 values of oseltamivir for live and inactivated virus isolates remained within the 95% confidence interval of interassay variability for the MUNANA assay, demonstrating that NA enzyme inhibition by oseltamivir was not affected by Triton X-100 or formalin treatment (Fig. 3B and C).
Since Triton X-100 treatment is a rapid and easy-to-use inactivation protocol that in addition enhances the NA activities of influenza virus isolates (Fig. 3A), Triton X-100-treated supernatants were subsequently tested with the commercial NA-Star assay. The NA-Star assay performed similarly to the MUNANA assay, demonstrating that NAI susceptibility testing using 1% Triton X-100-treated supernatants works well with both assays (Fig. 3C). Subsequent parallel NAI susceptibility testing of 12 Triton X-100-treated A(H1N1)v virus isolates in the MUNANA and NA-Star assays showed the NAI-sensitive phenotype of current A(H1N1)v viruses, for which IC50 values of both oseltamivir and zanamivir were <1 nM (Fig. 3D and Table 3).
TABLE 3.
Phenotypic NAI susceptibilities of 12 clinical A(H1N1)v virus isolatesa
| NAI assay | Virus | Mean IC50 (nM) (−SD, +SD)b of: |
|
|---|---|---|---|
| Oseltamivir | Zanamivir | ||
| NA-Star assay | A(H1N1)v | 0.28 (0.25, 0.31) | 0.21 (0.19, 0.23) |
| MUNANA assay | A(H1N1)v | 0.67* (0.58, 0.79) | 0.29** (0.27, 0.32) |
| A(H1N1)a | 0.85* (0.68, 1.05) | 0.68** (0.49, 0.92) | |
Baseline NAI susceptibilities of Dutch seasonal A(H1N1) influenza viruses are given (17).
The NAI susceptibilities of pandemic A(H1N1)v virus were greater than those of seasonal A(H1N1) virus. *, P = 0.002; **, P < 0.001.
B-cell responses to inactivated influenza A(H1N1) and A(H1N1)v viruses.
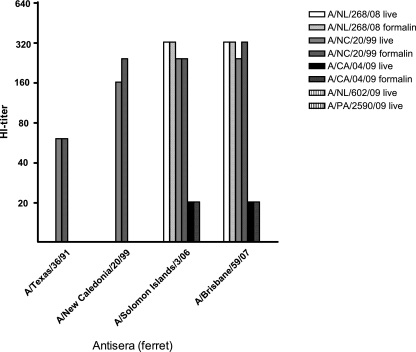
To verify the preservation of B-cell response antigenic epitopes after influenza virus inactivation, we used the HI assay. Due to the reduction of HA activity after Triton X-100 treatment, only the formalin-treated influenza viruses, except for formalin-treated influenza A(H1N1)v viruses A/Netherlands/602/09 and A/Paris/2590/09 (Fig. 2A and C), demonstrated sufficient hemagglutination titers to be used in the HI assay. The HI titers of four ferret antisera for live- and formalin-treated seasonal influenza A/New Caledonia/20/99 and A/Netherlands/268/08 viruses and pandemic influenza A/California/04/09 virus were similar, illustrating that epitopes on HA to which B-cell responses are directed are preserved after formalin treatment (Fig. 4). In contrast to A/California/04/09 virus, pandemic A/Netherlands/602/09 and A/Paris/2590/09 viruses did not show any cross-reactivity with ferret sera against recent seasonal A(H1N1) viruses.
FIG. 4.
HI titers of four ferret antisera with live or formalin-treated influenza A(H1N1) viruses. Only one of the A(H1N1)v pandemic strains (A/California/04/09) showed some cross-reactivity with ferret antisera against recent seasonal A(H1N1) influenza viruses. Formalin-treated A/Netherlands/602/09 and A/Paris/2590/09 viruses had too-low hemagglutination titers to be included in the HI assay (Fig. 2C). HI titers of <20 are not shown.
T-cell responses to inactivated seasonal influenza virus.
Since the highest degree of preservation of HA and NA activities was achieved by inactivating influenza A(H3N2) and A(H7N3) viruses using formalin, the preservation of T-cell epitope antigenicity was assessed using formalin-inactivated influenza viruses. To determine the magnitudes and kinetics of T-cell responses to formalin-treated influenza viruses, PBMC from six donors were infected with live influenza A/Wisconsin/67/2005 (H3N2) virus or pulsed with formalin-treated A/Wisconsin/67/2005 (H3N2) virus or a negative-control sample, after which T-cell responses were measured by flow cytometry at 24, 48, 72, and 168 h poststimulation.
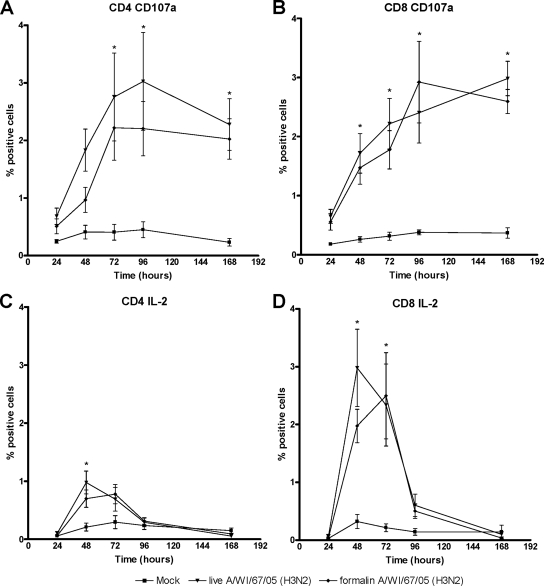
Infection of PBMC with live influenza A/Wisconsin/67/2005 (H3N2) virus resulted in a significant increase (P < 0.05; Newman-Keuls analysis of variance) in CD4+ and CD8+ T cells expressing CD107a and IL-2, indicating cytotoxic activity and cellular proliferation, respectively (Fig. 5). A similar expression pattern among PBMC pulsed with formalin-treated influenza A/Wisconsin/67/2005 (H3N2) virus was observed. The highest percentages of CD107a+ and IL-2+ T cells in response to formalin-treated virus were observed approximately 24 h later than the highest level of the response to live virus. Furthermore, the proportions of CD107a+ and IL-2+ T cells found after pulsing with formalin-treated virus were lower than those found after infection with live virus, but the difference was not statistically significant (P > 0.05). Pulsing PBMC with twice the amount of formalin-treated virus did not result in significantly higher T-cell responses (P > 0.05). Similar results were obtained with A/New Caledonia/20/99 (H1N1) virus (Fig. 6). Together, these results indicate that T-cell epitope antigenicity of seasonal influenza viruses is preserved after formalin treatment.
FIG. 5.
T-cell response kinetics after PBMC stimulation with live or formalin-treated influenza virus A/Wisconsin/67/05 (H3N2). (A) Percentages of CD4+ CD107a+ T cells; (B) percentages of CD8+ CD107a+ T cells; (C) percentages of CD4+ IL-2+ T cells; (D) percentages of CD8+ IL-2+ T cells. Mock, culture supernatant from uninfected cells; *, P < 0.05 for live and formalin-inactivated A(H3N2) virus samples compared to mock-treated samples.
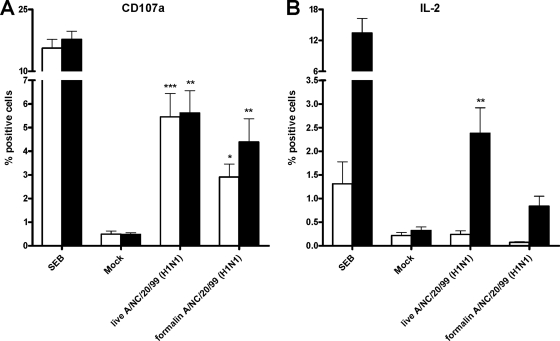
FIG. 6.
Detection of T-cell responses against live or formalin-treated A/New Caledonia/20/99 (H1N1) virus demonstrates significant T-cell epitope antigenicity 72 h poststimulation. (A) Percentages of CD4+ CD107a+ T cells (white bars) and CD8+ CD107a+ T cells (black bars); (B) percentages of CD4+ IL-2+ T cells (white bars) and CD8+ IL-2+ T cells (black bars). SEB, positive control using S. aureus enterotoxin B; mock, culture supernatant from uninfected cells. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 for live and formalin-inactivated A(H1N1) virus samples compared to mock-treated samples.
T-cell responses to inactivated 2009 pandemic influenza A(H1N1)v virus.
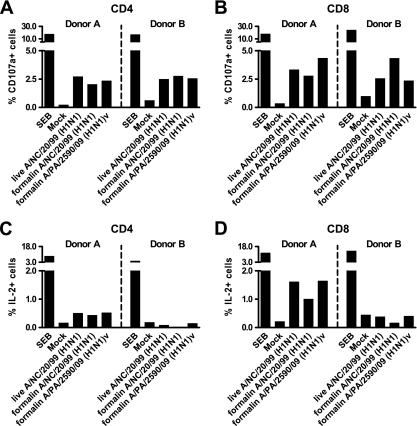
Since formalin treatment effectively inactivated seasonal influenza viruses while preserving T-cell epitope antigenicity, we determined whether the T-cell epitope antigenicity of the pandemic A(H1N1)v virus was also preserved. To this end, (H1N1)v virus-specific T-cell responses by PBMC isolated from two individuals with laboratory-confirmed A(H1N1)v virus infections were evaluated. In vitro stimulation of these PBMC with inactivated influenza A/Paris/2590/2009 (H1N1)v virus demonstrated increases in the percentages of CD107a+ T cells in the CD4+ and CD8+ T-cell subsets from both donors (Fig. 7). As a control, PBMC were infected with live seasonal influenza A/New Caledonia/20/99 (H1N1) virus or pulsed with formalin-treated A/New Caledonia/20/99 (H1N1) virus, showing a similar increase in CD107a+ cells. Furthermore, live and formalin-treated seasonal influenza A(H1N1) virus samples and formalin-treated influenza A(H1N1)v virus led to increases in IL-2+ T cells in PBMC from donor A, which were most apparent in the CD8+ T-cell subset. The stimulation of PBMC from donor B did not produce an increase in IL-2+ T cells in both the CD4+ and CD8+ T-cell subsets, indicating donor variability in the T-cell responses.
FIG. 7.
Detection of T-cell responses against formalin-treated pandemic influenza A/Paris/2590/09 (H1N1)v virus demonstrates significant T-cell epitope antigenicity. (A) Percentages of CD4+ CD107a+ T cells; (B) percentages of CD8+ CD107a+ T cells; (C) percentages of CD4+ IL-2+ T cells; (D) percentages of CD8+ IL-2+ T cells. SEB, positive control using S. aureus enterotoxin B; mock, culture supernatant from uninfected cells.
Together, these data demonstrate that the influenza A(H1N1)v virus retains T-cell-stimulatory capacity after formalin treatment, indicating that T-cell epitope antigenicity is also preserved for this pandemic influenza virus.
DISCUSSION
Our study provides validated virus inactivation protocols that allow implementation of phenotypic NAI susceptibility testing, HI assessment (for serology as well antigenic characterization of viruses), and T-cell response characterization using avian, swine, and human influenza viruses under BSL-2 containment conditions. Using pandemic influenza A(H1N1)v virus strains, we illustrate the ease of carrying out Triton X-100 and formalin virus inactivation protocols prior to the assessment of A(H1N1)v virus susceptibility to antivirals and the characterization of B- and T-cell responses, respectively, outside the BSL-3 high-containment facility. These inactivation protocols facilitate the diagnostic examination of pandemic influenza viruses by applying standard laboratory conditions at BSL-2.
Considering results from documented studies and taking ease of use under BSL-3 conditions into account, therefore excluding, e.g., irradiation protocols, we evaluated human A(H3N2) and avian A(H7N3) virus inactivation by means of heat and the virucidal compounds β-PL, Triton X-100, and formalin (5-7, 14, 18). The most optimal virus inactivation protocols were used to demonstrate the proof of principle with seasonal and pandemic A(H1N1) viruses.
Heat inactivation of influenza A virus can be very efficient, as demonstrated by studies of thermal processing of meat from influenza virus-infected chickens (36, 39), and can be achieved in less than 5 s at temperatures above 70°C (38). Reproducible heat inactivation, however, should be carried out under controlled conditions, e.g., by using small volumes in a thermocycler (37). We used a 30-min incubation period with temperatures below 70°C in an effort to preserve protein function. Unfortunately, infectivity and glycoprotein function disappeared almost simultaneously at temperatures above 55.6°C. Interestingly, the avian A(H7N3) virus NA enzyme demonstrated better heat resistance than the human A(H3N2) virus NA (ΔT, 14°C). Nevertheless, heat treatment appeared not to be suitable for our purposes.
Although β-PL treatment resulted in complete inactivation of influenza virus, it negatively affected both HA and NA functions of the tested A(H3N2) virus, as was demonstrated previously by Goldstein and Tauraso (14). During β-PL hydrolysis, the pH can decrease, resulting in a conformational change in HA (28, 31). Furthermore, β-PL is highly reactive with nucleic acids and proteins and can alter RNA/DNA structure, as well as form RNA/DNA-protein complexes (24, 26). As pH measurements during β-PL treatment demonstrated that the pH remained at physiologic values (data not shown), we hypothesize that the formation of RNA/DNA-protein complexes compromised glycoprotein function of the tested A(H3N2) virus. Even though β-PL is widely used by the vaccine industry for vaccine preparation (1), our results indicate that β-PL inactivation is preferentially not to be used in combination with virological and immunological assays.
Treatment with 1% Triton X-100 appeared to be an easy-to-perform procedure for complete influenza virus inactivation and subsequent determination of NAI susceptibility. However, for validation of virus inactivation in cell cultures by Triton X-100 treatment, the detergent had to be removed by passing the supernatants over a detergent-absorbing column to prevent solubilization of MDCK cell membranes in the cell monolayer. As the capacity of the column used is 15 mg detergent and the amount of Triton X-100 at 1% in 500 ml is about 5 mg, complete removal of Triton X-100 was assumed and culture validation results were eventually not affected by residual Triton X-100. Although the column itself had no significant effect on log TCID50, the removal of Triton X-100 resulted in variable functional glycoprotein recovery depending on the Triton X-100 concentration used (Fig. 1). Without the column-based removal of Triton X-100, however, NA activity increased, allowing the straightforward application of phenotypic NA inhibition assays (the NA-Star assay and the MUNANA assay) (Fig. 3A). These effects may be explained by the mode of action of Triton X-100, solubilization of the lipid membrane of influenza virus and formation of micelles in which the HA and NA proteins are trapped. The concentration of Triton X-100 in a 1% (vol/vol) solution (17 mM) is well above the critical micelle concentration (0.2 to 0.9 mM) at which detergent molecules begin to accumulate in a lipid bilayer membrane, guaranteeing complete solubilization and, therefore, inactivation of influenza virus. However, a note of caution should be made. As the critical micelle concentration of Triton X-100 depends on temperature and complete solubilization at a given concentration of Triton X-100 depends on the amounts of membranes and protein in the solution, our protocol is validated for the given temperature and duration (room temperature for 1 h) and for tissue culture supernatants containing infection medium without FCS that have been cleared of cell debris. The use of higher-protein or lipid-content matrixes, e.g., allantoic fluid with influenza virus, may adversely affect inactivation, and a higher concentration of Triton X-100 should probably be used. Increased NA activity in 1% Triton X-100 solution may be explained by better access of the substrate to the NA in micelles than in the intact virus. The removal of Triton X-100 will cause clumping of NA and HA molecules as well, due to the hydrophobic action of the tails of NA and HA, which are normally located in the lipid bilayer of the virus membrane, resulting in decreased NA activity and inconsistent hemagglutination.
Nevertheless, using 1% Triton X-100 inactivation without detergent removal, we demonstrated that the baseline susceptibilities of A(H1N1)v virus to both oseltamivir (P = 0.002; Student's t test) and zanamivir (P < 0.001; Student's t test) were greater than those of seasonal influenza A(H1N1) viruses (Table 3). However, since phenotypic NAI susceptibility assays such as the MUNANA and NA-Star assays are conducted with virus isolates, this approach still obstructs rapid antiviral susceptibility profiling of an influenza virus infection. The NA activity-enhancing mechanism of Triton X-100 treatment may increase the sensitivity of phenotypic NAI susceptibility assays and may allow direct testing of low-protein-content clinical specimens without the need of virus isolation in the near future.
Formalin treatment appeared to be superior to the other treatments for antigenic characterization and measurement of B- and T-cell responses. The assessment of the B-cell epitope integrity of formalin-treated influenza virus by the HI assay showed similar HI titers for live and formalin-treated influenza virus samples. Although the ferret antiserum HI assay is able to distinguish major antigenic variants, minor changes in antigenicity are difficult to define reliably (34). Nevertheless, we demonstrated that HA-specific antibodies still bind to formalin-inactivated viruses, suggesting preservation of B-cell epitopes. The HI titers further confirmed the major antigenic difference between recent seasonal influenza A(H1N1) viruses and pandemic A(H1N1)v viruses, corresponding with published data (12). Additionally, we detected antigenic differences among A(H1N1)v viruses. Significant HI titers of the ferret sera for recent seasonal A(H1N1) viruses were found by using live and formalin-inactivated influenza A/California/04/09 H1N1v virus samples, while HI titers of the same sera tested using live A/Netherlands/602/09 H1N1v and A/Paris/2590/09 H1N1v viruses were <20. Influenza virus sequence data obtained from GenBank demonstrates the presence of amino acid substitutions P83S, I191L, T197A, and I321V in HAs of A/Netherlands/602/09 and A/Paris/2590/09 viruses, compared to HA in A/California/04/09 virus. As HA substitutions P83S, I191L, and T197A are located on HA1 antigenic sites, this may explain the differences in HI titers obtained with the three influenza A(H1N1)v viruses. Whether this antigenic change was induced by human immune pressure and correlates with B-cell escape is unclear.
Furthermore, we have demonstrated that formalin treatment of both seasonal and pandemic influenza viruses preserves significant T-cell epitope antigenicity for the detection of cellular immune responses. Upon infection of PBMC with live influenza virus, the numbers of cells expressing CD107a and IL-2 increased rapidly. Interestingly, the responses induced by formalin-treated influenza virus started to increase after a 24-h delay and were reduced compared to the levels induced by live influenza virus. The delay may be due to decreased HA integrity, which we observed in the hemagglutination assays with some formalin-treated viruses, ultimately resulting in less efficient binding and uptake of virus by antigen-presenting cells and subsequent activation of T cells (20, 33). However, a more likely explanation is the difference in antigen presentation routes using live and inactivated viruses. During a live-virus infection, antigen is presented on major histocompatibility complex class I (MHC-I) molecules mainly via the endogenous pathway, whereas antigens from inactivated virus are presented on MHC-I molecules by the alternative cross-presentation pathway, which may affect the induction of CD8+ T-cell responses (15). Furthermore, formalin-inactivated influenza virus is not able to replicate and cannot lead to de novo synthesis of viral particles (30). This may result in fewer viral particles and a lower magnitude of the T-cell response (20). However, the amount of inactivated virus did not limit the T-cell response, since pulsing of PBMC with a double amount of inactivated influenza virus did not result in increased T-cell responses (data not shown). Taken together, these findings suggest that differences in antigen processing or activation of different signaling pathways are more likely to affect the T-cell response than the amount of available antigens.
Similarly, differences in IL-2+ T cells specific for pandemic influenza A(H1N1)v virus in PBMC from recently infected individuals may depend on the differentiation status of the T cells, since the PBMC from the two donors were isolated at different time points after the start of symptoms, 13 and 19 days (21, 32). Nevertheless, the significance of this finding is that the assay is also applicable to the pandemic A(H1N1)v virus.
In conclusion, we have shown that the standard repertoire of virological and immunological assays using pandemic influenza virus strains can be performed at BSL-2 when appropriate virus inactivation protocols are applied. Depending on the type of assay, different virus inactivation protocols may be preferred. Rapid antiviral susceptibility profiling with inactivated influenza viruses is possible using 1% Triton X-100 treatment, while 18 h of 0.02% formalin treatment and subsequent dialysis are more time-consuming but enable the implementation of a variety of both virological and immunological assays, including the detection of T-cell responses. Both inactivation procedures allow studies of highly pathogenic or pandemic influenza viruses without the requirement for a BSL-3 facility and greatly expand diagnostic and research possibilities. A limiting factor, however, remains the initial isolation and expansion of these viruses at BSL-3. Conclusively, the timely determination of the public health impact and the development of protective measures when a new influenza virus is introduced in humans are assisted by the implementation of our influenza virus inactivation protocols.
Acknowledgments
We are grateful to S. van der Werf (Institute Pasteur, Paris, France) for providing influenza viruses A/New Caledonia/20/99 (H1N1), A/Wisconsin/67/2005 (H3N2), and A/Paris/2590/2009 (H1N1)v, to R. A. M. Fouchier (Erasmus Medical Centre, Rotterdam, Netherlands) for providing influenza viruses A/Mallard/NL/12/2000 (H7N3) and A/Netherlands/602/2009 (H1N1)v, and to I. Jankovics (National Center of Epidemiology, Budapest, Hungary) for providing influenza virus A/California/04/2009 (H1N1)v.
Marcel Jonges was funded by the Impulse Veterinary Avian Influenza Research in the Netherlands program of the Economic Structure Enhancement Fund. The work of Wai Ming Liu, Ronald Jacobi, Inge Pronk, Claire Boog, and Ernst Soethout was funded by IMECS. IMECS has been made possible by contributions of the European Commission DG Research and the participating member states. The work was performed at the National Institute for Public Health and the Environment, Center for Infectious Disease Control, Laboratory for Infectious Diseases and Screening, P.O. Box 1, 3720 BA Bilthoven, Netherlands, and the Netherlands Vaccine Institute, P.O. Box 457, 3720 AL Bilthoven, Netherlands.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Bardiya, N., and J. H. Bae. 2005. Influenza vaccines: recent advances in production technologies. Appl. Microbiol. Biotechnol. 67:299-305. [DOI] [PubMed] [Google Scholar]
- 2.Besselaar, T. G., D. Naidoo, A. Buys, V. Gregory, J. McAnerney, J. M. Manamela, L. Blumberg, and B. D. Schoub. 2008. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 14:1809-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bot, A., A. Reichlin, H. Isobe, S. Bot, J. Schulman, W. M. Yokoyama, and C. A. Bona. 1996. Cellular mechanisms involved in protection and recovery from influenza virus infection in immunodeficient mice. J. Virol. 70:5668-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claas, E. C., A. D. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 5.De Benedictis, P., M. S. Beato, and I. Capua. 2007. Inactivation of avian influenza viruses by chemical agents and physical conditions: a review. Zoonoses Public Health 54:51-68. [DOI] [PubMed] [Google Scholar]
- 6.De Flora, S., and G. Badolati. 1973. Inactivation of A2-Hong Kong influenza virus by heat and by freeze-thawing. Comparison of untreated and gamma-irradiated preparations. Boll. Ist. Sieroter. Milan. 52:293-305. [PubMed] [Google Scholar]
- 7.De Flora, S., and G. Badolati. 1973. Thermal inactivation of untreated and gamma-irradiated A2-Aichi-2-68 influenza virus. J. Gen. Virol. 20:261-265. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, J. C., A. M. Palache, W. E. Beyer, G. F. Rimmelzwaan, A. C. Boon, and A. D. Osterhaus. 2003. Haemagglutination-inhibiting antibody to influenza virus. Dev. Biol. (Basel) 115:63-73. [PubMed] [Google Scholar]
- 9.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, K. St. George, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 10.Doherty, P. C., D. J. Topham, R. A. Tripp, R. D. Cardin, J. W. Brooks, and P. G. Stevenson. 1997. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol. Rev. 159:105-117. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier, R. A., P. M. Schneeberger, F. W. Rozendaal, J. M. Broekman, S. A. Kemink, V. Munster, T. Kuiken, G. F. Rimmelzwaan, M. Schutten, G. J. Van Doornum, G. Koch, A. Bosman, M. Koopmans, and A. D. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U. S. A. 101:1356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goddard, N., P. Zucs, B. Ciancio, F. Plata, O. Hungnes, A. Mazick, A. Meijer, A. Hay, R. Daniels, A. Nicoll, and M. Zambon. 2009. Start of the influenza season 2008-9 in Europe—increasing influenza activity moving from West to East dominated by A(H3N2). Euro Surveill. 14:pii=19097. [PubMed] [Google Scholar]
- 14.Goldstein, M. A., and N. M. Tauraso. 1970. Effect of formalin, β-propiolactone, merthiolate, and ultraviolet light upon influenza virus infectivity, chicken cell agglutination, hemagglutination, and antigenicity. Appl. Microbiol. 19:290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groothuis, T. A., and J. Neefjes. 2005. The many roads to cross-presentation. J. Exp. Med. 202:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, K. Takahashi, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonges, M., I. M. van der Lubben, F. Dijkstra, L. Verhoef, M. Koopmans, and A. Meijer. 2009. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008. Antiviral Res. 83:290-297. [DOI] [PubMed] [Google Scholar]
- 18.King, D. J. 1991. Evaluation of different methods of inactivation of Newcastle disease virus and avian influenza virus in egg fluids and serum. Avian Dis. 35:505-514. [PubMed] [Google Scholar]
- 19.Koopmans, M., B. Wilbrink, M. Conyn, G. Natrop, H. van der Nat, H. Vennema, A. Meijer, J. van Steenbergen, R. Fouchier, A. Osterhaus, and A. Bosman. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587-593. [DOI] [PubMed] [Google Scholar]
- 20.La Gruta, N. L., K. Kedzierska, K. Pang, R. Webby, M. Davenport, W. Chen, S. J. Turner, and P. C. Doherty. 2006. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc. Natl. Acad. Sci. U. S. A. 103:994-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallard, E., F. Vernel-Pauillac, T. Velu, F. Lehmann, J. P. Abastado, M. Salcedo, and N. Bercovici. 2004. IL-2 production by virus- and tumor-specific human CD8 T cells is determined by their fine specificity. J. Immunol. 172:3963-3970. [DOI] [PubMed] [Google Scholar]
- 22.McElhaney, J. E., D. Xie, W. D. Hager, M. B. Barry, Y. Wang, A. Kleppinger, C. Ewen, K. P. Kane, and R. C. Bleackley. 2006. T cell responses are better correlates of vaccine protection in the elderly. J. Immunol. 176:6333-6339. [DOI] [PubMed] [Google Scholar]
- 23.Meijer, A., A. Lackenby, O. Hungnes, B. Lina, S. van-der-Werf, B. Schweiger, M. Opp, J. Paget, J. van-de-Kassteele, A. Hay, and M. Zambon. 2009. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg. Infect. Dis. 15:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nietert, W. C., L. M. Kellicutt, and H. Kubinski. 1974. DNA-protein complexes produced by a carcinogen, beta-propiolactone. Cancer Res. 34:859-864. [PubMed] [Google Scholar]
- 25.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 26.Perrin, P., and S. Morgeaux. 1995. Inactivation of DNA by beta-propiolactone. Biologicals 23:207-211. [DOI] [PubMed] [Google Scholar]
- 27.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 28.Puri, A., F. P. Booy, R. W. Doms, J. M. White, and R. Blumenthal. 1990. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J. Virol. 64:3824-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed, L., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 30.Ronni, T., T. Sareneva, J. Pirhonen, and I. Julkunen. 1995. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J. Immunol. 154:2764-2774. [PubMed] [Google Scholar]
- 31.Ruigrok, R. W., E. A. Hewat, and R. H. Wade. 1992. Low pH deforms the influenza virus envelope. J. Gen. Virol. 73(Pt. 4):995-998. [DOI] [PubMed] [Google Scholar]
- 32.Sallusto, F., and A. Lanzavecchia. 2001. Exploring pathways for memory T cell generation. J. Clin. Invest. 108:805-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 34.Smith, D. J. 2003. Applications of bioinformatics and computational biology to influenza surveillance and vaccine strain selection. Vaccine 21:1758-1761. [DOI] [PubMed] [Google Scholar]
- 35.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 36.Swayne, D. E. 2006. Microassay for measuring thermal inactivation of H5N1 high pathogenicity avian influenza virus in naturally infected chicken meat. Int. J. Food Microbiol. 108:268-271. [DOI] [PubMed] [Google Scholar]
- 37.Swayne, D. E., and J. R. Beck. 2004. Heat inactivation of avian influenza and Newcastle disease viruses in egg products. Avian Pathol. 33:512-518. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, C., D. J. King, and D. E. Swayne. 2008. Thermal inactivation of avian influenza and Newcastle disease viruses in chicken meat. J. Food Prot. 71:1214-1222. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, C., and D. E. Swayne. 2007. Thermal inactivation of H5N1 high pathogenicity avian influenza virus in naturally infected chicken meat. J. Food Prot. 70:674-680. [DOI] [PubMed] [Google Scholar]
- 40.Ward, C. L., M. H. Dempsey, C. J. Ring, R. E. Kempson, L. Zhang, D. Gor, B. W. Snowden, and M. Tisdale. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]