Abstract
In response to epidemic levels of serogroup B meningococcal disease in Cuba during the 1980s, the VA-MENGOC-BC vaccine was developed and introduced into the National Infant Immunization Program in 1991. Since then the incidence of meningococcal disease in Cuba has returned to the low levels recorded before the epidemic. A total of 420 Neisseria meningitidis strains collected between 1983 and 2005 in Cuba were analyzed by multilocus sequence typing (MLST). The set of strains comprised 167 isolated from disease cases and 253 obtained from healthy carriers. By MLST analysis, 63 sequence types (STs) were identified, and 32 of these were reported to be a new ST. The Cuban isolates were associated with 12 clonal complexes; and the most common were ST-32 (246 isolates), ST-53 (86 isolates), and ST-41/44 (36 isolates). This study also showed that the application of VA-MENGOC-BC, the Cuban serogroup B and C vaccine, reduced the frequency and diversity of hypervirulent clonal complexes ST-32 (vaccine serogroup B type-strain) and ST-41/44 and also affected other lineages. Lineages ST-8 and ST-11 were no longer found during the postvaccination period. The vaccine also affected the genetic composition of the carrier-associated meningococcal isolates. The number of carrier isolates belonging to hypervirulent lineages decreased significantly after vaccination, and ST-53, a sequence type common in carriers, became the predominant ST.
Neisseria meningitidis is a major cause of meningitis and septicemia worldwide, and meningococcal disease (MD) remains one of the most aggressive bacterial infectious diseases in humans. Only 5 of the 13 distinct N. meningitidis serogroups, which are determined on the basis of the immunochemistry of their capsular polysaccharides, are associated with invasive disease; of these, serogroups A, B, and C account for approximately 90% of all cases (42). The disease mainly affects infants and young children, with case fatality rates being 10% to 15%, despite the availability of effective antibiotics. Although early diagnosis and antibiotic treatment greatly enhance survival, prevention through vaccination constitutes the best way to control meningococcal disease (21).
Traditionally, isolates of N. meningitidis have been identified and characterized phenotypically on the basis of the recognition of meningococcal surface structures by specific monoclonal or polyclonal antibodies (2, 19). While serogrouping, serotyping, and serosubtyping, based on the detection of variants of capsular polysaccharide, PorB and PorA, respectively, are still used extensively, they have often been found to lack resolution, especially when they are used to work with endemic strains (11, 51).
The polysaccharide capsule is the major virulence factor of N. meningitidis; unencapsulated meningococci are essentially avirulent (52). The development of comprehensive capsule-based vaccines has, however, been constrained by the poor immunogenicity of the serogroup B capsule (53) and by concerns over the antigenic similarity of this molecule with host sialic acids. Although the focus of vaccine development efforts for serogroup B has consequently been switched toward the meningococcal outer membrane proteins, the large degree of antigenic variation exhibited by these antigens as a result of the selection pressure imposed by the immune system has become the most important challenge to both serogroup B vaccine development and epidemiological studies. Understanding this variation in terms of the genetic structure of meningococcal populations is, therefore, of paramount importance for steering vaccine research and analyzing the present and future performance of existing vaccination approaches.
Meningococcal populations are known to be genetically highly diverse. Many genotypes have been identified by the examination of housekeeping genes that are subject to selection for conserved metabolic function (8, 24, 28, 55). These genotypes have been identified as electrophoretic types by multilocus enzyme electrophoresis (MLEE) (44) and, more recently, as sequence types (STs) by multilocus sequence typing (MLST) (28). Population studies that exploit MLEE and MLST have identified equivalent groups of related genotypes that are referred to as clonal complexes (49); which are now named after a predominant, or central, ST. Isolate collections corresponding to populations of asymptomatically carried meningococci show the greatest genetic diversity, while most disease-associated meningococci belong to a limited number of clonal complexes known as hyperinvasive lineages (5, 7, 13, 24, 55).
MD in Cuba has not been considered a public health problem since the introduction of an outer membrane vesicle (OMV)-based vaccine (VA-MENGOC-BC) in the 1980s (16, 41, 46). The incidence of MD in the country slowly increased from a reported figure of 0.1 cases per 100,000 inhabitants in 1962 to 1.8 cases per 100,000 inhabitants in 1978, with most cases caused by serogroup C isolates. In spite of nationwide vaccination in 1979 with a capsular polysaccharide A-C vaccine (Merieux Laboratories), the incidence rate continued to rise and was accompanied by a shift to predominantly serogroup B among disease-associated isolates. The peak of the epidemic occurred during 1983 and 1984, reaching a maximum incidence of 14.4 cases per 100,000 inhabitants; most cases at this point were caused by a single serogroup B clone. In the 1980s, research started in Cuba on an OMV-based serogroup B-C vaccine developed from the outer membrane antigens of the epidemic type strain (strain CU385) combined with serogroup C polysaccharide. The resulting formulation (VA-MENGOC-BC) was shown to be safe and effective, and in 1991 the vaccine was included in the National Immunization Program (NIP) for all infants (45, 46). After its introduction in the vaccination campaign in 1989 (in which the population between 3 months and 24 years old was vaccinated), the overall incidence decreased to below 0.4 per 100,000 inhabitants by 2001 and to 0.1 cases per 100,000 inhabitants by 2008 (34).
Epidemiological studies conducted in Cuba have used typing methods based on phenotypic traits (30, 31), which lack discriminatory power for the study of the genetic structure of bacterial populations or for monitoring of the changes during outbreaks and epidemics. The present study, therefore, was aimed at obtaining further information about the Cuban epidemic on the basis of the characterization by MLST of N. meningitidis strain isolates collected on the island during a 22-year period (1983 to 2005) and to use this information to begin trying to understand the impact of mass vaccination with the Cuban VA-MENGOC-BC vaccine on the genetic structure of the N. meningitidis population isolated from cases and carriers.
(Part of this study was presented at the 16th International Pathogenic Neisseria Conference, Rotterdam, The Netherlands, September 2008.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 420 meningococcal isolates from patients with MD and healthy carriers were included in the present study, and all strains belonged to the Finlay Institute culture collection. The isolates were collected between 1983 and 2005 in different regions of Cuba, and they were preserved at −70°C. A complete strain list, including their phenotypic characteristics, sources, and places of isolation, is available under request. The set of strains comprised 167 isolated from disease cases and 253 obtained from healthy carriers (Table 1).
TABLE 1.
Sequence types and clonal complexes observed per incidence period and partition of the data into disease and carrier isolates
| CC and STb | No. of disease strains/no. of carrier strains per perioda of isolation |
|||
|---|---|---|---|---|
| 1983-1989 | 1990-1992 | 1993-2005 | Total | |
| CC ST-32 | ||||
| 32 | 1/0 | 1/0 | 2/0 | |
| 33 | 52/13 | 37/68 | 13/30 | 102/111 |
| 34 | 0/2 | 0/2 | ||
| 1880 | 1/0 | 1/0 | ||
| 2857 | 1/0 | 1/0 | ||
| 2858 | 3/0 | 0/1 | 3/0 | 6/1 |
| 3584 | 1/0 | 1/0 | ||
| 5682 | 1/0 | 1/0 | ||
| New STs | 5/0 | 4/3 | 0/5 | 9/8 |
| CC ST-41/44 | ||||
| 41 | 1/3 | 1/3 | ||
| 44 | 5/0 | 5/0 | ||
| 159 | 1/0 | 1/0 | ||
| 303 | 1/1 | 2/3 | 3/4 | |
| 409 | 0/1 | 0/1 | ||
| 414 | 2/0 | 2/0 | ||
| 883 | 2/0 | 2/0 | ||
| 1103 | 0/4 | 0/4 | ||
| 1255 | 1/0 | 1/0 | ||
| 1790 | 2/0 | 2/0 | ||
| 1947 | 0/2 | 0/2 | ||
| New STs | 3/1 | 1/0 | 3/1 | |
| CC ST-53 | 0/7 | 1/22 | 0/56 | 1/85 |
| CC ST-103 | ||||
| 103 | 0/5 | 0/5 | ||
| 5171 | 3/0 | 2/1 | 5/1 | |
| CC ST-22 | 0/7 | 0/7 | ||
| CC ST-8 | ||||
| 153 | 5/0 | 5/0 | ||
| CC ST-198 | ||||
| 823 | 0/5 | 0/5 | ||
| CC ST-254 | ||||
| 2844 | 0/1 | 0/1 | ||
| New STs | 0/2 | 0/2 | ||
| CC ST-269 | ||||
| 352 | 1/0 | 1/0 | ||
| New STs | 0/1 | 0/1 | 0/2 | |
| CC ST-23 | 1/1 | 1/1 | ||
| CC ST-167 | ||||
| 1624 | 0/1 | 0/1 | ||
| New STs | 0/1 | 0/1 | ||
| CC ST-11 | 1/0 | 1/0 | ||
| UA | 7/0 | 2/1 | 1/4 | 10/5 |
| Total | 88/23 | 59/103 | 20/127 | 167/253 |
Periods were defined according to the incidence of meningococcal disease in Cuba (34). The incidences during the periods 1983 to 1989, 1990 to 1992, and 1993 to 2005 were >5.0, 5.0 to 1.0, and <1.0 per 100,000 population, respectively.
Data for clonal complexes (CCs) ST-11, ST-22, ST-23, and ST-53, with each containing only one ST (corresponding to the central ST of the clonal complex), are tabulated on the same row.
Carrier strains were isolated in the course of several studies with healthy children or young adults ages 0 to 12 and 14 to 22 years, respectively, attending either kindergarten, primary school, high school, military schools, or university in Cuba at the time of recruitment. The selection criteria excluded individuals receiving any antimicrobial therapy during the 7 days before the sampling date or under any immunomodulatory treatment, such as immunosuppressive medication, and a signed informed-consent form was obtained beforehand from all participants or their parents, in the case of underage subjects. The samples were taken from the posterior nasopharynx and larynx with a sterile cotton swab and were immediately inoculated into Mueller-Hinton plates supplemented with 5% defibrinated goat blood and vancomycin-nystatin-colistin (VCN). The plates were transferred to a 37°C incubator within 2 h and remained at that temperature under a water-saturated 5% CO2 atmosphere. Readings were performed at 24 and 48 h. Bacterial taxonomic identification was performed by conventional methods and by use of the API NH system (bioMérieux). The design and implementation of the study complied with the ethical principles for medical research involving humans.
Serogrouping, serotyping, and serosubtyping.
The isolates were serogrouped by latex agglutination with specific antisera (Difco). Serotypes and serosubtypes were determined by whole-cell enzyme-linked immunosorbent assay (1) with specific monoclonal antibodies provided by NIBSC (United Kingdom).
Preparation of chromosomal DNA.
Meningococcal chromosomal DNA was prepared from bacterial suspensions by using an Isoquick nucleic acid extraction kit (Orca Research), following the instructions from the manufacturer.
MLST.
All the isolates were characterized by MLST by the method previously described by Maiden et al. (28). The primers used for amplification and sequencing of the housekeeping genes are reported on the Neisseria MLST website (http://neisseria.org/nm/typing/mlst/). All nucleotide sequences were determined directly from the PCR products. Termination products were generated by cycle sequencing with the appropriate primers and BigDye Terminators (Applied Biosystems), and the products were separated with an ABI Prism 377 XL automated DNA sequencer. Housekeeping alleles and sequence types were assigned by interrogating the MLST database (http://mlst.zoo.ox.ac.uk).
Data analysis.
STs were assigned to lineages with the program eBURST (18), which resolved lineages, defined as groups of strains in which each member shares at least four of the seven loci in common with a central ST. The overall mean distances for each of the seven genes and concatenates were calculated by using the Kimura two-parameter model in the Molecular Evolutionary Genetics Analysis software package, version 3.1 (MEGA 3.1) (25). By using MEGA 3.1, pairwise comparisons were performed with each set of aligned sequences to identify distinct alleles and segregating sites. The number of synonymous and nonsynonymous nucleotide substitutions and the p distances (pS and pN, respectively) were calculated as overall means by the method of Nei and Gojobori (37) in the MEGA 3.1 program. Statistical analyses of the results obtained by MLST were performed by using the START software application (23). The relationships among the STs were determined by constructing a distance matrix of allelic mismatches. Each locus difference was treated identically, in that no relationships among the different alleles were assumed. The different lineages in the sample were then resolved from the clusters obtained when this distance matrix was visualized by split decomposition analysis with the program SplitsTree, version 4 (20).
RESULTS
ST assignment and diversity of MLST genes.
We analyzed 420 isolates: 167 isolated from cases with MD and 253 from carriers (Table 1). A total of 63 STs were identified by MLST analysis within this sample of Cuban isolates (Fig. 1). Thirty-two of the STs identified in this study have not previously been found in the MLST database (http://pubmlst.org/neisseria) and were registered as ST-6365 to ST-6376, ST-6378 to ST-6381, ST-6389, ST-6437 to ST-6440, ST-7085, and ST-7086. Fifty of the total STs were successfully assigned to 12 clonal complexes; 13 could not be assigned to any known clonal complex and are denoted here as unassigned (UA) STs. The four most frequent clonal complexes were ST-32 with 246 isolates (58.6%), ST-53 with 86 isolates (20.5%), ST-41/44 with 36 isolates (8.6%), and ST-103 with 11 isolates. Each of the eight remaining clonal complexes occurred at a frequency of less than 2.0%.
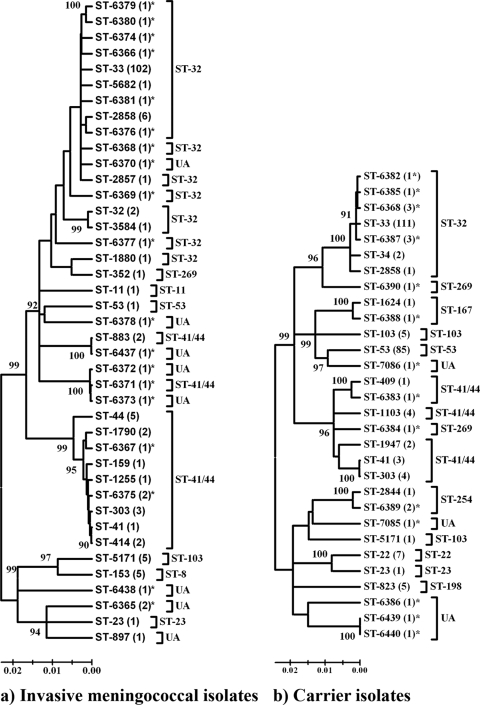
FIG. 1.
Genetic relationships among STs of Cuban isolates by neighbor-joining analysis. (a) Phylogenetic relationships of 167 invasive meningococcal isolates; (b) phylogenetic relationships of 253 carrier isolates (the numbers of strains are given in parentheses). New STs are marked by asterisks, and the clonal complexes are indicated by brackets. Nodes with bootstrap support higher than 90% are indicated.
The total number of alleles present at each locus for the set of 420 isolates, separated according to the source of isolation, is shown in Table 2, together with the number of polymorphic sites present at each locus, the number of alleles, the mean p distance, and the proportion of nonsynonymous to synonymous nucleotide substitutions (pN/pS). A comparison of the variables reported in Table 2 between disease-associated and healthy carrier isolates showed some differences in the mean p distance and the nonsynonymous-to-synonymous nucleotide substitution ratio for some alleles. The number of alleles present at each locus for the disease isolates ranged from 9 for adk to 14 for abcZ, pdhC, gdh, and pgm; and the numbers were relatively similar for the carrier isolates (from 8 for fumC to 16 for abcZ). While the diversity of the alleles present at each locus was similar for both invasive and carrier isolates, they did not represent the same population, as there were multiple alleles unique to each data set (Table 2). The mean p distances confirmed the higher degree of sequence diversity for carrier isolates than for bacteria that caused disease.
TABLE 2.
Genetic diversity in MLST loci among Cuban meningococcal population, 1983 to 2005
| Locus | Size (bp) | Disease isolates (n = 167) |
Carrier isolates (n = 253) |
Total no. of alleles | No. (%) of shared alleles | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of alleles | No. of polymorphic sites | Mean p distance | pN/pSa | No. of alleles | No. of polymorphic sites | Mean p distance | pN/pSa | ||||
| abcZ | 432 | 14 | 61 | 0.025 | 0.139 | 16 | 74 | 0.046 | 0.088 | 22 | 8 (36.4) |
| adk | 465 | 9 | 36 | 0.006 | 0.042 | 11 | 28 | 0.012 | 0.021 | 14 | 6 (42.9) |
| aroE | 489 | 13 | 126 | 0.049 | 0.308 | 10 | 130 | 0.073 | 0.359 | 16 | 7 (43.7) |
| fumC | 465 | 13 | 25 | 0.013 | 0.043 | 8 | 25 | 0.017 | 0.050 | 15 | 6 (40.0) |
| gdh | 501 | 14 | 25 | 0.007 | 0.037 | 12 | 23 | 0.015 | 0.032 | 18 | 8 (44.4) |
| pdhC | 480 | 14 | 60 | 0.016 | 0.036 | 15 | 69 | 0.024 | 0.063 | 23 | 6 (26.1) |
| pgm | 450 | 14 | 49 | 0.012 | 0.147 | 11 | 75 | 0.038 | 0.101 | 19 | 6 (31.6) |
| Concatenate | 3,282 | 41b | 382 | 0.018 | 0.135 | 30b | 424 | 0.031 | 0.132 | 63b | 8 (12.7) |
Calculated by using the MEGA 3.1 program.
The number of alleles for the concatenates corresponds to the STs.
The pN/pS ratio was calculated as a measure of the degree of selection in the population. The pN/pS ratio was calculated for all seven loci, and for most of them it was <0.15 (Table 2). However, the pN/pS ratios for the aroE locus (mean 0.32) could indicate that the degree of selection is not as strong as it is for typical housekeeping genes.
Phenotype diversity among the clonal complexes.
Among the 342 strains that were phenotypically characterized in this study, 149 were isolated from cases with meningococcal disease and 193 were isolated from asymptomatic carriers (Table 3). A group of strains (19%) was not classified by serological phenotype due to a lack of viability of the preserved bacteria. Among the isolates assessed, 255 (74.6%) were serogrouped as B, 5 (1.5%) as C, 2 (0.6%) as W135, and 1 (0.3%) as Z; the remaining 79 strains were nonserogroupable (NG) (23.1%). The distribution of serogroups across the total sample of invasive isolates was dominated by serogroup B, with 144 strains (96.6%), followed by serogroup C, with 5 strains (3.4%). Serogroup C strains were detected only between 1983 and 1989 (i.e., during the prevaccination period), and as expected for invasive isolates, no NG strains were identified in this subset. The phenotypic characteristics of the 193 strains obtained from healthy individuals are also shown in Table 3. Serogroup B was also predominant among carriers, with 111 isolates (57.5%), followed by NG strains with 79 isolates (40.9%). Serogroup W135 and Z isolates were obtained only from carriers. These results corroborate previous findings that serogroup B was dominant in Cuba before and after the introduction of a serogroup B and C vaccine (31, 41).
TABLE 3.
Phenotypic diversity within the lineages represented among the Cuban isolates
| Phenotypic characteristic | No. of disease strains/no. of carrier strains in clonal complex: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ST-32 | ST-41/44 | ST-53 | ST-103 | ST-22 | ST-8 | ST-198 | ST-269 | Other STs | Total | |
| Serogroup | ||||||||||
| B | 123/106 | 5/1 | 0/2 | 4/0 | 5/0 | 1/0 | 6/2 | 144/111 | ||
| C | 4/0 | 1/0 | 5/0 | |||||||
| W135 | 0/2 | 0/2 | ||||||||
| Z | 0/1 | 0/1 | ||||||||
| NG | 0/7 | 0/4 | 0/48 | 0/2 | 0/5 | 0/5 | 0/1 | 0/7 | 0/79 | |
| Serotype | ||||||||||
| 4 | 123/109 | 4/1 | 0/1 | 4/0 | 5/0 | 1/0 | 3/3 | 140/114 | ||
| 15 | 3/0 | 0/1 | 0/5 | 3/3 | 6/9 | |||||
| Others | 0/6 | 0/6 | ||||||||
| NT | 0/4 | 2/4 | 0/42 | 0/3 | 0/7 | 0/1 | 1/3 | 3/64 | ||
| Serosubtype | ||||||||||
| 5 | 5/0 | 0/4 | 5/4 | |||||||
| 5,2 | 4/0 | 2/0 | 6/0 | |||||||
| 6 | 2/0 | 0/5 | 0/5 | 2/10 | ||||||
| 7 | 1/0 | 0/8 | 2/0 | 3/8 | ||||||
| 12 | 0/7 | 0/7 | ||||||||
| 19,15 | 120/91 | 2/0 | 1/0 | 1/2 | 124/93 | |||||
| Others | 3/19 | 4/4 | 0/6 | 0/1 | 2/2 | 9/32 | ||||
| NST | 0/3 | 0/1 | 0/29 | 0/3 | 0/2 | 0/1 | 0/39 | |||
| Combinations (phenotypes) | ||||||||||
| B:4:P1.19,15 | 120/90 | 2/0 | 1/0 | 1/2 | 124/92 | |||||
| NG:NT:P1.NST | 0/2 | 0/1 | 0/27 | 0/2 | 0/32 | |||||
| Others | 3/21 | 7/4 | 0/23 | 4/1 | 0/7 | 5/0 | 0/5 | 0/1 | 6/7 | 25/69 |
N. meningitidis B:4:P1.19,15 was the phenotype the most frequently identified (Table 3), representing 63.2% of all typed strains; this is the same serologic classification of type strain CU385, used to produce the Cuban serogroup B meningococcal vaccine VA-MENGOC-BC. NG:NT:P1.NST (where NG is nongroupable, NT is nontypeable, and NST is nonsubtypeable) strains, found only in isolates from carriers, were the second (16.6%) in frequency. The remaining phenotypes were less prevalent among the set of typed isolates.
There was strong evidence for the association of certain clonal complexes with particular serogroups, although these associations were not absolute. For example, most of the ST-32-complex isolates were serogroup B (229 isolates, 93.5%); only 7 members of this clonal complex were nonserogroupable (2.9%) (Table 3). The ST-53 complex was predominantly nonserogroupable (48 isolates, 55.8%). The only two serogroup W135 strains and the serogroup Z strain that were collected from carriers belonged to the ST-22 and ST-103 clonal complexes, respectively. For the rest of the clonal complexes, the distribution of serogroups was highly heterogeneous; however, these complexes are represented by a significantly lower number of isolates.
STs and lineages among carrier and disease isolates.
A total of 30 different STs were detected among carrier strains, of which 14 corresponded to new STs. In the case of disease isolates, there were 41 STs, 19 of which corresponded to novel STs. Eight STs (ST-23, ST-33, ST-41, ST-53, ST-303, ST-2858, ST-5171, and ST-6368) were shared between carrier and disease-associated isolates, of which only one (ST-6368) has not been described previously. The genetic relationships between STs from disease-associated and carrier isolates are shown in Fig. 1. The dendrograms of the 41 STs from disease isolates (Fig. 1a) and the 30 STs from carrier isolates (Fig. 1b) illustrate the genetic variety of the Cuban meningococcal population. As can be appreciated in the figure, the complexity of the carrier isolate sample is lower than that of the disease-associated strains (Fig. 1b). Remarkably, some of the new STs clustered with those of the hypervirulent ST-41/44 lineage (e.g., ST-6373 and ST-6372 with ST-6371 and ST-6437 with ST-883 in Fig. 1a). Another ST classified as UA also clustered with a hypervirulent clonal lineage (ST-6370 to ST-32).
Temporal distribution of Cuban STs and impact of vaccination.
In this work and on the basis of the incidence of meningococcal disease in Cuba, we chose to partition our data into three periods (Table 1). As it is shown in Table 1, there is a marked temporal structuring in the distribution of some STs and clonal complexes during the whole period under study. These differences could be due to the introduction of a serogroup B-C vaccine in the late 1980s. However, it was difficult to define a precise unique year that could be used to divide our data (into pre- and postvaccination periods, for example) and to make accurate inferences about the impact of the vaccination, since nationwide mass vaccination in Cuba with VA-MENGOC-BC was gradually implemented from 1989 to 1990 (95% coverage for the population between 3 months and 24 years old) until the current routine vaccination of infants started in 1991 (46). In order to characterize the potential epidemiological impact of the vaccination with VA-MENGOC-BC and on the basis of previous results of Martínez et al. (31), who characterized the phenotypes of N. meningitidis strains isolated from healthy carriers during a 20-year period (1982 to 2002) in Cuba, the first two periods described in Table 1 were combined into a single stage comprising the period from 1983 to 1992, denominated as the prevaccination and peak vaccination period. Accordingly, the years from 1993 to 2005 were included in the postvaccination stage.
Forty-five of the STs found in our collection were represented among strains collected between 1983 and 1992, split into 37 STs from disease-associated cases and 14 from carriers. Twenty-three of the STs were represented among postvaccination period isolates (1993 to 2005), split into 6 STs from cases and 17 from carriers (Table 1). Five STs (ST-33, ST-53, ST-2858, ST-6368, and ST-6387) were shared between both periods, of which two (ST-6368 and ST-6387) correspond to previously unknown STs.
Complex ST-32 was the most common lineage among the 273 strains from the period from 1983 to 1992, comprising 193 isolates (70.7%) assigned to 18 STs. ST-33 was the predominant ST within this hypervirulent lineage, with 170 isolates (88.1%). Other frequent complexes, in addition to ST-32, were ST-53 (30 strains, 11.0%) and ST-41/44 (26 strains, 9.5%). Other hypervirulent lineages were also detected among the isolates from this period; these were ST-8 (5 strains) and ST-11 (1 strain), both detected exclusively in disease-associated cases. Twenty-five novel STs were represented among strains from this period, of which 11 were associated with the ST-32 complex and 4 with the ST-41/44 complex.
As mentioned above, the collection from the postvaccination period (1993 to 2005) included 147 strains, 20 isolated from cases and 127 from carriers. Of these 147 strains, 135 were assigned to 14 STs and 12 isolates belonged to 9 novel STs (Fig. 1). A high proportion of the strains isolated during this period belonged to the ST-32 and ST-53 complexes (52 and 56 strains, respectively). Six STs were detected among isolates from the ST-32 complex, and three of these were previously unknown. ST-33 was the most common ST among ST-32-complex strains, with 43 isolates. However, in the ST-53 complex, represented by a similar number of isolates, we detected only one ST, and almost all strains from this period were collected from carriers.
Although 62 of the isolates from the postvaccination period belonged to two hyperinvasive lineages strongly associated with endemic and epidemic meningococcal disease (the ST-32 and ST-41/44 complexes), most of these hyperinvasive clones were obtained from healthy carriers (35 of 52 for the ST-32 complex and 9 of 10 for the ST-41/44 complex). Interestingly, only three ST-41/44-complex STs were found after the implementation of mass vaccination; none of them were found among isolates from the previous period.
Within-lineage variability.
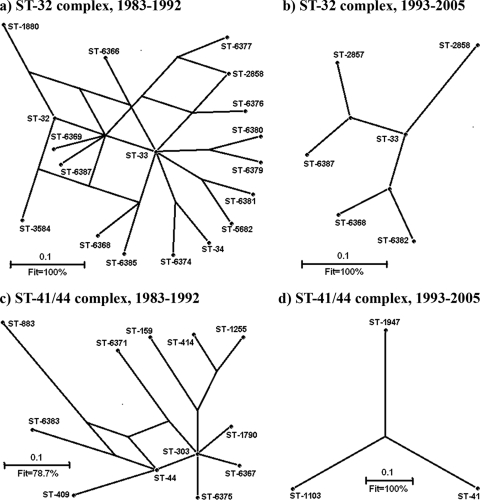
Among the hypervirulent lineages reported by Maiden et al. (28), those most frequently found in our collection were ST-32 and ST-41/44. The relationships of the members of both complexes are illustrated by the splits graph in Fig. 2. In order to analyze the possible influence of the Cuban antimeningococcal mass vaccination on lineage diversity, the samples were grouped into STs collected during all the periods under study. This analysis placed the most common ST (ST-33) at the center of the graph for the ST-32 complex in all periods, positioning this ST as a possible ancestor for the strains isolated between 1983 and 1992 and also for the few ST-32-complex STs remaining after vaccination with VA-MENGOC-BC. Before vaccination, 14 of the STs belonging to the ST-32 complex (ST-32, ST-34, ST-2858, ST-5682, ST-6366, ST-6368, ST-6369, ST-6374, ST-6376, ST-6379, ST-6380, ST-6381, ST-6385, and ST-6387) (Fig. 2a) were single-locus variants of ST-33 and 3 were double-locus variants (ST-1880, ST-3584, and ST-6377). All of the few STs belonging to complex ST-32 that were found after vaccination (Fig. 2b) were single-locus variants of ST-33.
FIG. 2.
Splits graphs showing the relationships among STs in the two most common and diverse hypervirulent lineages present in Neisseria meningitidis in Cuba: ST-32 (246 isolates) and ST-41/44 (36 isolates) clonal complexes. (a and b) ST-32 complex; (c and d) ST-41/44 complex.
For the next most common hypervirulent lineage found during the period between 1983 and 1992 (complex ST-41 to 44, Fig. 2c), ST-303 occupied the central position with four single-locus variants (ST-44, ST-1790, ST-6367, and ST-6375), five double-locus variants (ST-159, ST-409, ST-414, ST1255, and ST-6371), and one triple-locus variant (ST-6383, a double-locus variant of ST-44). There was a singleton in this complex (ST-883), as detected by eBURST analysis. We were unable to detect any central ST within the few remaining members of the ST-41/44 complex (Fig. 2d) from the postvaccination period.
DISCUSSION
In 1980, meningococcal disease was considered the main public health problem in Cuba, with the incidence reaching 5.9 per 100,000 population. The peak incidence was detected during 1983 and 1984, reaching a maximum of 14.4 per 100,000 population (46). A nationwide intervention with VA-MENGOC-BC vaccination started in 1989, and since 1991 this vaccine has been included in the NIP for children 3 months of age. The mass vaccination campaign and the subsequent immunization of all infants have resulted in a sharp and sustained decline in the incidence of the disease, which was reduced to 0.1 per 100,000 population by 2008 (34). Although VA-MENGOC-BC is a vaccine that was developed more than 20 years ago (46), its influence on the genetic structure of the population of circulating meningococci was unknown. To our knowledge, this is the first report of a study that has used MLST to type N. meningitidis strains isolated in Cuba. Although the sample of N. meningitidis strains used in this study cannot be claimed to accurately represent the Cuban meningococcal population throughout the history of meningococcal disease in that country, it does, however, contain samples from two significant periods within the context of the most recent Cuban epidemic.
The epidemic in Cuba was largely caused, according to previous studies (31, 45), by a single serogroup B clone with phenotype B:4:P1.19,15 (electrophoretic type 5 [ET-5] complex). MLST analysis revealed that in spite of the presence of a large number of B:4:P1.19,15 strains belonging to the ST-32 complex, there were phenotypically identical isolates that by sequence typing classified either in the ST-41/44, ST-167, or ST-269 complexes or as UA. Within the ST-32 complex, strains with the phenotype B:4:P1.19,15 were assigned to 17 different STs, of which 10 had not been previously described. Most of the novel STs described in this work arose by allelic replacement of the original clone (Fig. 2), reflecting the genetic variability of the B:4:P1.19,15 clones isolated in Cuba, which possibly facilitated their emergence during the epidemic period and/or their persistence after vaccination. Our results confirm that the findings obtained by phenotyping must, whenever possible, be complemented by genotyping methods if the full characterization of the available isolates is desired, as demonstrated previously (28, 48).
The epidemiological impact of VA-MENGOC-BC vaccination has previously been reported to be a reduction in morbidity from meningococcal disease in Cuba (41). Additionally, Martínez et al. (31) made an important and significant contribution to the epidemiological understanding of meningococcal disease in Cuba and the impact of vaccination, by characterizing the phenotypes of N. meningitidis strains isolated from healthy carriers during a 20-year period (1982 to 2002), including the epidemic stage and the period of mass vaccination with VA-MENGOC-BC starting in 1989. The main finding was that during the postepidemic stage, there was a significant decrease in the incidence of the epidemic strain (B:4:P1.19,15) among carriers, a total absence of N. meningitidis serogroup C strains, and a predominance of strains not associated with epidemic outbreaks. We illustrate here the changes in the structure of the meningococcal population that occurred during the same period of time, in terms of the variation of MLST genes and its associations. In agreement with the results obtained by the phenotypic characterization of carrier strains (31), our molecular study demonstrates that VA-MENGOC-BC affected the genetic composition of the carrier-associated meningococcal isolates. The number of isolates belonging to hypervirulent lineages decreased significantly after vaccination, and ST-53, a sequence type common in healthy carriers (12, 24, 55), became the predominant ST. The few hypervirulent STs detected in carriers after vaccination were mostly collected during the earlier years of the introduction of the serogroup B and C vaccine. These results are important for assessing the effects of the introduction of meningococcal vaccines, since carriage studies are essential for understanding meningococcal spread and developing public health policy (9, 24).
It is important to note that this is a preliminary examination of the impact of vaccination in Cuba with VA-MENGOC-BC on the N. meningitidis population structure, and further studies should be conducted to define the precise time frame in which the vaccine redrew the meningococcal scenario in Cuba. For example, sequence characterization of the main epitopes of outer membrane proteins PorA, PorB, and FetA is being conducted with the same strain panel. However, some general conclusions deserve to be discussed. Among the strains collected from patients, there was a significant decline in ST diversity after the introduction of the vaccine; hypervirulent lineages ST-8 and ST-11 were no longer found during this period. From the splits-graph analysis of the association of hypervirulent STs during the postvaccination period (Fig. 2), we showed that complex ST-32, the predominant complex responsible for the Cuban epidemic, underwent a reduction in the number of its ST variants. We also demonstrated that the ST-41/44 complex is completely different than it was during the period between 1983 and 1992 in terms of the associated STs and the lack of a defined relationship between them (Fig. 2c), with only one disease isolate being detected after vaccination was introduced. The impact of the introduction of the vaccine on lineages other than complex ST-32 corroborates that VA-MENGOC-BC can confer protection against some serogroup B strains different from the vaccine type strain, as previously demonstrated by seroepidemiological and case-control studies (14, 32, 35, 38). Importantly, most of the small number of cases that occurred after vaccination were caused by ST-33 strains (13 of 20 strains) with the same phenotype (B:4:P1.19,15) as the VA-MENGOC-BC vaccine strain. The prevalence of this phenotype after the introduction of the Cuban vaccine has previously been reported in Cuba (40) and Brazil (3, 13). However, in Cuba, where VA-MENGOC-BC is used systematically, the vaccine has had a dramatic impact on the morbidity and mortality from the disease (41). In Brazil, only a proportion of the population was vaccinated in response to specific epidemiological situations (14, 33, 38), a fact that could explain the persistence of antigenic variants of the ST-33 epidemic strain. The availability of additional information about the PorA, PorB, and FetA variable regions (22, 50) and pulsed-field gel electrophoresis patterns (10) will increase the discriminatory power of our analysis for the sample of invasive meningococci, in particular, for those collected after vaccination.
Fine typing has been employed to evaluate the impact of mass anti-serogroup C vaccination on the risk of emergence of escape variants within the ST-11 clonal complex between the pre- and postvaccination periods in France (26). MLST and phenotyping have also been used to help evaluate the impact of serogroup C conjugate vaccines on meningococcal carriage in the United Kingdom (29). That combined study demonstrated that the composition of the population of meningococcal isolates recovered from carriers remained essentially unchanged, with the exception of serogroup C ST-11-complex bacteria. The strong impact of vaccination on the carriage of ST-11 complex serogroup C could be attributed to high levels of capsule expression. In the United Kingdom, the prevalence of carriage of the meningococci most commonly associated with serogroup B disease was unaffected by the introduction of vaccine (29). Our results are the first obtained by use of the MLST approach to evaluate the impact of an OMV-based vaccine on the genetic structure of a meningococcal population. This type of vaccine has recently been used to combat a clonal epidemic in New Zealand (39), and OMV-based vaccines are still in development by either the use of genetically modified meningococci or the addition of recombinant antigens (27). The epidemic of disease in New Zealand was attributable to the clonal expansion of strain type B:4:P1.7-2,4, which belongs to the ST-41/44 clonal complex (17). The stability of the P1.7-2,4 PorA protein (15) and the results of our study suggest that the epidemic in New Zealand may be controlled by a strain-specific OMV vaccine.
A comparison of our results with those from other studies deserves further comments. First, data collected in several European countries indicate that carrier populations are more diverse than isolates from patients and have relatively low levels of representation of hyperinvasive lineages (24, 55). However, during the prevaccination and peak vaccination period (1983 to 1992), the number of alleles per MLST locus was greater for disease-associated isolates than for strains from healthy carriers (data not shown). The greater diversity during this period of disease-associated isolates was probably due to extensive recombination which occurred among circulating strains, as suggested by the split decomposition analysis (Fig. 2a and c). Second, the most frequent hypervirulent clonal complexes identified here, i.e., ST-32 (ET-5 complex) and ST-41/44 (lineage III), have also been identified to be the most abundant among the meningococcal population from other Latin American countries (13). Complex ST-32 has been reported to cause numerous outbreaks worldwide and has a tendency to cause hyperendemic disease, which has an increased incidence of septicemia and a high death rate (36). Strains of lineage III are known to be responsible for the increased incidence of meningococcal disease in European countries (6, 43) and for outbreaks in China (54) and Japan (47). Therefore, the continuous monitoring of circulating strains of these complexes in Cuba, mainly in healthy carriers, is mandatory if eventual meningococcal outbreaks are to be prevented.
To our knowledge, this is the first analysis of Cuban meningococcal isolates by molecular methods and the first application of MLST for the preliminary analysis of the epidemiological impact of extensive and systematic immunization with a serogroup B vaccine. Furthermore, our results can be used to improve the continuous monitoring of the distribution of the circulating meningococcal strains in Cuba in the postvaccination era. This study also confirms the original concept that sequence typing methods are important for evolutionary analyses and for examining meningococcal transmission and evolution and the impacts of vaccines on meningococci (4, 28).
Acknowledgments
This work was supported in part by the British Council United Kingdom-Cuba Science Links Programme.
We thank Lilia López-Cánovas from the Cuban Center for Neuroscience/CNIC for critical review of the manuscript.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Definition of meningococcal class 1 OMP subtyping antigens by monoclonal antibodies. FEMS Microbiol. Immunol. 1:139-144. [DOI] [PubMed] [Google Scholar]
- 2.Abdillahi, H., and J. T. Poolman. 1988. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J. Med. Microbiol. 26:177-180. [PubMed] [Google Scholar]
- 3.Baethgen, L. F., L. Weidlich, C. Moraes, C. Klein, L. S. Nunes, P. I. Cafrune, A. P. Lemos, S. S. Rios, M. F. Abreu, C. Kmetzsch, A. F. Sperb, L. W. Riley, M. L. Rossetti, and A. Zaha. 2008. Epidemiology of meningococcal disease in southern Brazil from 1995 to 2003, and molecular characterization of Neisseria meningitidis using multilocus sequence typing. Trop. Med. Int. Health 13:31-40. [DOI] [PubMed] [Google Scholar]
- 4.Brehony, C., K. A. Jolley, and M. C. Maiden. 2007. Multilocus sequence typing for global surveillance of meningococcal disease. FEMS Microbiol. Rev. 31:15-26. [DOI] [PubMed] [Google Scholar]
- 5.Buckee, C. O., K. A. Jolley, M. Recker, B. Penman, P. Kriz, S. Gupta, and M. C. Maiden. 2008. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 105:15082-15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caugant, D. A., P. Bol, E. A. Hoiby, H. C. Zanen, and L. O. Froholm. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958-1986. J. Infect. Dis. 162:867-874. [DOI] [PubMed] [Google Scholar]
- 7.Caugant, D. A., B. E. Kristiansen, L. O. Froholm, K. Bovre, and R. K. Selander. 1988. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect. Immun. 56:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caugant, D. A., L. F. Mocca, C. E. Frasch, L. O. Froholm, W. D. Zollinger, and R. K. Selander. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J. Bacteriol. 169:2781-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caugant, D. A., G. Tzanakaki, and P. Kriz. 2007. Lessons from meningococcal carriage studies. FEMS Microbiol. Rev. 31:52-63. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, C. S., J. C. Liao, T. L. Liao, C. C. Li, C. Y. Chou, H. L. Chang, S. M. Yao, and Y. S. Lee. 2006. Molecular epidemiology and emergence of worldwide epidemic clones of Neisseria meningitidis in Taiwan. BMC Infect. Dis. 6:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, S. C., M. A. Diggle, and G. F. Edwards. 2001. Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis. J. Clin. Microbiol. 39:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claus, H., M. C. Maiden, R. Maag, M. Frosch, and U. Vogel. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813-1819. [DOI] [PubMed] [Google Scholar]
- 13.de Filippis, I., and A. C. Vicente. 2005. Multilocus sequence typing and repetitive element-based polymerase chain reaction analysis of Neisseria meningitidis isolates in Brazil reveal the emergence of 11 new sequence types genetically related to the ST-32 and ST-41/44 complexes and high prevalence of strains related to hypervirulent lineages. Diagn. Microbiol. Infect. Dis. 53:161-167. [DOI] [PubMed] [Google Scholar]
- 14.de Moraes, J. C., B. A. Perkins, M. C. Camargo, N. T. Hidalgo, H. A. Barbosa, C. T. Sacchi, I. M. Landgraf, V. L. Gattas, H. G. Vasconcelos, B. D. Plikaytis, J. D. Wenger, and C. V. Broome. 1992. Protective efficacy of a serogroup B meningococcal vaccine in Sao Paulo, Brazil. Lancet 340:1074-1078. [DOI] [PubMed] [Google Scholar]
- 15.Devoy, A. F., K. H. Dyet, and D. R. Martin. 2005. Stability of PorA during a meningococcal disease epidemic. J. Clin. Microbiol. 43:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson, F. O., and A. E. Perez. 2005. Bacterial meningitis in children and adolescents: an observational study based on the national surveillance system. BMC Infect. Dis. 5:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyet, K. H., and D. R. Martin. 2006. Clonal analysis of the serogroup B meningococci causing New Zealand's epidemic. Epidemiol. Infect. 134:377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frasch, C. E., W. D. Zollinger, and J. T. Poolman. 1985. Serotype antigens of Neisseria meningitidis and a proposed scheme for designation of serotypes. Rev. Infect. Dis. 7:504-510. [DOI] [PubMed] [Google Scholar]
- 20.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 21.Jodar, L., I. M. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359:1499-1508. [DOI] [PubMed] [Google Scholar]
- 22.Jolley, K. A., C. Brehony, and M. C. Maiden. 2007. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol. Rev. 31:89-96. [DOI] [PubMed] [Google Scholar]
- 23.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 24.Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek, P. Kriz, and M. C. Maiden. 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.Lancellotti, M., A. Guiyoule, C. Ruckly, E. Hong, J. M. Alonso, and M. K. Taha. 2006. Conserved virulence of C to B capsule switched Neisseria meningitidis clinical isolates belonging to ET-37/ST-11 clonal complex. Microbes Infect. 8:191-196. [DOI] [PubMed] [Google Scholar]
- 27.Lewis, S., M. Sadarangani, J. C. Hoe, and A. J. Pollard. 2009. Challenges and progress in the development of a serogroup B meningococcal vaccine. Expert. Rev. Vaccines 8:729-745. [DOI] [PubMed] [Google Scholar]
- 28.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiden, M. C., A. B. Ibarz-Pavon, R. Urwin, S. J. Gray, N. J. Andrews, S. C. Clarke, A. M. Walker, M. R. Evans, J. S. Kroll, K. R. Neal, D. A. Ala'Aldeen, D. W. Crook, K. Cann, S. Harrison, R. Cunningham, D. Baxter, E. Kaczmarski, J. MacLennan, J. C. Cameron, and J. M. Stuart. 2008. Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J. Infect. Dis. 197:737-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez, I., O. Lopez, F. Sotolongo, M. Mirabal, and A. Bencomo. 2003. Carriers of Neisseria meningitidis among children from a primary school. Rev. Cubana Med. Trop. 55:162-168. (In Spanish.) [PubMed] [Google Scholar]
- 31.Martínez, I., G. Sierra, N. Nuñez, L. Izquierdo, Y. Climent, and M. Mirabal. 2006. Characterization of Neisseria meningitidis strains isolated from carriers in Cuba during 20 years. Rev. Cubana Med. Trop. 58:8-17. (In Spanish.) [PubMed] [Google Scholar]
- 32.Milagres, L. G., M. C. Gorla, C. T. Sacchi, and M. M. Rodrigues. 1998. Specificity of bactericidal antibody response to serogroup B meningococcal strains in Brazilian children after immunization with an outer membrane vaccine. Infect. Immun. 66:4755-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milagres, L. G., S. R. Ramos, C. T. Sacchi, C. E. Melles, V. S. Vieira, H. Sato, G. S. Brito, J. C. Moraes, and C. E. Frasch. 1994. Immune response of Brazilian children to a Neisseria meningitidis serogroup B outer membrane protein vaccine: comparison with efficacy. Infect. Immun. 62:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MINSAP. 2008. Incidence and mortality of meningococcal disease 1980-2008. Anuario Estadístico de Salud de Cuba. http://www.sld.cu/servicios/estadisticas/. (In Spanish.)
- 35.Morley, S. L., M. J. Cole, C. A. Ison, M. A. Camaraza, F. Sotolongo, N. Anwar, I. Cuevas, M. Carbonero, H. C. Campa, G. Sierra, and M. Levin. 2001. Immunogenicity of a serogroup B meningococcal vaccine against multiple Neisseria meningitidis strains in infants. Pediatr. Infect. Dis. J. 20:1054-1061. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, K. M., K. A. O'Donnell, A. B. Higgins, C. O'Neill, and M. T. Cafferkey. 2003. Irish strains of Neisseria meningitidis: characterisation using multilocus sequence typing. Br. J. Biomed. Sci. 60:204-209. [DOI] [PubMed] [Google Scholar]
- 37.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 38.Noronha, C. P., C. J. Struchiner, and M. E. Halloran. 1995. Assessment of the direct effectiveness of BC meningococcal vaccine in Rio de Janeiro, Brazil: a case-control study. Int. J. Epidemiol. 24:1050-1057. [DOI] [PubMed] [Google Scholar]
- 39.Oster, P., D. Lennon, J. O'Hallahan, K. Mulholland, S. Reid, and D. Martin. 2005. MeNZB: a safe and highly immunogenic tailor-made vaccine against the New Zealand Neisseria meningitidis serogroup B disease epidemic strain. Vaccine 23:2191-2196. [DOI] [PubMed] [Google Scholar]
- 40.Perez, A., and F. Dickinson. 1998. Results of the meningococcal BC National Immunization Program in Cuba in children under 1 year of age. Rev. Cubana. Pediatr. 70:133-140. (In Spanish.) [Google Scholar]
- 41.Rodriguez, A. P., F. Dickinson, A. Baly, and R. Martinez. 1999. The epidemiological impact of antimeningococcal B vaccination in Cuba. Mem. Inst. Oswaldo Cruz 94:433-440. [DOI] [PubMed] [Google Scholar]
- 42.Rosenstein, N. E., B. A. Perkins, D. S. Stephens, T. Popovic, and J. M. Hughes. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378-1388. [DOI] [PubMed] [Google Scholar]
- 43.Scholten, R. J., J. T. Poolman, H. A. Valkenburg, H. A. Bijlmer, J. Dankert, and D. A. Caugant. 1994. Phenotypic and genotypic changes in a new clone complex of Neisseria meningitidis causing disease in The Netherlands, 1958-1990. J. Infect. Dis. 169:673-676. [DOI] [PubMed] [Google Scholar]
- 44.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sierra, G. V., H. C. Campa, N. M. Varcacel, I. L. Garcia, P. L. Izquierdo, P. F. Sotolongo, G. V. Casanueva, C. O. Rico, C. R. Rodriguez, and M. H. Terry. 1991. Vaccine against group B Neisseria meningitidis: protection trial and mass vaccination results in Cuba. NIPH Ann. 14:195-207. [PubMed] [Google Scholar]
- 46.Sotolongo, F., C. Campa, G. V. Casanueva, E. M. Fajardo, I. Cuevas, and N. Gonzalez. 2007. Cuban meningococcal BC vaccine: experiences & contributions from 20 years of application. MEDICC Rev. 9:16-22. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi, H., T. Kuroki, Y. Watanabe, H. Tanaka, H. Inouye, S. Yamai, and H. Watanabe. 2004. Characterization of Neisseria meningitidis isolates collected from 1974 to 2003 in Japan by multilocus sequence typing. J. Med. Microbiol. 53:657-662. [DOI] [PubMed] [Google Scholar]
- 48.Tzanakaki, G., R. Urwin, M. Musilek, P. Kriz, J. Kremastinou, A. Pangalis, C. C. Blackwell, and M. C. Maiden. 2001. Phenotypic and genotypic approaches to characterization of isolates of Neisseria meningitidis from patients and their close family contacts. J. Clin. Microbiol. 39:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urwin, R., and M. C. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 50.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel, U., H. Claus, M. Frosch, and D. A. Caugant. 2000. Molecular basis for distinction of the ET-15 clone within the ET-37 complex of Neisseria meningitidis. J. Clin. Microbiol. 38:941-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 53.Wyle, F. A., M. S. Artenstein, B. L. Brandt, E. C. Tramont, D. L. Kasper, P. L. Altieri, S. L. Berman, and J. P. Lowenthal. 1972. Immunologic response of man to group B meningococcal polysaccharide vaccines. J. Infect. Dis. 126:514-521. [DOI] [PubMed] [Google Scholar]
- 54.Yang, L., Z. Shao, X. Zhang, L. Xu, J. Peng, X. Xu, X. Liang, Y. Qi, and Q. Jin. 2008. Genotypic characterization of Neisseria meningitidis serogroup B strains circulating in China. J. Infect. 56:211-218. [DOI] [PubMed] [Google Scholar]
- 55.Yazdankhah, S. P., P. Kriz, G. Tzanakaki, J. Kremastinou, J. Kalmusova, M. Musilek, T. Alvestad, K. A. Jolley, D. J. Wilson, N. D. McCarthy, D. A. Caugant, and M. C. Maiden. 2004. Distribution of serogroups and genotypes among disease-associated and carried isolates of Neisseria meningitidis from the Czech Republic, Greece, and Norway. J. Clin. Microbiol. 42:5146-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]