Abstract
Whole-cell fingerprinting by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) in combination with a dedicated bioinformatic software tool (MALDI Biotyper 2.0) was used to identify 152 staphylococcal strains corresponding to 22 staphylococcal species. Spectra of the 152 isolates, previously identified at the species level using a sodA gene-based oligonucleotide array, were analyzed against the main spectra of 3,030 microorganisms. A total of 151 strains out of 152 (99.3%) were correctly identified at the species level; only one strain was identified at the genus level. The MALDI-TOF MS method revealed different clonal lineages of Staphylococcus epidermidis that were of either human or environmental origin, which suggests that the MALDI-TOF MS method could be useful in the profiling of staphylococcal strains. The topology of the dendrogram generated by the MALDI Biotyper 2.0 software from the spectra of 120 Staphylococcus reference strains (representing 36 species) was in general agreement with that inferred from the 16S rRNA gene-based analysis. Our findings indicate that the MALDI-TOF MS technology, associated with a broad-spectrum reference database, is an effective tool for the swift and reliable identification of Staphylococci.
Most staphylococci are harmless and reside normally on the skin and mucous membranes of humans and other organisms (16, 22, 34). Staphylococcal strains are isolated from various food products in which they are involved in fermentation (18, 29). Staphylococcus species can cause a wide variety of diseases in humans and other animals (2, 22, 30-32, 35). S. aureus is a major pathogen in human infections (31). Several other Staphylococcus species have also been implicated in human infections, notably S. saprophyticus, S. epidermidis, S. lugdunensis, and S. schleiferi (4, 16, 31, 34). Coagulase-negative staphylococci (CoNS) have emerged as predominant pathogens in hospital-acquired infections (4, 16, 31, 34). One of the major challenges of daily diagnostic work is therefore to identify Staphylococcus species.
Several manual and automated methods based on phenotypic characteristics have been developed for the identification of Staphylococci (12, 24). Unfortunately, these systems have their limitations, mostly due to phenotypic differences between strains from the same species (6, 10, 19, 21). Over the last 10 years, many genotypic methods based on the analysis of selected DNA targets have been designed for species-level identification of most common isolated CoNS (20, 26, 33). The sequence polymorphism of the sodA gene has significant discriminatory power (20) and allows the development of assays based on DNA chip technologies (“Staph array”) (8). Recently, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) using protein “fingerprints” was used for the identification of microorganisms (1, 3, 5, 9, 11, 14, 25, 36). In the present study, we assessed the ability of the MALDI Biotyper system (Bruker Daltonique, Wissembourg, France) to identify Staphylococcus species of clinical and environmental origins previously identified by sodA gene-based oligonucleotide array (8).
MATERIALS AND METHODS
Bacterial strains.
A total of 152 blind-coded Staphylococcus strains from our collection, isolated from clinical (n = 60) and food and plant samples (n = 92) and representing 22 species, were studied (Table 1). All strains were previously identified as belonging to a specific species by the oligonucleotide “Staph array” (8). This system associates PCR amplification of the sodA gene with an oligonucleotide-based array to efficiently discriminate between the Staphylococcus species. Identifications found with this array were identical to those obtained by sequencing the internal fragment sodA (sodAint). In addition, the Vitek 2 system has been used with the Gram-positive (GP) identification card (bioMérieux, Marcy l'Etoile, France) to identify Staphylococcus strains. If the GP card indicated the possibility of two or three species, supplementary tests (pigmentation, hemolysis, or novobiocin resistance) were then performed to determine the identification.
TABLE 1.
Clinical and environmental staphylococcal strains used in this study
| Species (“Staph array” identification) | No. of isolates |
||
|---|---|---|---|
| Clinical | Environmental | Total | |
| S. arlettae | 0 | 1 | 1 |
| S. aureus | 6 | 1 | 7 |
| S. capitis | 5 | 2 | 7 |
| S. caprae | 1 | 0 | 1 |
| S. carnosus | 0 | 7 | 7 |
| S. cohnii subsp. ureal | 2 | 2 | 4 |
| S. delphini | 1 | 0 | 1 |
| S. epidermidis | 11 | 8 | 19 |
| S. equorum | 0 | 13 | 13 |
| S. fleurettii | 0 | 2 | 2 |
| S. haemolyticus | 5 | 1 | 6 |
| S. hominis | 5 | 1 | 6 |
| S. lugdunensis | 5 | 0 | 5 |
| S. pasteuri | 0 | 8 | 8 |
| S. saprophyticus | 6 | 10 | 16 |
| S. schleiferi | 2 | 0 | 2 |
| S. sciuri | 1 | 2 | 3 |
| S. simulans | 4 | 0 | 4 |
| S. succinus | 0 | 9 | 9 |
| S. vitulinus | 0 | 5 | 5 |
| S. warneri | 6 | 6 | 12 |
| S. xylosus | 0 | 14 | 14 |
| Total | 60 | 92 | 152 |
Sample preparation.
Sample preparation for MALDI-TOF MS was performed as previously described (14). Briefly, a few colonies of a fresh overnight culture grown on Columbia blood agar at 37°C under aerobic conditions were suspended in 300 μl distilled water to which 900 μl absolute ethanol was added. The mixture was centrifuged at 12,000 × g for 2 min, and the supernatant was discarded. Ten microliters of formic acid (70%) was added to the pellet and mixed thoroughly by pipetting before the addition of 10 μl of acetonitrile to the mixture. The mixture was centrifuged at 12,000 × g for 2 min. One microliter of the supernatant was placed onto a steel target plate and air dried at room temperature. Each sample was overlaid with 2 μl saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile-2.5% trifluoroacetic acid and air dried at room temperature. MALDI-TOF MS measurements were performed with a Bruker Ultraflex II MALDI-TOF/TOF (tandem TOF) instrument equipped with 200-Hz Smartbeam laser technology. Spectra were recorded in the positive linear mode within a mass range of 2,000 to 20,000 Da. Five hundred laser shots were recorded for each spectrum.
Data analysis.
Raw spectra of the Staphylococcus strains were analyzed by MALDI Biotyper 2.0 software (Bruker Daltonique) with default settings. The whole process from MALDI-TOF MS measurement to identification was performed automatically without any user intervention. The peak lists generated were used for matches against the reference library directly using the integrated pattern-matching algorithm of the software. The software provides a log score, and the cutoff log score of 2 was used to validate identification at the species level, as recommended by the manufacturer. For strain classification, the creation of the dendrogram is based on cross-wise minimum spanning tree (MSP) matching. Similar MSPs result in a high matching score value. Each MSP is matched against all MSPs of the analyzed set. The list of score values is used to calculate normalized distance values between the analyzed species, resulting in a matrix of matching scores. The visualization of the respective relationship between the MSPs is displayed in a dendrogram using the standard settings of the MALDI Biotyper 2.0 software. Species with distance levels under 500 have been described as reliably classified (25).
RESULTS AND DISCUSSION
Classification of staphylococcal reference strains.
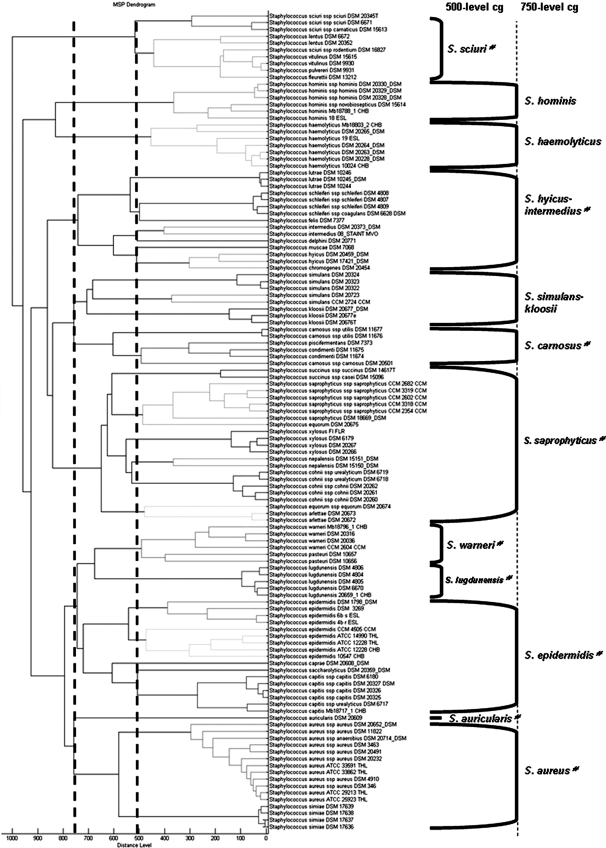
A score-oriented dendrogram was generated on the basis of the 120 reference strains in the Bruker MALDI Biotyper 2.0 database (Fig. 1). This dendrogram revealed, with the default critical distance level of 500, 4 of the 11 cluster groups defined by Poyart et al. from the phylogenetic analysis based on the sodA gene (20): S. sciuri, S. warneri, S. lugdunensis, and S. auricularis (Fig. 1). These cluster groups were also in accordance with those established by the phylogenetic analysis of the 16S rRNA gene (28). With a critical distance level of 750, the dendrogram revealed overall the cluster groups of 16S rRNA genes, with some variations (Fig. 1) (17, 28). The 16S rRNA gene-based cluster group S. haemolyticus including S. haemolyticus and S. hominis was divided into branches which linked at a distance level of 850. Unlike in other phylogenetic analyses, in which S. kloosii belonged to S. saprophyticus cluster group, this species was clustered within the S. simulans group in the MALDI-TOF MS-based dendrogram (13, 20, 28). Inside the S. saprophyticus cluster group, the two strains S. equorum subsp. equorum DSM 20675 and DSM 20674 belonged to two distinct clusters. It is noteworthy that the S. lutrae and S. schleiferi species each constituted a well-defined clade inside the S. hyicus-S. intermedius cluster group, like S. succinus, S. xylosus, S. cohnii, and S. arlettae in the S. saprophyticus cluster group and S. capitis in the S. epidermidis cluster group.
FIG. 1.
Classification of staphyloccal reference strains. Shown is a score-oriented dendrogram of staphylococcal reference strains included in the database. The terms “500-level cg” and “750-level cg” define cluster groups (cg) based on the branching pattern using critical distance levels of 500 and 750, respectively. #, cluster groups as defined by the phylogenetic analysis of 16S rRNA genes.
Although some differences were observed, the topology of the MALDI-TOF MS-based phylogenetic tree was in more general agreement with that inferred from the analysis of the 16S rRNA gene sequences than that based on the sodA gene (20, 28). The proteins analyzed by MALDI Biotyper correspond to predominant proteins, such as ribosomal proteins. The probable coevolution of the ribosomal proteins and rRNAs may explain the similarities between 16S rRNA gene and MALDI-TOF-based dendrograms (23).
MALDI-TOF MS could thus be a valuable tool in phyloproteomics. It might serve as a technique for protein profiling, which is recommended as an additional test for the description of new staphylococcal species (7).
Species identification of staphylococcal isolates.
Our dendrogram showed that the 36 reference Staphylococcus species were correctly distinguished. S. vitulinus and S. pulvereri, which have recently been shown to be one single species (27), were not differentiated by the MALDI-TOF MS technique. This technique seemed to be suitable for the differentiation of Staphylococcus isolates at the species level.
MALDI-TOF MS spectra were then obtained for all clinical and environmental staphylococcal isolates. A total of 151 strains out of 152 (99.3%) were identified at the species level by the MALDI Biotyper. One S. saprophyticus strain out of 16 was identified as S. xylosus, with a log score of 1.995. It was therefore identified at the genus level. The system identified both clinical (14 different species) and environmental (17 different species) strains without any complementary tests.
Carbonnelle et al. used the MALDI-TOF technology to identify a clinical collection of Staphylococcus strains constituted by the four major clinically relevant Staphylococcus species (S. aureus, S. epidermidis, S. warneri, and S. haemolyticus) (3). Their reference database contained 22 staphylococci reference strains, including 17 staphylococcal species. In our study, we identified 22 Staphylococcus species from the MALDI Biotyper database, which comprised the spectra of 3,030 microorganisms, corresponding to Gram-negative and Gram-positive bacteria, whose 120 staphylococcal strains represented 36 staphylococcus species. A fortiori, the MALDI Biotyper did not require any initial assessment such as Gram staining or a catalase test, unlike the technology developed by Carbonnelle et al., which requires a presumptive identification of Staphylococcus at the genus level.
The MALDI Biotyper database comprised 36 staphylococcal species. The Staphylococcus genus comprises 39 validly described species (J. P. Euzéby, List of bacterial names with standing in nomenclature, 5 November 2008, posting date; http://www.bacterio.cict.fr/). The three species S. pettenkoferi, S. pseudintermedius, and S. gallinarum were absent from the MALDI Biotyper database. The MALDI Biotyper database contains four species (S. pasteuri, S. delphini, S. fleurettii, and S. succinus) absent from the Vitek 2 Gram-positive (GP) card database (bioMérieux, La-Balme les Grottes, France). Moreover, according to the previously published data from our strain collection, the Vitek 2 system had misidentified or not identified 1 out of 7 S. capitis, 2 out of 7 S. carnosus, 11 out of 13 S. equorum, 1 out of 4 S. simulans, and 1 out of 5 S. vitulinus strains (6). This suggests that the MALDI-TOF MS technique is better fitted to the identification of staphylococcal strains that are rarely isolated in clinical bacteriology than automated biochemical methods. In addition, sample preparation and analysis with the MALDI-TOF MS technique are easier and more time-saving (approximately 20 min for one identification) than with genotypical methods, and result acquisition is faster than with biochemical methods.
Biodiversity of S. epidermidis species.
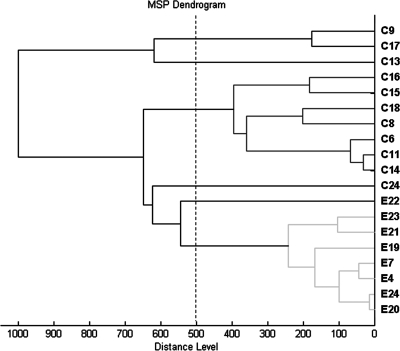
The dendrogram of Fig. 2 shows protein “fingerprint” heterogeneity of clinical and environmental (salt meat originated) S. epidermidis strains. Clinical isolates are gathered within two cluster groups and two isolated strains, whereas meat origin isolates displayed one distinct cluster group and one isolated strain. Diversity within the S. epidermidis species has been observed for human origin S. epidermidis and animal origin S. epidermidis (15). The MALDI-TOF MS method might have enough discriminatory power to group isolates below the species level.
FIG. 2.
Score-oriented dendrogram of clinical (C) and environmental (E) S. epidermidis isolates.
Conclusion.
MALDI-TOF MS associated with MALDI Biotyper software appears a reliable and accurate tool for the identification of Staphylococcus species. However, further studies are required to test this technology with a large collection of staphylococci of diverse origins. The speed and the simplicity of sample preparation and result acquisition associated with minimal consumable costs make this method well suited for routine and high-throughput use. Hence, it is an excellent alternative to traditional methods in food processing and medical care laboratories. It may also be used for the analysis of clonal and/or taxonomic relationships.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid identification and typing of Listeria species using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergonier, D., R. de Cremoux, R. Rupp, G. Lagriffoul, and X. Berthelot. 2003. Mastitis of dairy small ruminants. Vet. Res. 34:689-716. [DOI] [PubMed] [Google Scholar]
- 3.Carbonnelle, E., J. L. Beretti, S. Cottyn, G. Quesne, P. Berche, X. Nassif, and A. Ferroni. 2007. Rapid identification of staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 45:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, S. H., J. H. Woo, J. Y. Jeong, N. J. Kim, M. N. Kim, Y. S. Kim, and J. Ryu. 2006. Clinical significance of Staphylococcus saprophyticus identified on blood culture in a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 56:337-339. [DOI] [PubMed] [Google Scholar]
- 5.Claydon, M. A., S. N. Davey, V. Edwards-Jones, and D. B. Gordon. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584-1586. [DOI] [PubMed] [Google Scholar]
- 6.Delmas, J., J. P. Chacornac, F. Robin, P. Giammarinaro, R. Talon, and R. Bonnet. 2008. Evaluation of the Vitek 2 system with a variety of Staphylococcus species. J. Clin. Microbiol. 46:311-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freney, J., W. E. Kloos, V. Hajek, J. A. Webster, M. Bes, Y. Brun, and C. Vernozy-Rozand. 1999. Recommended minimal standards for description of new staphylococcal species. Subcommittee on the taxonomy of staphylococci and streptococci of the International Committee on Systematic Bacteriology. Int. J. Syst. Bacteriol. 49:489-502. [DOI] [PubMed] [Google Scholar]
- 8.Giammarinaro, P., S. Leroy, J. P. Chacornac, J. Delmas, and R. Talon. 2005. Development of a new oligonucleotide array to identify staphylococcal strains at species level. J. Clin. Microbiol. 43:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosse-Herrenthey, A., T. Maier, F. Gessler, R. Schaumann, H. Bohnel, M. Kostrzewa, and M. Kruger. 2008. Challenging the problem of clostridial identification with matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14:242-249. [DOI] [PubMed] [Google Scholar]
- 10.Ieven, M., J. Verhoeven, S. R. Pattyn, and H. Goossens. 1995. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 33:1060-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keys, C. J., D. J. Dare, H. Sutton, G. Wells, M. Lunt, T. McKenna, M. McDowall, and H. N. Shah. 2004. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 4:221-242. [DOI] [PubMed] [Google Scholar]
- 12.Kim, M., S. R. Heo, S. H. Choi, H. Kwon, J. S. Park, M. W. Seong, D. H. Lee, K. U. Park, J. Song, and E. C. Kim. 2008. Comparison of the MicroScan, VITEK 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative staphylococci. BMC Microbiol. 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwok, A. Y., S. C. Su, R. P. Reynolds, S. J. Bay, Y. Av-Gay, N. J. Dovichi, and A. W. Chow. 1999. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int. J. Syst. Bacteriol. 49:1181-1192. [DOI] [PubMed] [Google Scholar]
- 14.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagase, N., A. Sasaki, K. Yamashita, A. Shimizu, Y. Wakita, S. Kitai, and J. Kawano. 2002. Isolation and species distribution of staphylococci from animal and human skin. J. Vet. Med. Sci. 64:245-250. [DOI] [PubMed] [Google Scholar]
- 16.Otto, M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantucek, R., I. Sedlacek, P. Petras, D. Koukalova, P. Svec, V. Stetina, M. Vancanneyt, L. Chrastinova, J. Vokurkova, V. Ruzickova, J. Doskar, J. Swings, and V. Hajek. 2005. Staphylococcus simiae sp. nov., isolated from South American squirrel monkeys. Int. J. Syst. Evol. Microbiol. 55:1953-1958. [DOI] [PubMed] [Google Scholar]
- 18.Papamanoli, E., P. Kotzekidou, N. Tzanetakis, and E. Litopoulou-Tzanetakis. 2002. Characterization of Micrococcaceae isolated from dry fermented sausage. Food Microbiol. 19:441-449. [DOI] [PubMed] [Google Scholar]
- 19.Perl, T. M., P. R. Rhomberg, M. J. Bale, P. C. Fuchs, R. N. Jones, F. P. Koontz, and M. A. Pfaller. 1994. Comparison of identification systems for Staphylococcus epidermidis and other coagulase-negative Staphylococcus species. Diagn. Microbiol. Infect. Dis. 18:151-155. [DOI] [PubMed] [Google Scholar]
- 20.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhoden, D. L., and J. M. Miller. 1995. Four-year prospective study of STAPH-IDENT system and conventional method for reference identification of Staphylococcus, Stomatococcus, and Micrococcus spp. J. Clin. Microbiol. 33:96-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rich, M. 2005. Staphylococci in animals: prevalence, identification and antimicrobial susceptibility, with an emphasis on methicillin-resistant Staphylococcus aureus. Br. J. Biomed. Sci. 62:98-105. [DOI] [PubMed] [Google Scholar]
- 23.Ryzhov, V., and C. Fenselau. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746-750. [DOI] [PubMed] [Google Scholar]
- 24.Sampimon, O. C., R. N. Zadoks, S. De Vliegher, K. Supre, F. Haesebrouck, H. W. Barkema, J. Sol, and T. J. Lam. 2009. Performance of API Staph ID 32 and Staph-Zym for identification of coagulase-negative staphylococci isolated from bovine milk samples. Vet. Microbiol. 136:300-305. [DOI] [PubMed] [Google Scholar]
- 25.Sauer, S., A. Freiwald, T. Maier, M. Kube, R. Reinhardt, M. Kostrzewa, and K. Geider. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sivadon, V., M. Rottman, S. Chaverot, J. C. Quincampoix, V. Avettand, P. de Mazancourt, L. Bernard, P. Trieu-Cuot, J. M. Feron, A. Lortat-Jacob, P. Piriou, T. Judet, and J. L. Gaillard. 2005. Use of genotypic identification by sodA sequencing in a prospective study to examine the distribution of coagulase-negative Staphylococcus species among strains recovered during septic orthopedic surgery and evaluate their significance. J. Clin. Microbiol. 43:2952-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svec, P., M. Vancanneyt, I. Sedlacek, K. Engelbeen, V. Stetina, J. Swings, and P. Petras. 2004. Reclassification of Staphylococcus pulvereri Zakrzewska-Czerwinska et al. 1995 as a later synonym of Staphylococcus vitulinus Webster et al. Int. J. Syst. Evol. Microbiol. 54:2213-2215. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 29.Talon, R., S. Leroy-Setrin, and S. Fadda. 2002. Bacterial starters involved in the quality of fermented meat products, p. 175-191. In F. Toldra (ed.), Research advances in quality of meat and meat products. Research Signpost, Trivandrum, Kerala, India.
- 30.Taponen, S., and S. Pyorala. 2009. Coagulase-negative staphylococci as cause of bovine mastitis—not so different from Staphylococcus aureus? Vet. Microbiol. 134:29-36. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F. C., and R. P. Gayne. 2000. The epidemiology of Staphylococcus infections, p. 414-421. In J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, DC.
- 32.van Duijkeren, E., D. J. Houwers, A. Schoormans, M. J. Broekhuizen-Stins, R. Ikawaty, A. C. Fluit, and J. A. Wagenaar. 2008. Transmission of methicillin-resistant Staphylococcus intermedius between humans and animals. Vet. Microbiol. 128:213-215. [DOI] [PubMed] [Google Scholar]
- 33.Vliegen, I., J. A. Jacobs, E. Beuken, C. A. Bruggeman, and C. Vink. 2006. Rapid identification of bacteria by real-time amplification and sequencing of the 16S rRNA gene. J. Microbiol. Methods 66:156-164. [DOI] [PubMed] [Google Scholar]
- 34.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 35.Weese, J. S., J. Rousseau, J. L. Traub-Dargatz, B. M. Willey, A. J. McGeer, and D. E. Low. 2005. Community-associated methicillin-resistant Staphylococcus aureus in horses and humans who work with horses. J. Am. Vet. Med. Assoc. 226:580-583. [DOI] [PubMed] [Google Scholar]
- 36.Winkler, M. A., J. Uher, and S. Cepa. 1999. Direct analysis and identification of Helicobacter and Campylobacter species by MALDI-TOF mass spectrometry. Anal. Chem. 71:3416-3419. [DOI] [PubMed] [Google Scholar]