Abstract
Analysis of 316 outbreaks of gastroenteritis in France from September 2007 through March 2009 showed that genogroup II.4 (GGII.4) noroviruses were predominant and mostly belonged to the 2006b variant. However, the new GGII.4 variants, variant 2008 and the newly discovered Cairo variant from the Middle East, were also detected. The epidemiological survey suggests that these new variants might become the next predominant strains.
Human noroviruses (NoVs) are causative agents of gastroenteritis worldwide and are increasingly incriminated in outbreaks of gastroenteritis. The molecular typing of NoV is now routinely performed in laboratories involved with surveys of enteric viruses. The recent establishment of networks (e.g., the FBVE and noronet networks) to monitor enteric viruses showed the large predominance of genogroup II.4 (GGII.4) NoVs (11). Over the past 10 years, NoVs and especially GGII.4 variants have increasingly been studied. It has been shown for the first time that GGII.4 NoV variant US95/96 was predominant worldwide during the winter of 1995-1996 (8). NoV variant US95/96 was detected from outbreaks for the next 7 years, until 2002, when a new type of GGII.4 strain, the 2002 variant, named Farmington Hills, was detected and became predominant (7). The main feature of this strain was the insertion of 1 amino acid in the hypervariable region of the capsid (3). Since 2002, a new GGII.4 variant has become predominant every 2 or 3 years, as shown previously. Indeed, the 2002 variant was replaced in 2004 by the 2004 variant (1), which was then replaced by the 2006a and 2006b variants, which cocirculated in 2006 and 2007 (13). In France, the 2006b variant has been predominant since 2008. Previous studies showed that the heterogeneity of the amino acid sequences among GGII.4 variants was no more than 10%. However, Lindesmith et al. showed that minor changes in the capsid between GGII.4 variants were biologically relevant and corresponded to changing patterns in the binding of the glycosaccharides (6).
Moreover, it is worth mentioning that not all GGII.4 variants are circulating throughout the world. For example, the Sakai variants have mostly been detected in Asia. For the Cairo variants that we recently described (4), it was difficult to predict whether they would circulate locally or worldwide. During the last year, the national French survey network and other laboratories in the world detected emerging GGII.4 isolates, called 2008 variants, which might be the new predominant strain. Here we report on the emergence of new GGII.4 isolates, including the 2008 variants, which have been collected and characterized by the French National Reference Center (NRC) for Enteric Viruses.
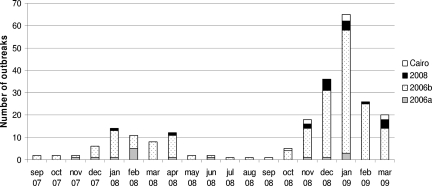
During a period that included two consecutive winter seasons, from 1 September 2007 through 31 March 2009, 1,422 stool specimens from 316 outbreaks were sent to the NRC for analysis. Sixty-seven percent of the outbreaks were reported during the second winter season (September 2008 through March 2009), thanks to the countrywide improvement in the outbreak reporting system. The NoV outbreaks were reported predominantly from nursing homes (n = 251), but also from hospitals (n = 21), schools (n = 22), and adult meetings (n = 22). RNA was routinely extracted from stool specimens from individuals involved in each outbreak prior to amplification with capsid-specific primers (5). For each outbreak, the PCR products from up to three samples were purified and sequenced for typing, as described previously (4). The presence of at least one virus was detected in 85% of the outbreaks (n = 268), and 254 of these were NoV related. For 14 NoV-negative outbreaks, rotavirus (n = 9), astrovirus (n = 3), sapovirus (n = 1), and Aichi virus (n = 1) were detected by PCR. Twenty-one outbreaks were positive for non-GGII.4 NoVs. The other GGII NoVs belonged to GGII.2 (n = 2), GGII.6 (n = 6), and GGIIb/GGII.3 (n = 4). For nine outbreaks, GGI.2 (n = 2), GGI.3 (n = 3), GGI.4 (n = 1), GGI.8 (n = 2), and GGI.10 (n = 1) were detected. GGII.4 NoV represented the bulk of the strains (229 outbreaks, 72%) (Fig. 1). GGII.4 NoVs were incriminated in 17 of 24 mixed infections. The predominant 2006b variants were identified in 192 outbreaks, 13 of which were mixed infections. The 2006a accounted for 4.7% (n = 14) of the total number of outbreaks (three outbreaks were mixed infections). For 27 outbreaks, the GGII.4 NoVs were not related to other known GGII.4 subtypes. Preliminary typing based on the first 227 bases of open reading frame 2 (ORF2) showed that 9 and 18 of those outbreaks were caused by NoVs that belonged to the Cairo and 2008 variants, respectively (Table 1). Two 2008 variant isolates were documented in January and April 2008, but these variants were mostly detected during the 2008-to-2009 winter season (n = 16) (Fig. 1). The Cairo variants clearly emerged in France during the last winter season, with the first outbreak being documented in September 2008. A panel of these variants (n = 10) was selected from the typing results obtained by use of the capsid region to undertake sequence analysis of the entire capsid gene. After RNA extraction, the ORF2-coding sequence was amplified by reverse transcription-PCR before it was cloned into the pGEM-Teasy vector, as described previously (4). The entire ORF2 sequence was then determined. Eighty-two sequences (Table 2) corresponding to the complete ORF2 from previously described GGII.4 variants and the sequences from this study (Table 1) were analyzed with the MEGA (version 4) package and the MrBayes (version 3.1) program (9, 12). For the latter, the same settings described previously were used (6), except that the amino acid model was set to “mixed.” The amino acid differences mostly occurred in the P2 domain (data not shown). The residues contributing to the attachment of the H type 1 antigen were conserved for the Cairo and the 2008 variants (data not shown) (2). However, the triplet S396-R397-N398, located at the N-terminal part of the P2 domain, was shared only by the 2006b, Cairo, and 2008 variants. Similarly, the triplet D376-A377-N378 was found only for the 2008 variants (data not shown). Interestingly, the threonine inserted at position 395 had been replaced by an alanine residue for all but one of the 2008 variants. Further studies will be required to verify whether these amino acid residues correlate with an increased pathogenicity of the new circulating strains. The trees were generated by the neighbor-joining method by use of the MEGA (version 4) program suite (Fig. 2A) and by Bayesian inference of phylogeny by use of the MrBayes program (Fig. 2B) (9, 12). The topologies of the two trees were similar and showed that the 2008 and the Cairo variants were closely related to the currently predominant 2006b variant (Fig. 2). By combining data from epidemiological surveys with phylogenetic analysis, we clearly showed that during the last season, the emerging variant 2008 and Cairo strains were cocirculating with the predominant 2006b variants (Fig. 1 and 2). It is likely that the cocirculation of variants probably occurred very often in the past, and the increasing efficacies of surveillance systems worldwide are contributing to the detection of emerging GGII.4 variants. The results of our phylogenetic analysis suggest that the 2008 and Cairo variants might have derived from the 2006b variants and support the idea that genetic drift is a major force in the evolution of GGII.4, as described previously (6).
FIG. 1.
Monthly distribution of GGII.4 variants during the study period (September 2007 through March 2009). The GGII.4 variants are shaded in gray, and the key is indicated on the right.
TABLE 1.
The 2008 variants and Cairo variants from this study
| Isolatea | Date of onset (mo/yr) | Setting | Variantb | GenBank accession no.c |
|---|---|---|---|---|
| E2271 | 01/08 | Retirement home | 2008 | |
| E2763 | 04/08 | Nursing home | 2008 | |
| E3020 | 09/08 | Nursing home | Cairo | GQ246791 |
| E3060 | 10/08 | Retirement home | Cairo | |
| E3138 | 11/08 | Retirement home | Cairo | |
| E3165 | 11/08 | Nursing home | 2008 | GQ246792 |
| E3184 | 11/08 | Nursing home | 2008 | |
| E3203 | 11/08 | High school | Cairo | |
| E3367 | 12/08 | Nursing home | 2008 | |
| E3379 | 12/08 | Community | 2008 | GQ246793 |
| E3436 | 12/08 | Retirement home | 2008 | GQ246794 |
| E3487 | 12/08 | Nursing home | 2008 | GQ246795 |
| E3550 | 12/08 | Hospital (intensive care) | 2008 | GQ246796 |
| E3594 | 01/09 | Retirement home | Cairo | |
| E3642 | 01/09 | Nursing home | 2008 | GQ246797 |
| E3743 | 01/09 | Nursing home | 2008 | GQ246798 |
| E3809 | 01/09 | Nursing home | Cairo | |
| E3880 | 01/09 | Nursing home | Cairo | GQ246800 |
| E3956 | 01/09 | Retirement home | 2008 | |
| E3959 | 01/09 | Sporting event | 2008 | |
| E4032 | 02/09 | Hospital (geriatric) | 2008 | GQ246801 |
| E4114 | 03/09 | Hospital (geriatric) | Cairo | |
| E4126 | 03/09 | Retirement home | 2008 | |
| E4134 | 03/09 | Retirement home | 2008 | |
| E4141 | 03/09 | Retirement home | 2008 | |
| E4158 | 03/09 | Nursing home | Cairo | |
| E4173 | 03/09 | Retirement home | 2008 |
Each isolate corresponds to one outbreak.
Typing of GGII.4 NoV variants was based on phylogenetic analysis of partial nucleotide sequences of the capsid region, as indicated in the Materials and Methods section.
Strains for which the complete ORF2 sequence was determined are referenced in GenBank. The corresponding plasmid constructs are available upon request.
TABLE 2.
GGII.4 isolates from GenBank used for the analysis
| Isolate | Variant | GenBank accession no. |
|---|---|---|
| MD145 | Bristol | AY032605 |
| Camberwell | Bristol | AF145896 |
| MD134 | Bristol | AY030098 |
| Lordsdale | Bristol | X86557 |
| Bristol | Bristol | X76716 |
| UK317 | Bristol | AF414417 |
| Symgreen95 | US95/96 | AJ277619 |
| Yuri | US95/96 | AB083781 |
| Tiel001 | US95/96 | AB303922 |
| Parkroyal | US95/96 | AJ277613 |
| Altenkirchen140 | US95/96 | AF425765 |
| DOUG4770 | US95/96 | AF406793 |
| Oder170 | US95/96 | AF427114 |
| Miami-Beach | US95/96 | AF414424 |
| Ludwigslust221 | US95/96 | AF427116 |
| Burwash Landing | US95/96 | AF414425 |
| Bochum339 | US95/96 | AY532127 |
| VA98387 | US95/96 | AY038600 |
| Dijon171 | US95/96 | AF472623 |
| Narita104 | US95/96 | AB078336 |
| Grimsby | US95/96 | AJ004864 |
| Mora97 | US95/96 | AY081134 |
| Dillingen 259 | US95/96 | AF425766 |
| Dresden153 | US95/96 | AY532115 |
| Beeskow124 | US95/96 | AF427120 |
| Erlangen195 | US95/96 | AY532114 |
| Lanzhou | Henry | DQ364459 |
| Houston-TCH186 | Henry | EU310927 |
| Anchorage | Farmington | AY502019 |
| MD-2004 | Farmington | DQ658413 |
| E872-Dijon | Farmington | FJ538900 |
| Farmington Hill | Farmington | AY502023 |
| Apeldoorn317 | Farmington | AB445395 |
| Carlow | Farmington | DQ415279 |
| Langen | Farmington | AY485642 |
| Germanton | Farmington | AY502017 |
| CruiseG1 | Farmington | AY502020 |
| Witney | Farmington | AY588030 |
| Oxford B5S22 | Farmington | AY581254 |
| Oxford B2S16 | Farmington | AY587989 |
| Chipping Norton | Farmington | AY588028 |
| E1057 | Hunter | EU876890 |
| 0317/NL | Hunter | AY883096 |
| Dongen46 | Hunter | EF126961 |
| Nijmegen | Hunter | AB303941 |
| Hunter | Hunter | DQ078794 |
| DenHaag54 | Hunter | EF126962 |
| Ehime | Sakai | AB220923 |
| Guangzhou | Sakai | EF535854 |
| CR2932 | Sakai | DQ419908 |
| Chiba | Sakai | AB220921 |
| NVgz01 | Sakai | DQ369797 |
| Sakai | Sakai | AB220922 |
| E1501-Dijon | 2006a | EU876894 |
| Cairo 6 | 2006a | EU876886 |
| Cairo 3 | 2006a | EU876883 |
| Cairo 7 | 2006a | EU876887 |
| Cairo 9 | 2006a | EU876889 |
| Cairo 5 | 2006a | EU876886 |
| Yerseke | 2006a | EF126963 |
| RotterdamP7D0 | 2006a | AB385639 |
| Aomori1 | 2006a | AB447432 |
| Aomori2 | 2006a | AB447433 |
| Isumi | 2006a | AB294790 |
| Terneuzen | 2006a | EF126964 |
| Duan-China | 2006b | EU366113 |
| Kobe034 | 2006b | AB291542 |
| Cairo 1 | 2006b | EU876892 |
| E1267-Dijon | 2006b | EU876895 |
| E2703-Dijon | 2006b | EU876891 |
| E3808-Dijon | 2006b | GQ246799 |
| Nijmegen115 | 2006b | EF126966 |
| DenHagg89 | 2006b | EF126965 |
| SSCS | Cairo | FJ411171 |
| OC07138 | Cairo | AB434770 |
| RIS | Cairo | FJ411172 |
| Cairo 8 | Cairo | EU876888 |
| Cairo 2 | Cairo | EU876882 |
| Cairo 4 | Cairo | EU876884 |
| Apeldoorn317 | 2008 | AB445395 |
| Stockholm | 2008 | AB492092 |
| Erfurt | Outlier | AF427117 |
FIG. 2.
Phylogenetic analysis of the amino acid sequences corresponding to the entire ORF2 of representative GGII.4 NoVs from GenBank (n = 82) and this study (n = 10). The isolates sequenced for this study are indicated on the tree. The groups of variants are color coded on the basis of the period of circulation of these strains (e.g., the Cairo and 2008 variants) and/or their predominance (e.g., the Bristol, US95/96, 2002, 2004, 2006b, and Sakai variants). The color code is indicated on the bottom right. E, Erfurt outlier isolate (GenBank accession number AF427117). (A) Neighbor-joining tree constructed with noncorrected distances; (B) Bayesian inference tree constructed with previously described settings (6).
Since 2002, the predominant circulating GGII.4 NoVs have contained an extra amino acid, inserted into the hypervariable region of the capsid, with ongoing genetic drift being observed for this genotype. For the 2007-to-2008 and 2008-to-2009 winter seasons, the 2006a variant and the predominant 2006b variant were circulating in France. During an epidemiological survey conducted in the conurbation of Cairo, Egypt, in 2006 and 2007, we observed that the 2006a variant strains were predominant. We also detected new GGII.4 variants that we called Cairo variants. At this stage, it was unclear whether these new GGII.4 isolates were restricted to the Middle East or could disseminate throughout the world. During the same period, no Cairo variant-like GGII.4 NoV were detected in a large epidemiological survey that we conducted in Tunisia, where only 2004 variants were found (10). The survey of the GGII.4 strains detected by the NRC showed that the 2008 and Cairo variants emerged in France during the 2008-to-2009 winter season. These data show that NoV GGII.4 strains are circulating within Mediterranean countries. For the Cairo variants, we may also hypothesize that new GGII.4 variants might spread from the Middle East to Europe and the Maghreb. Additional studies will be required to further evaluate the circulation of these variants in North Africa. Our data also emphasize the need for a careful survey of enteric viruses that are not routinely screened for by many clinical laboratories.
Plotting of the epidemiological data on the radial tree (Fig. 2) showed that several GGII.4 variants circulated at the same time in the world (e.g., the 2006a, 2006b, 2008, and Cairo variants) and that one type of variant was more often detected locally (e.g., Sakai variants in Asia) or worldwide (e.g., the Farmington Hills variant from 2002 to 2004). Even though the mechanism by which the GGII.4 NoV strains evolved remains unknown, the emergence of these new GGII.4 isolates in France might be a precursor to a larger disease burden during coming winter seasons due to these variants.
Acknowledgments
This study has been funded by the public hospital of Dijon, France, and the French National Reference Center for Enteric Viruses. A. H. Kamel received a fellowship from the ASRT and the IMHOTEP program from Egide (Paris, France).
We thank the NRC staff and Philip Bastable for technical and editorial assistance.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingle, K. E. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamel, A. H., M. A. Ali, H. G. El-Nady, A. de Rougemont, P. Pothier, and G. Belliot. 2009. Predominance and circulation of enteric viruses in the region of greater Cairo, Egypt. J. Clin. Microbiol. 47:1037-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 6.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinje, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682-688. [DOI] [PubMed] [Google Scholar]
- 8.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 10.Sdiri-Loulizi, K., K. Ambert-Balay, H. Gharbi-Khelifi, N. Sakly, M. Hassine, S. Chouchane, M. N. Guediche, P. Pothier, and M. Aouni. 2009. Molecular epidemiology of norovirus gastroenteritis investigated using samples collected from children in Tunisia during a four-year period: detection of the norovirus variant GGII.4 Hunter as early as January 2003. J. Clin. Microbiol. 47:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siebenga, J. J., H. Vennema, D. P. Zheng, J. Vinje, B. E. Lee, X. L. Pang, E. C. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. B. Rasool, J. Hewitt, G. E. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. A. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 12.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 13.Tu, E. T., R. A. Bull, G. E. Greening, J. Hewitt, M. J. Lyon, J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 46:413-420. [DOI] [PubMed] [Google Scholar]