Abstract
The hospital epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) has changed in the past few years due to the encroachment of community-associated MRSA (CA-MRSA) strains into health care settings. MRSA strains that were isolated during a 2-year period from patients of the Luzerner Kantonsspital were analyzed to elucidate their epidemiology. Moreover, extended surveillance of individuals who were contacts of those patients was carried out for 6 months to identify the routes of spread and to assess the quality of the infection control measures used in our setting. Patient data were collected to distinguish CA-MRSA strains from health care-associated MRSA (HA-MRSA) strains by epidemiological criteria, as defined by the Centers for Disease Control and Prevention (CDC). On the basis of the CDC definition, the majority of the strains were considered to be HA-MRSA. However, 87% of them belonged to staphylococcal cassette chromosome mec (SCCmec) types IV and V, which are traditionally associated with CA-MRSA. Surprisingly, classical nosocomial SCCmec types I and II represented a minority, whereas SCCmec type III was completely absent. By PFGE analysis, four predominant clonal lineages and 21 highly variable sporadic genotypes were detected. Twenty-eight percent of the MRSA strains studied carried the genes encoding the Panton-Valentine leukocidin (PVL), of which 21% and 83% were associated with SCCmec types IV and V, respectively. Among 289 contact individuals screened for MRSA carriage throughout the extended surveillance, a single secondary patient was discovered. The possibility of nosocomial transmission could be excluded. The high proportions of SCCmec type IV and V strains as well as PVL-positive strains suggest strong infiltration of CA-MRSA into our institution. Moreover, the low endemic prevalence of MRSA demonstrates that current infection control measures are sufficient to limit its spreading and the emergence of large epidemic outbreaks.
Methicillin-resistant Staphylococcus aureus (MRSA), one of the most common nosocomial pathogens, usually carries genetic elements that confer resistance to a broad range of antibiotics (2, 25). Methicillin resistance in staphylococci is based on the expression of a modified penicillin-binding protein (PBP), PBP 2a, which is encoded by the mecA gene. This gene is located on the staphylococcal cassette chromosome mec (SCCmec), a mobile genetic element integrated in the chromosome (6, 23) which also carries the genes for specific recombinases (ccr) necessary for its excision and integration (24). On the basis of the mec and ccr complex sequences, SCCmec is classified into types I to VII (1, 25). Previous studies showed that health care-associated (HA) MRSA (HA-MRSA) infections are generally caused by multidrug-resistant strains harboring SCCmec types I, II, and III but rarely SCCmec type IV (26, 38, 47, 52). On the other hand, community-associated (CA) MRSA (CA-MRSA) strains carry SCCmec type IV, V, or VII; are commonly susceptible to the majority of non-β-lactam antibiotics; frequently produce the Panton-Valentine leukocidin (PVL); and differ in their pulsed-field gel electrophoresis (PFGE) patterns (38). Recent studies have reported that CA-MRSA strains are spreading in hospital settings and are replacing traditional HA-MRSA strains (44, 46, 47, 52).
Although the average prevalence of MRSA strains in Switzerland has been lower than that in the surrounding countries—between 0 and 6% of the first isolates of S. aureus recovered, except in Geneva, where the rate is 20% (2, 17, 19)—the incidence of MRSA infections is increasing. Even though there are epidemiological data for MRSA in Switzerland (2, 17, 20, 44, 47), the extent of the problem in the central part of Switzerland is unknown. In this study, we aimed at assessing the frequency, diversity, and clonal distribution of MRSA strains that were isolated at the Luzerner Kantonsspital (LUKS) between January 2007 and December 2008, in order to elucidate their origin and ways of circulation within our institution. Moreover, in a 6-month prospective study, we evaluated the effects of extended health care-associated hygienic measures on nosocomial MRSA transmission.
MATERIALS AND METHODS
Retrospective study. (i) Institutional review board approval.
The study was approved by the independent Ethics Committee of the Canton of Luzern in 2008 (approval no. 805).
(ii) Hospital setting and MRSA policy.
LUKS is the largest nonuniversity hospital in Switzerland and has a low average prevalence of MRSA (5% of the first isolates of S. aureus). It includes three hospitals at different geographical locations (Lucerne, Wolhusen, and Sursee) within a radius of 50 km and provides stationary and ambulatory care services for over 130,000 patient visits per year. The LUKS at Lucerne accounts for 600 beds, the LUKS at Sursee for 150 beds, and the LUKS at Wolhusen for 110 beds. Since 2004, LUKS has followed a search-and-destroy policy based on the guidelines issued by the Dutch Society of Infection Prevention and Control (49, 51). Briefly, this policy requires all patients transferred from geographical areas with a high prevalence of MRSA, the contacts of MRSA carriers, and other patients at high risk for MRSA infection to be isolated at the time of hospital admission until screening cultures (from nasal and inguinal swabs) have proven negative (“search”). The MRSA carriers are then isolated and decontaminated (“destroy”) (49, 51).
(iii) Epidemiological data.
The following data were collected from the discharge records for each MRSA case: demographics, dates of admission and discharge, hospitalization within the past 12 months, past or present MRSA colonization or infection, and underlying diagnosis. MRSA infection was defined as any clinical infection with a positive culture for MRSA.
The strains were classified as either CA- or HA-MRSA, according to the standard epidemiological definitions of the Centers for Disease Control and Prevention (CDC) (5). Such strains were defined as CA-MRSA if MRSA was detected in an outpatient setting or by a positive culture for MRSA within 48 h after hospital admission. Moreover, CA-MRSA carriers should not have had (i) a medical history of MRSA colonization or infection; (ii) a history of hospitalization, admission to a nursing home or hospice, dialysis, or surgery during the previous year; or (iii) permanent indwelling percutaneous catheters or medical devices. Strains that did not meet the definition of CA-MRSA were classified as HA-MRSA. Seventy-eight MRSA strains that were isolated from various clinical specimens from inpatients and outpatients between 2007 and 2008 were included in this study. All strains were unique isolates from different patients. Nosocomial clusters were defined as two or more patients who were hospitalized on the same ward within a 6-month period and who harbored identical MRSA isolates (2).
(iv) Bacterial strains, culture, identification, and susceptibility testing.
The identification of S. aureus was performed by standard laboratory methods and was confirmed by analysis with a Vitek 2 instrument (bioMérieux, Marcy l'Etoile, France). Resistance to methicillin was detected by screening on commercial chromID MRSA agar medium (Mueller-Hinton oxacillin; bioMérieux) (42) and was confirmed by PBP 2a agglutination (Slidex MRSA detection assay; bioMérieux). Antimicrobial susceptibility testing (see Table 2) was performed by the disk diffusion method on Mueller-Hinton agar, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (7). mecA was detected from swabs or S. aureus colonies by PCR with an Xpert-MRSA detection kit (Cepheid, Sunnyvale, CA), according to the instructions of the manufacturer or as described previously (45).
TABLE 2.
Characteristics of the MRSA isolates grouped by SCCmec type
| SCCmec type | No. of MRSA strains | No. (%) of HA-MRSA strainsa | No. of dendrogram profiles | No. (%) of PVL-positive strains | No. (%) of strains resistant tob: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | CLI | ERY | MLS | GEN | RIF | SXT | TOB | VAN | |||||
| I | 3 | 3 (100) | 2 | 0 | 3 (100) | 2 (67) | 1 (33) | 0 | 1 (33) | 0 | 0 | 2 (67) | 0 |
| II | 5 | 5 (100) | 1 | 0 | 5 (100) | 4 (80) | 5 (100) | 1 (20) | 0 | 0 | 0 | 5 (100) | 0 |
| IV | 58 | 46 (79) | 25 | 12 (21) | 37 (64) | 3 (5) | 19 (33) | 11 (19) | 3 (5) | 0 | 0 | 4 (7) | 0 |
| V | 12 | 6 (50) | 3 | 10 (83) | 2 (17) | 0 | 1 (8) | 1 (8) | 10 (83) | 0 | 0 | 10 (83) | 0 |
On the basis of the CDC definition (5).
CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; MLS, macrolide-lincosamide-streptogramin B; GEN, gentamicin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin; VAN, vancomycin.
(v) Pulsed-field gel electrophoresis.
PFGE following SmaI digestion was performed according to the HARMONY protocol (8, 37). Intra- and intergel PFGE runs were normalized with S. aureus strain BB255 (39). Band assignments were manually adjusted after automatic band detection; bands ranging from 36 kb to 600 kb were considered for analysis in the study. Comparison and grouping of the PFGE patterns were performed with BioNumerics (version 5.1) software (Applied Maths, St-Martens-Latem, Belgium). PFGE types and subtypes were defined by groups formed at 80% and 95% Dice similarity cutoffs (4, 13, 33, 37) on a dendrogram constructed by the unweighted-pair group method using average linkages (UPGMA), as described by Faria et al. (16). The groups defined at these thresholds were previously shown to approximate those defined by use of the criteria of Tenover and others for visual PFGE type definition (4, 37, 48). Types were labeled with a number, and subtypes were labeled with a letter. Each type and its subtypes were considered to belong to the same clone. If more than five isolates showed an indistinguishable pattern, they were considered members of a predominant clone (2, 48).
(vi) SCCmec typing and detection of PVL genes.
SCCmec types were determined by a multiplex PCR strategy which generates a specific amplification pattern for SCCmec types I to VI (34, 35, 40). Untypeable strains were further analyzed by ccr and mec gene complex typing (27). PCR detection of the PVL genes (lukS-PV and lukF-PV) was performed as described previously (30).
Prospective study.
A 6-month prospective study was carried out between January and June 2009. Index patients were defined as nonisolated patients who were hospitalized >24 h before MRSA was detected. For each index patient discovered, contact patients and health care workers on the same ward were screened for MRSA colonization by the use of nasal swabs.
RESULTS
Retrospective study. (i) Clinical and epidemiological data.
Seventy-eight MRSA strains were isolated from patients at the three LUKS hospitals (Table 1). According to the CDC definition (5), 60 MRSA strains were HA-MRSA and 18 were CA-MRSA. At the major hospital setting (LUKS at Lucerne), most MRSA strains were isolated from patients in the Department of Surgery (35%), followed by patients in the Department of Internal Medicine (19%) and the Children's Hospital (19%). Five MRSA-positive patients were detected at the LUKS at Sursee and seven were detected at the LUKS at Wolhusen; all patients at those locations were hospitalized in either the Department of Internal Medicine, the Department of Surgery, or the Department of Gynecology and Obstetrics. The ages of the patients ranged from 10 days to 92 years (mean ± standard deviation age, 48 ± 29 years; median age, 58 years). Fifty-six patients (72%) were on an inpatient service and 22 (28%) were outpatients at the time of the first MRSA detection. Sixty-seven (86%) MRSA isolates were isolated from skin and soft tissue specimens and 11 (13%) were isolated from other body sites. A relevant proportion of the MRSA isolates were associated with infection (53% of the HA-MRSA isolates and 72% of the CA-MRSA isolates), mostly abscesses (33%) or wound infections (31%) (Table 1). In 10 cases (13%), bacteremia followed infection. MRSA carriage was associated with a previous hospitalization in Switzerland (33%), transfer from a hospital in an area at high risk for MRSA (such as southern Switzerland, Spain, Italy, or France; 9%), residence in a nursing home (17%), or, in general, a history of migration or travel (31%).
TABLE 1.
Demographics, clinical background, type of MRSA carriage, and type of specimens for the 78 MRSA-positive patients detected at the three acute-care hospitals of LUKS between 2007 and 2008
| Characteristic | Value |
|---|---|
| Demographic data | |
| Mean ± SD (median) age (yr) | 48 ± 29 (58) |
| No. (%) of patients | |
| Male | 47 (60) |
| Of foreign origin | 24 (31) |
| With previous hospitalization (12 mo) | 33 (42a) |
| With previous institutionalization (nursing home, prison) | 13 (17) |
| Transferred from a hospital in a high-prevalence area | 7 (9) |
| Clinical background (no. [%] of patients) | |
| Outpatients | 22 (28) |
| Inpatients | 56 (72) |
| LUKS location | |
| Lucerne | 66 (85) |
| Surgery | 27 (35) |
| Internal medicine | 15 (19) |
| Pediatrics | 15 (19) |
| Gynecology and obstetrics | 8 (10) |
| Otorhinolaryngology | 1 (1) |
| Sursee | 5 (6) |
| Wolhusen | 7 (9) |
| Type of MRSA carriage (no. [%] of patients) | |
| HA-MRSAb | 60 (77) |
| CA-MRSAb | 18 (23) |
| Specimen type | |
| Associated with infection | 45 (58) |
| Abscess/tissue | 15 (19) |
| Wound | 14 (18) |
| Skin or subcutaneous lesions | 4 (5) |
| Urine | 6 (8) |
| Other body sites | 6 (8) |
| Associated with colonization | 33 (42) |
The value of 42% includes all hospitalizations in Switzerland as well as abroad.
According to the CDC definition (5).
(ii) Characterization of MRSA strains.
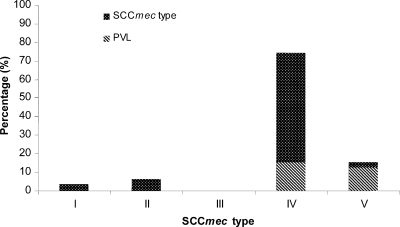
The MRSA isolates were analyzed by SCCmec typing. Four different SCCmec types were detected. Surprisingly, only very few isolates showed the PCR patterns of the traditional nosocomial SCCmec types I (n = 3) and II (n = 5), with those isolates representing 10% of all isolates, whereas SCCmec type III isolates were completely absent. The majority of the MRSA isolates carried SCCmec type IV (n = 58; 74%) or type V (n = 12; 15%) (Fig. 1). Symptomatic infection with MRSA strains was, to various extents, associated with the four SCCmec types (67% for type I, 83% for type II, 48% for type IV, and 92% for type V).
FIG. 1.
Prevalence of SCCmec types at the LUKS hospitals over a 2-year period (2007 and 2008).
MRSA strains carrying SCCmec type I were resistant to ciprofloxacin (100%), clindamycin and tobramycin (67%), erythromycin (33%), and gentamicin (33%). Strains carrying SCCmec type II were characterized by 100% resistance to ciprofloxacin, erythromycin, and tobramycin and by 80% resistance to clindamycin. A high proportion of the SCCmec type IV isolates showed resistance to clindamycin (64%) and erythromycin (33%). Finally, SCCmec type V MRSA strains were characterized by frequent resistance to aminoglycosides. Without exception, all MRSA isolates were susceptible to vancomycin, rifampin, and trimethoprim-sulfamethoxazole (Table 2).
The MRSA strains were also screened for the presence of the PVL genes (lukS-PV and lukF-PV). Twenty-two isolates (28%) carried the genes for PVL, which was strongly associated with SCCmec type IV and V MRSA strains (of which 21% and 83%, respectively, carried the PVL genes) (Fig. 1). A high proportion of the PVL-positive isolates showed resistance to gentamicin (45%), tobramycin (41%), ciprofloxacin (27%), and erythromycin (23%), whereas 23% were pansusceptible (except to the β-lactams) (Table 2). Ninety-one percent (n = 20) of all PVL-positive MRSA strains were associated with infection (55% with abscesses/furunculosis, 25% with skin or soft tissue infections, and 20% with wound infections).
Molecular typing by means of PFGE indicated that 59% (n = 46) of all MRSA strains belonged to four epidemiologically dominant genetic lineages: PFGE patterns 1 (n = 10), 11 (n = 7), 18 (n = 23), and 24 (n = 6) (Fig. 2). Three strains showed PFGE pattern 6B, which was indistinguishable from the pattern of CA-MRSA strain USA300. PFGE type 18 carried SCCmec type IV and was detected in HA-MRSA strains only, while PFGE type 11 carried SCCmec type IV and was mostly detected (86%) in HA-MRSA strains. PFGE type 1A carried SCCmec type V, often exhibited PVL (67%), and was found in both HA-MRSA and CA-MRSA strains. As expected, PFGE type 1B-SCCmec type II was exclusively found among HA-MRSA isolates. MRSA strain PFGE type 24-SCCmec type V was characterized by the uniform presence of PVL (100%) and was found in both CA-MRSA and HA-MRSA strains. A high degree of genetic diversity was observed among sporadic clones (41%), which accounted for clusters of one to three patients and which represented 21 PFGE types (Fig. 2).
FIG. 2.
Dendrogram showing the PFGE patterns of MRSA strains representative of each type and subtype detected. Percent similarity was calculated by use of the Dice coefficient (with a tolerance of 1% and an optimization of 0.5%) and is represented by UPGMA. Bands larger than 600 kb and smaller than 36 kb were not analyzed, due to poor resolution. (a) PFGE pattern; (b) SCCmec type; (c) number of isolates sharing the same PFGE pattern and SCCmec type. Pattern 6B represents that for reference strain USA300.
(iii) Epidemiological results.
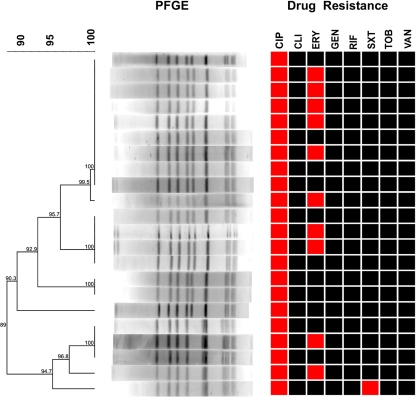
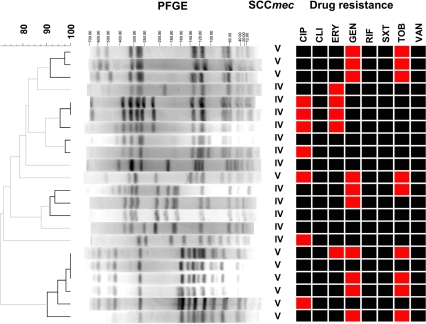
MRSA strains showing the predominant genotype, PFGE type 18-SCCmec type IV, were isolated at all study sites (LUKS at Lucerne, Sursee, and Wolhusen) (Fig. 3). The presence of this particular strain had already been recorded at the LUKS at Lucerne in 2005 (Department of Surgery), and the same strain was also isolated from patients transferred from hospitals in regions at high risk for MRSA. Moreover, several residents of three different nursing homes in our area carried this genotype. The nosocomial transmission of this strain was verified in five cases. Three of them involved concomitant hospitalization in the Department of Surgery in the LUKS at Lucerne, whereas two cases involved residents of the nursing homes mentioned above who were eventually hospitalized in the LUKS at Wolhusen. Vertical mother-to-child and nosocomial PFGE type 11-SCCmec type IV MRSA transmission was observed during a small outbreak in the intensive care unit of the Children's Hospital (LUKS at Lucerne) and involved three neonates and one mother. After 5 months, the same strain was isolated from a neonate on the same ward. Moreover, this clone was previously found in a health care worker of the Department of Gynecology and Obstetrics of the LUKS at Lucerne who had given birth at the same clinic 5 months prior to the outbreak mentioned above. Another cluster comprised four patients from Macedonia and Bosnia-Herzegovina who were infected with a PVL-producing clone (PFGE type 1A-SCCmec type V) showing resistance to aminoglycosides (Fig. 4). Another PVL-producing clone (PFGE type 24-SCCmec type V) was isolated from three infected patients originating from Albania and Serbia and from a child with a heel wound after returning from holidays in Spain (Fig. 4). This clone also caused nosocomial postoperative wound infections in two patients who were hospitalized on the same ward within a period of 2 months (Department of Surgery, LUKS at Lucerne). More cases that were epidemiologically linked were identified among family members or residents of nursing homes. We detected one case of vertical transmission of USA300 (PFGE type 6B-SCCmec type IV) from a mother of African origin to her child. In addition, this strain was also found in an HIV-positive woman originating from Cameroon (Fig. 4).
FIG. 3.
Dendrogram of the predominant cluster of MRSA isolates showing the pattern PFGE type 18 and SCCmec type IV, together with the antibiograms. Susceptibility is depicted in black, and resistance is depicted in red (CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin; VAN, vancomycin). Percent similarity was calculated by use of the Dice coefficient (with a tolerance of 1% and an optimization of 0.5%) and is represented by UPGMA. Bands larger than 600 kb and smaller than 36 kb were not analyzed, due to poor resolution.
FIG. 4.
Dendrogram of PVL-positive MRSA strains. Specific PFGE patterns are shown, as are the SCCmec types and antibiograms. Susceptibility is depicted in black, and resistance is depicted in red (CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TOB, tobramycin; VAN, vancomycin). Percent similarity was calculated by use of the Dice coefficient (tolerance of 1% and an optimization of 0.5%) and is represented by UPGMA. Bands larger than 600 kb and smaller than 36 kb were not analyzed, due to poor resolution.
Prospective study.
Between January and June 2009, for each new index patient the whole ward was screened for MRSA colonization by collecting nasal swabs from patients and medical and nursing staff. In total, four index patients were discovered during this study period. (i) The first index patient was a 5-year-old child originating from Albania who had a labial abscess and who was hospitalized at the Children's Hospital. Screening of 10 patients and 48 health care workers was performed. No MRSA transmission could be detected. (ii) The second index patient was a 61-year-old patient with cancer who was transferred from Spain and hospitalized at the Clinic of Internal Medicine. Fifteen patients and 28 members of the hospital staff were screened, again with negative results. (iii) The third index patient was a 78-year-old man hospitalized in the rehabilitation ward for 27 days prior to the isolation of MRSA from urine. Fifty-seven employees from the rehabilitation ward, 25 employees from the dialysis ward, and 35 patients were screened. One patient was colonized with a MRSA strain resistant only to β-lactams. Molecular characterization of the two strains by PFGE showed two different patterns. (iv) The final index patient was a 50-year-old man with a diabetic foot infection who was found to be MRSA positive during surgery. Thirty patients and 44 hospital staff members were monitored, and all yielded negative results.
In total, 289 extended screenings were performed. Only a single MRSA carrier was discovered, but nosocomial transmission could safely be excluded.
DISCUSSION
In the present report, we provide retrospective epidemiological data and data on the molecular characterization of 78 consecutive single-patient MRSA isolates discovered during a 2-year period in the major nonuniversity academic teaching hospital in Switzerland, in order to elucidate the genetic pool and potential emergence of unexpected MRSA strains. The epidemiological data for the MRSA isolates revealed a prevalence of 5% in our institution with a 77% presence of HA-MRSA, according to the CDC definition. SCCmec types I to III have been reported to be the most frequent nosocomial MRSA strains in the United States (10, 32), Europe (8, 10), and Switzerland, i.e., 86% in Geneva (type I; 2003) and 45% in Basel (types I, II, and III; 2004) (20, 47). Surprisingly, these types were either absent (type III) or present in very low proportions (10%; types I and II) in our hospitals, whereas molecular typing showed a clear overall predominance of MRSA strains carrying SCCmec types IV and V (90%), which are traditionally attributed to CA-MRSA strains (52). These data confirm a tendency seen in previous studies from the University Hospitals of Zurich (45%; type IV) and Basel (58%; type IV) (44, 46) and in international studies that reported the spread of CA-MRSA SCCmec type IV strains in hospital settings in both Europe and the United States (12, 14, 21, 26, 32, 44, 47). To our knowledge, such high proportions of SCCmec types IV and V as were found in our institution have not been reported before. Further epidemiological classification of the typed MRSA strains revealed that 87% of the HA-MRSA strains found in our hospitals carried SCCmec type IV or V. The predominance of HA-MRSA strains carrying a CA genotype in our hospitals is significantly higher than the values reported from previous studies performed in the United States (49% to 56% CA genotypes causing HA infections) (11, 32, 43) and Switzerland (43% of HA-MRSA strains showing SCCmec type IV) (47).
Antimicrobial resistance patterns have been used to distinguish between CA-MRSA and HA-MRSA strains, with CA-MRSA strains showing greater susceptibility to several antimicrobial agents (usually gentamicin, clindamycin, and trimethoprim-sulfamethoxazole) than HA-MRSA (11, 14, 32, 44). In our study, CA-MRSA strains showed greater susceptibility than HA-MRSA strains to clindamycin (0% and 15%, respectively), ciprofloxacin (39% and 67%, respectively), and erythromycin (17% and 38%, respectively). On the other hand, CA-MRSA strains had higher rates of resistance to gentamicin (33% and 13%, respectively). However, by taking into account the SCCmec types as well, the differences in the antimicrobial resistance patterns were negligible. In fact, HA-MRSA strains (n = 6) and CA-MRSA strains (n = 6) carrying SCCmec type V did not show any differences in antibiotic susceptibility, and HA-MRSA (n = 46) and CA-MRSA (n = 12) carrying SCCmec type IV showed slight differences (for ciprofloxacin, 67% and 50%, respectively; for clindamycin, 6% and 0%, respectively; for erythromycin, 35% and 25%, respectively; for gentamicin, 4% and 8%, respectively; for tobramycin, 9% and 0%, respectively). A relevant proportion of the MRSA strains were associated with infection (58%), mostly abscesses (33%) or wound infections (31%). As a whole, 93% of all infections were caused by MRSA strains carrying SCCmec types IV and V, demonstrating that the majority of infections in our hospital settings were caused by strains with the CA genotype.
Another classical feature of CA-MRSA is the production of PVL, a bicomponent exotoxin that is frequently associated with suppurative skin and soft tissue infections (15, 19, 22). Surprisingly, we identified much higher rates of PVL-positive MRSA strains (28%) than the ones reported from other European countries (3 to 15%) (28, 50) or from other Swiss urban areas (Basel, 1%; Zurich, 5%) (44, 47). The only study recording high proportions (35%) of PVL-positive strains comes from Geneva (2002 to 2005), but it was performed with a highly selective collection of non-multidrug-resistant CA-MRSA strains (17). In our study, PVL was strongly associated with SCCmec type V (83%) and, to a lesser extent, type IV (21%). Moreover, we could confirm that the PVL-positive strains were strongly associated with infection (91%) and predominantly caused abscesses or skin or soft tissue infections. The highly infectious CA-MRSA strain USA300 (21, 26) was only rarely detected in our institution.
In addition to the genetic and phenotypic characterization of MRSA strains, the retrospective part of our study aimed at monitoring the circulation of MRSA within the LUKS hospitals. Molecular characterization of the MRSA strains revealed that the majority of all strains isolated (59%) belonged to four predominant genetic lineages, whereas the remaining sporadic isolates showed a high degree of genetic diversity, suggesting that new strains, which are likely to be imported from the expanding community reservoir or from other countries, are adding up in the LUKS pool. Although no large epidemic outbreaks were observed, the circulation of MRSA strains and their local spread within the LUKS hospitals could be partly disclosed, as several nosocomial clusters were retrospectively identified. Interestingly, the predominant clones PFGE type 18-SCCmec type IV, PFGE type 1B-SCCmec type II, and PFGE type 1A-SCCmec type V were isolated from patients hospitalized at all three locations of LUKS. This reinforces the hypothesis of HA spread of these genotypes as patients circulate inside one of the LUKS hospitals and are also transferred from one LUKS hospital to another. However, in an endemic situation in which MRSA is spread to the surrounding institutions and to the community, proven clonality does not provide compelling evidence of recent nosocomial transmission. Thus, the main value of strain typing still lies in the exclusion of nosocomial transmission by the disclosure of different strains.
Taken together, the high proportions of HA-MRSA strains carrying SCCmec types IV and V, together with the considerable occurrence of PVL-positive MRSA strains and the negligible differences in the antibiotic resistance profiles of the CA- and HA-MRSA strains, confirm the hypothesis of the extensive infiltration of CA-MRSA in our hospital settings. These results are in agreement with those provided by mathematical models that predict the replacement of traditional HA-MRSA strains by CA-MRSA strains, due to their higher growth rate and greater fitness (3, 9). This is based on the observation that CA-MRSA strains carrying small SCCmec type IV or V grow faster and achieve higher infectious burdens than nosocomial MRSA strains and, hence, have a selective advantage (29, 43). The strong presence of CA-MRSA in hospital settings has been predicted to have several important implications for infection control, as the effectiveness of infection-control guidelines for the control of CA-MRSA is unknown and data from outbreaks of CA-MRSA infections suggest that skin-to-skin and skin-to-fomite contacts represent important routes of acquisition of the infecting strains (3, 36, 38, 43, 46).
The unique constellation with a very high prevalence of MRSA strains carrying a CA genotype disclosed in our institution enabled us to test the adequacy of current hospital hygiene measures to control outbreaks of in-hospital CA-MRSA strains in a 6-month prospective study. Since 2004, LUKS has followed a search-and-destroy strategy (49, 51), which has been successfully implemented in Northern and Western European countries with both low and high prevalences of MRSA (28, 31, 41) and in the United States (18). From January to June 2009, 4 index patients were discovered in our institution and 289 patients and health care staff members were screened for MRSA carriage, but no nosocomial transmission could be verified. However, our prospective study should be interpreted in light of the limitations of the study. First, the observational period was short and the MRSA incidence rate during this time frame was low. Second, as the transmission pathways of CA-MRSA are still unknown (34, 41), we might have missed colonized individuals by performing only nasal screening. Despite these concerns, it must be stated that epidemic outbreaks did not emerge throughout the 2.5-year time frame of the study and nosocomial transmission was not detected during the 6-month prospective study. Therefore, the search-and-destroy strategy seems to be adequate for limiting the spread of these fitter CA-MRSA strains in an environment with a low prevalence of MRSA.
Acknowledgments
We thank B. Berger-Bächi (Institute of Medical Microbiology, University of Zurich) for kindly providing the reference strains for the PFGE and SCCmec typing assays, the nurses of the LUKS Division of Infectious Disease Control for the collaboration, the team of the LUKS Department of Medical Microbiology and, in particular, G. Sägesser for technical support.
This study was entirely funded by the Bonizzi-Theler Foundation, Switzerland.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Berglund, C., T. Ito, M. Ikeda, X. X. Ma, B. Soderquist, and K. Hiramatsu. 2008. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52:3512-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc, D. S., D. Pittet, C. Ruef, A. F. Widmer, K. Mühlemann, C. Petignat, S. Harbarth, R. Auckenthaler, J. Bille, R. Frei, R. Zbinden, P. Moreillon, P. Sudre, and P. Francioli. 2002. Molecular epidemiology of predominant clones and sporadic strains of methicillin resistant Staphylococcus aureus in Switzerland and comparison with European epidemic clones. Clin. Microbiol. Infect. 8:419-426. [DOI] [PubMed] [Google Scholar]
- 3.Bootsma, M. C. J., O. Diekmann, and M. J. M. Bonten. 2006. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc. Natl. Acad. Sci. U. S. A. 103:5620-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrico, J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazao, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2005, posting date. Community associated MRSA information for clinicians. Infection control topics. Centers for Disease Control and Prevention, Atlanta, GA. www.cdc.gov/ncidod/dhqp/ar_mrsa_ca_clinicians.html.
- 6.Chambers, H. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing, 18th informational supplement, vol. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeeuwen, A. van Belkum, A. Vindel, J. Garaizer, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45:1830-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Agata, E. M. C., G. F. Webb, M. A. Horn, J. Moellering, C. Robert, and S. Ruan. 2009. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clin. Infect. Dis. 48:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, T. A., W. Shang, K. M. Amsler, S. Bajaksouzian, M. R. Jacobs, and K. Bush. 2009. Molecular characterisation of meticillin-resistant Staphylococcus aureus isolates from two ceftobiprole phase 3 complicated skin and skin-structure infection clinical trials. Int. J. Antimicrob. Agents 34:166-168. [DOI] [PubMed] [Google Scholar]
- 11.Davis, S. L., M. J. Rybak, M. Amjad, G. W. Kaatz, and P. S. McKinnon. 2006. Characteristics of patients with healthcare-associated infection due to SCCmec type IV methicillin-resistant Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 27:1025-1031. [DOI] [PubMed] [Google Scholar]
- 12.Denis, O., A. Deplano, R. De Ryck, C. Nonhoff, and M. J. Struelens. 2003. Emergence and spread of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in Belgian hospitals. Microb. Drug Resist. 9:61-71. [DOI] [PubMed] [Google Scholar]
- 13.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 14.Donnio, P.-Y., L. Preney, A.-L. Gautier-Lerestif, J.-L. Avril, and N. Lafforgue. 2004. Changes in staphylococcal cassette chromosome type and antibiotic resistance profile in methicillin-resistant Staphylococcus aureus isolates from a French hospital over an 11 year period. J. Antimicrob. Chemother. 53:808-813. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, M. W., M. E. Griffith, J. H. Jorgensen, D. R. Hospenthal, K. Mende, and J. E. Patterson. 2009. Presence and molecular epidemiology of virulence factors in methicillin-resistant Staphylococcus aureus strains colonizing and infecting soldiers. J. Clin. Microbiol. 47:940-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 43:1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francois, P., S. Harbarth, A. Huyge, G. Renzi, M. Bento, A. Gervaix, D. Pittet, and J. Schrenzel. 2008. Methicillin-resistant Staphylococcus aureus, Geneva, Switzerland, 1993-1995. Emerg. Infect. Dis. 14:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacek, D. M., S. M. Paule, R. B. Thomson, Jr., A. Robicsek, and L. R. Peterson. 2009. Implementation of a universal admission surveillance and decolonization program for methicillin-resistant Staphylococcus aureus (MRSA) reduces the number of MRSA and total S. aureus isolates reported by the clinical laboratory? J. Clin. Microbiol. 47:3749-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbarth, S., P. François, J. Schrenzel, C. Fankhauser-Rodriquez, S. Hugonnet, T. Koessler, A. Huyge, and D. Pittet. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Switzerland. Emerg. Infect. Dis. 11:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbarth, S., Y. Martin, P. Rohner, N. Henry, R. Auckenthaler, and D. Pittet. 2000. Effect of delayed infection control measures on a hospital outbreak of methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 46:43-49. [DOI] [PubMed] [Google Scholar]
- 21.Healy, C. M., K. G. Hulten, D. L. Palazzi, J. R. Campbell, and C. J. Baker. 2004. Emergence of new strains of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Clin. Infect. Dis. 39:1460-1466. [DOI] [PubMed] [Google Scholar]
- 22.Issartel, B., A. Tristan, S. Lechevallier, F. Bruyere, G. Lina, B. Garin, F. Lacassin, M. Bes, F. Vandenesch, and J. Etienne. 2005. Frequent carriage of Panton-Valentine leucocidin genes by Staphylococcus aureus isolates from surgically drained abscesses. J. Clin. Microbiol. 43:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy, A. D., and F. R. DeLeo. 2009. Epidemiology and virulence of community-associated MRSA. Clin. Microbiol. Newsl. 31:153-160. [Google Scholar]
- 26.Klevens, R. M., J. R. Edwards, F. C. Tenover, L. C. McDonald, T. Horan, and R. Gaynes. 2006. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin. Infect. Dis. 42:389-391. [DOI] [PubMed] [Google Scholar]
- 27.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krziwanek, K., C. Luger, B. Sammer, S. Stumvoll, M. Stammler, S. Metz-Gercek, and H. Mittermayer. 2007. PVL-positive MRSA in Austria. Eur. J. Clin. Microbiol. Infect. Dis. 26:931-935. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S. M., M. Ender, R. B. Adhikari, J. M. Smith, B. Berger-Bächi, and G. M. Cook. 2007. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob. Agents Chemother. 51:1497-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 31.Lucet, J.-C., X. Paoletti, I. Lolom, C. Paugam-Burtz, J.-L. Trouillet, J.-F. Timsit, C. Deblangy, A. Andremont, and B. Regnier. 2005. Successful long-term program for controlling methicillin-resistant Staphylococcus aureus in intensive care units. Intensive Care Med. 31:1051-1057. [DOI] [PubMed] [Google Scholar]
- 32.Maree, C. L., R. S. Daum, S. Boyle-Vavra, K. Matayoshi, and L. G. Miller. 2007. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare associated infections. Emerg. Infect. Dis. 13:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 35.Milheiriço, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for the assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, L. G., and B. A. Diep. 2008. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46:752-760. [DOI] [PubMed] [Google Scholar]
- 37.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 39.Nair, S. P., M. Bischoff, M. M. Senn, and B. Berger-Bächi. 2003. The σB regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 71:4167-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan, A., G. Carnevale, P. Catenazzi, P. Colombini, L. Crema, L. Dolcetti, L. Ferrari, P. Mondello, L. Signorini, C. Tinelli, E. Q. Roldan, and G. Carosi. 2005. Trends in methicillin-resistant Staphylococcus aureus (MRSA) bloodstream infections: effect of the MRSA “search and isolate” strategy in a hospital in Italy with hyperendemic MRSA. Infect. Control Hosp. Epidemiol. 26:127-133. [DOI] [PubMed] [Google Scholar]
- 42.Perry, J. D., A. Davies, L. A. Butterworth, A. L. J. Hopley, A. Nicholson, and F. K. Gould. 2004. Development and evaluation of a chromogenic agar medium for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 42:4519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Popovich, K. J., R. A. Weinstein, and B. Hota. 2008. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin. Infect. Dis. 46:787-794. [DOI] [PubMed] [Google Scholar]
- 44.Qi, W., M. Ender, F. O'Brien, A. Imhof, C. Ruef, N. McCallum, and B. Berger-Bächi. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zurich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43:5164-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossney, A. S., C. M. Herra, G. I. Brennan, P. M. Morgan, and B. O'Connell. 2008. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 46:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saiman, L., M. O'Keefe, P. L. Graham III, F. Wu, B. Said-Salim, B. Kreiswirth, A. LaSala, P. M. Schlievert, and P. Della-Latta. 2003. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin. Infect. Dis. 37:1313-1319. [DOI] [PubMed] [Google Scholar]
- 47.Strandén, A., R. Frei, H. Adler, U. Flückiger, and A. Widmer. 2009. Emergence of SCCmec type IV as the most common type of methicillin-resistant Staphylococcus aureus in a university hospital. Infection 37:44-48. [DOI] [PubMed] [Google Scholar]
- 48.Tenover, F., R. Arbeit, R. Goering, P. Mickelsen, B. Murray, D. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandenbroucke-Grauls, C. M. J. E. 1996. Methicillin-resistant Staphylococcus aureus control in hospitals: the Dutch experience. Infect. Control Hosp. Epidemiol. 17:512-513. [DOI] [PubMed] [Google Scholar]
- 50.Wannet, W. J. B., E. Spalburg, M. E. O. C. Heck, G. N. Pluister, E. Tiemersma, R. J. L. Willems, X. W. Huijsdens, A. J. de Neeling, and J. Etienne. 2005. Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leucocidin genes in The Netherlands. J. Clin. Microbiol. 43:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wertheim, H. F. L., M. C. Vos, H. A. M. Boelens, A. Voss, C. M. J. E. Vandenbroucke-Grauls, M. H. M. Meester, J. A. J. W. Kluytmans, P. H. J. van Keulen, and H. A. Verbrugh. 2004. Low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) at hospital admission in The Netherlands: the value of search and destroy and restrictive antibiotic use. J. Hosp. Infect. 56:321-325. [DOI] [PubMed] [Google Scholar]
- 52.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]