Abstract
The species-level identification of sterile and/or arthroconidium-forming filamentous fungi presumed to be basidiomycetes based upon morphological or physiological features alone is usually not possible due to the limited amount of hyphal differentiation. Therefore, a reliable molecular approach capable of the unambiguous identification of clinical isolates is needed. One hundred sixty-eight presumptive basidiomycetes were screened by sequence analysis of the internal transcribed spacer (ITS) and D1/D2 ribosomal DNA regions in an effort to obtain a species identification. Through the use of this approach, identification of a basidiomycetous fungus to the species level was obtained for 167/168 of the isolates. However, comparison of the BLAST results for each isolate for both regions revealed that only 28.6% (48/168) of the isolates had the same species identification by use of both the ITS and the D1/D2 regions, regardless of the percent identity. At the less stringent genus-only level, the identities for only 48.8% (82/168) of the isolates agreed for both regions. Investigation of the causes for this low level of agreement revealed that 14% of the species lacked an ITS region deposit and 16% lacked a D1/D2 region deposit. Few GenBank deposits were found to be complete for either region, with only 8% of the isolates having a complete ITS region and 10% having a complete D1/D2 region. This study demonstrates that while sequence-based identification is a powerful tool for many fungi, sequence data derived from filamentous basidiomycetes should be interpreted carefully, particularly in the context of missing or incomplete GenBank data, and, whenever possible, should be evaluated in light of compatible morphological features.
The emergence of rare but clinically significant fungi has placed a growing diagnostic burden on clinical microbiologists. Nevertheless, the accurate identification of these etiologic agents remains critically important, despite the low frequency of some species that are encountered in clinical specimens (10, 21). For filamentous fungi, identification by the use of colonial and microscopic morphologies, the major identification method, largely depends on the production of reproductive structures. Although filamentous basidiomycetes rarely cause disease, they are increasingly recognized from clinical specimens (27). However, definitive identification can be problematic, with many isolates remaining sterile in culture (15, 23, 28). The inability to ascertain a genus or species due to the lack of observable reproductive structures can potentially increase the time to the reporting of an inconclusive result and, consequently, adversely affect treatment strategies (13, 26, 29). Therefore, there is a clear need for alternative methods for the identification of fungi that do not produce morphologically distinguishing features.
Sequencing of the ribosomal genes has emerged as a useful diagnostic tool for the rapid detection and identification of fungi, regardless of whether morphologically distinct structures are produced (6, 16, 32). One of the most common ribosomal targets for sequence identification is the internal transcribed spacer (ITS) region. This region contains two informative regions, ITS1 and ITS2, which are located between the 18S and 28S ribosomal subunits and which are separated by the 5.8S ribosomal subunit (8, 9). The ITS region can be amplified from a broad spectrum of fungi with primers ITS-1 and ITS-4 and can generally be recovered in a single PCR, since the amplicon is usually ∼400 to 700 bp in length (9, 11, 17). A second variable site within the ribosomal DNA (rDNA) cluster, called the D1/D2 region, can also be amplified from a broad spectrum of fungi with primers NL-1 and NL-4, although it is usually less variable than the ITS region (19). The D1/D2 region is located toward the 5′ end of the large ribosomal subunit (26S or 28S) and overlaps the ITS region at the ITS-4/NL-1 primer site. The combination of conserved and variable regions offers great flexibility for PCR sensitivity and specificity. The conserved sequences at the flanking ends of the D1/D2 and ITS regions provide universal PCR priming sites, while the variable internal regions provide species-specific sequences in many cases (4, 7, 19).
Although the ITS region displays enough sequence variability to allow the identification of many fungi to the species level, for some fungi the sequence of the ITS region alone is not sufficient for accurate identification to the species level (1, 24). In these cases, a second locus, such as that for β-tubulin or calmodulin, can be sequenced (2, 3). Unfortunately, universal priming sites, which are required to obtain an amplicon from an unknown fungus, are sacrificed for the more variable nature of these nonribosomal genes, which in turn requires enough knowledge of the strain identity to allow the selection of primers that will yield a PCR product. Since sterile molds could potentially be found in any phylum, it would not be possible to know for sure which gene-specific primer pair to select for use in a PCR assay, since the priming sites could be genus specific. Additionally, it is possible that sequencing of a second site could be even less informative than rDNA sequencing due to the fewer GenBank deposits for the target locus. Therefore, the goal of this study was to determine if combined sequencing of the ITS and D1/D2 regions of a large collection of mostly sterile filamentous molds, presumed to be basidiomycetes, could confirm this preliminary placement in the phylum Basidiomycota as well as provide an accurate species-level identification.
MATERIALS AND METHODS
Strains and media.
The isolates that were used in this study were from a large collection archived in the Fungus Testing Laboratory (http://strl.uthscsa.edu/fungus/) in the Department of Pathology at the University of Texas Health Science Center at San Antonio (UTHSCSA) (Table 1). The isolates were maintained on potato dextrose agar (PDA; Difco, Detroit, MI) slants and had previously been identified as probable basidiomycetes on the basis of their macroscopic morphology on potato flakes agar (25), their microscopic features (noted in Table 1), and their physiological features. All isolates demonstrated rapid, woolly growth that was white to cream or golden, had the ability to grow on agar containing 10 μg/ml benomyl (30), and failed to grow on medium containing 0.5 μg/ml cycloheximide (Mycobiotic agar; Remel, Inc., Lenexa, KS). These candidate isolates were plated onto PDA, grown at 25°C for 4 to 7 days, and then submitted for molecular characterization.
TABLE 1.
Strains used in this study
| Strain no. | Accession no. | Yra | Phenotypic featuresb | Source | H, A, or O sourcec |
|---|---|---|---|---|---|
| B-1 | 07-56 | 2007 | Sterile, chlamydoconidia, crystal-encrusted hyphae | BALd | H |
| B-2 | 07-31 | 2007 | Sterile, crystals | BAL | H |
| B-3 | 06-4454 | 2006 | Arthroconidia | Eye | H |
| B-4 | 06-4450 | 2006 | Arthroconidia | Scalp | H |
| B-5 | 06-4444 | 2006 | Arthroconidia | BAL | H |
| B-6 | 06-4410 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-7 | 06-4341 | 2006 | Sterile, chlamydoconidia | Sputum | H |
| B-8 | 06-4285 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-9 | 06-4161 | 2006 | Arthroconidia | Neck mass | H |
| B-10 | 06-4137 | 2006 | Sterile | BAL | H |
| B-11 | 06-4124 | 2006 | Sterile | BAL | H |
| B-12 | 06-4103 | 2006 | Arthroconidia, crystals | Bronchial wash | H |
| B-13 | 06-4057 | 2006 | Sterile, mushroom smell | Sputum | H |
| B-14 | 06-3994 | 2006 | Sterile, crystals, spathulate hyphae | Nasal | H |
| B-15 | 06-3970 | 2006 | Arthroconidia, brown diffusing pigment | BAL | H |
| B-16 | 06-3924 | 2006 | Arthroconidia | BAL | H |
| B-17 | 06-3906 | 2006 | Sterile, crystal-encrusted hyphae | Sputum | H |
| B-18 | 06-3888 | 2006 | Arthroconidia | BAL | H |
| B-19 | 06-3869 | 2006 | Sterile | Aspirate | H |
| B-20 | 06-3821 | 2006 | Arthroconidia, chlamydoconidia | Bronchial wash | H |
| B-21 | 06-3806 | 2006 | Sterile | Sputum | H |
| B-22 | 06-3795 | 2006 | Arthroconidia | Bronchial wash | H |
| B-23 | 07-312 | 2007 | Arthroconidia, chlamydoconidia | Axillary and lymph nodes | A |
| B-24 | 06-3621 | 2006 | Arthroconidia | BAL | H |
| B-25 | 06-3536 | 2006 | Curved conidia, gold | BAL | H |
| B-26 | 06-3497 | 2006 | Sterile | BAL | H |
| B-27 | 06-3349 | 2006 | Arthroconidia | Lung | H |
| B-28 | 06-3341 | 2006 | Arthroconidia | BAL | H |
| B-29 | 06-3321 | 2006 | Curved conidia, gold | BAL | H |
| B-30 | 07-315 | 2007 | Sterile | Draining tract | A |
| B-31 | 06-3788 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-32 | 06-3787 | 2006 | Sterile, chlamydoconidia | Lung | H |
| B-33 | 06-3769 | 2006 | Arthroconidia | BAL | H |
| B-34 | 06-3768 | 2006 | Sterile | Lung | H |
| B-35 | 06-3499 | 2006 | Sterile | BAL | H |
| B-36 | 06-3466 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-37 | 06-3460 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-38 | 06-3335 | 2006 | Sterile | BAL | H |
| B-39 | 06-3320 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-40 | 05-1243 | 2005 | Arthroconidia | BAL | H |
| B-41 | 05-1416 | 2005 | Arthroconidia, crystal-encrusted hyphae | Sputum | H |
| B-42 | 05-2219 | 2005 | Curved conidia | BAL | H |
| B-43 | 05-2239 | 2005 | Arthroconidia, chlamydoconidia | BAL | H |
| B-44 | 05-2353 | 2005 | Spicules | Sinus | H |
| B-45 | 05-2587 | 2005 | Arthroconidia, chlamydoconidia | BAL | H |
| B-46 | 05-2369 | 2005 | Sterile | BAL | H |
| B-47 | 07-551 | 2007 | Sterile | BAL | H |
| B-48 | 07-495 | 2007 | Sterile, chlamydoconidia, gold-brown | BAL | H |
| B-49 | 05-567 | 2005 | Sterile, setal hyphae | Carapace | A |
| B-50 | 05-459 | 2005 | Curved conidia | Aortic graft tissue | H |
| B-51 | 05-597 | 2005 | Clamp connections, crystals | BAL | H |
| B-52 | 05-679 | 2005 | Sterile, chlamydoconidia | BAL | H |
| B-53 | 05-1037 | 2005 | Sterile, chlamydoconidia | BAL | H |
| B-54 | 05-1063 | 2005 | Sterile, chlamydoconidia, crystals | BAL | H |
| B-55 | 05-1422 | 2005 | Sterile, crystals | Tissue | A |
| B-56 | 05-1553 | 2005 | Curved conidia | BAL | H |
| B-57 | 05-1560 | 2005 | Sterile, crystals | BAL | H |
| B-58 | 05-1575 | 2005 | Sterile, chlamydoconidia, crystals | Right lower lung tissue | H |
| B-59 | 05-1822 | 2005 | Sterile | BAL | H |
| B-60 | 05-1853 | 2005 | Arthroconidia, chlamydoconidia, crystals | BAL | H |
| B-61 | 05-1932 | 2005 | Sterile | Lung tissue | H |
| B-62 | 05-2034 | 2005 | Sterile | Respiratory | A |
| B-63 | 05-2061 | 2005 | Arthroconidia, chlamydoconidia | BAL | H |
| B-64 | 05-2112 | 2005 | Arthroconidia | Sputum | H |
| B-65 | 05-2164 | 2005 | Sterile, crystals | Sputum | H |
| B-66 | 05-2269 | 2005 | Arthroconidia, clamp connections | BAL | H |
| B-67 | 05-2308 | 2005 | Sterile | Sputum | H |
| B-68 | 05-2341 | 2005 | Sterile | BAL | H |
| B-69 | 05-2354 | 2005 | Sterile | BAL | H |
| B-70 | 05-2474 | 2005 | Sterile, crystals | BAL | H |
| B-71 | 05-2504 | 2005 | Arthroconidia | Cranium | H |
| B-72 | 05-2586 | 2005 | Sterile, crystals, yellow refractile hyphae | BAL | H |
| B-73 | 05-2588 | 2005 | Sterile, crystals | BAL | H |
| B-74 | 05-2641 | 2005 | Sterile, chlamydoconidia | BAL | H |
| B-75 | 06-3310 | 2006 | Sterile | BAL | H |
| B-76 | 06-3308 | 2006 | Sterile | BAL | H |
| B-77 | 06-3298 | 2006 | Sterile, frequently septate hyphae | BAL | H |
| B-78 | 06-3297 | 2006 | Arthroconidia | Sputum | H |
| B-79 | 06-3281 | 2006 | Arthroconidia | Sinus | H |
| B-80 | 06-3259 | 2006 | Arthroconidia | BAL | H |
| B-81 | 06-3223 | 2006 | Arthroconidia | BAL | H |
| B-82 | 06-3212 | 2006 | Curved conidia | BAL | H |
| B-83 | 06-3194 | 2006 | Sterile | CSF | H |
| B-84 | 06-3190 | 2006 | Sterile, skelatoid hyphae | BAL | H |
| B-85 | 06-3183 | 2006 | Sterile | BAL | H |
| B-86 | 06-3182 | 2006 | Arthroconidia | Bronchial biopsy | H |
| B-87 | 06-3176 | 2006 | Sterile | BAL | H |
| B-88 | 06-3159 | 2006 | Sterile | BAL | H |
| B-89 | 06-3094 | 2006 | Sterile | Sputum | H |
| B-90 | 06-3093 | 2006 | Sterile | BAL | H |
| B-91 | 06-3082 | 2006 | Sterile, crystals, yellow-orange | BAL | H |
| B-92 | 06-3080 | 2006 | Sterile | BAL | H |
| B-93 | 05-2738 | 2005 | Sterile, chlamydoconidia, crystals, brown | Sputum | H |
| B-94 | 05-2742 | 2005 | Basidiospores, phototropic | Pleural fluid | H |
| B-95 | 05-2777 | 2005 | Arthroconidia, chlamydoconidia, crystals | Sputum | H |
| B-96 | 05-2954 | 2005 | Sterile, crystals | BAL | H |
| B-97 | 05-3058 | 2005 | Sterile | BAL | H |
| B-98 | 05-3255 | 2005 | Sterile, chlamydoconidia | BAL | H |
| B-99 | 05-3313 | 2005 | Arthroconidia | BAL | H |
| B-100 | 05-3368 | 2005 | Sterile | BAL | H |
| B-101 | 05-738 | 2005 | Sterile, refractile brown hyphae, crystals | Right wrist | H |
| B-102 | 05-2582 | 2005 | Sterile, setal hyphae | BAL | H |
| B-103 | 05-2585 | 2005 | Arthroconidia | BAL | H |
| B-104 | 07-729 | 2007 | Sterile, chlamydoconidia | Urine | H |
| B-105 | 07-793 | 2007 | Arthroconidia | BAL | H |
| B-106 | 07-797 | 2007 | Arthroconidia | Sputum | H |
| B-107 | 05-2661 | 2005 | Sterile, skelatoid hyphae, crystals | BAL | H |
| B-108 | 05-2677 | 2005 | Sterile, crystals, clamp connections | Sputum | H |
| B-109 | 05-3095 | 2005 | Arthroconidia, golden brown | BAL | H |
| B-110 | 05-3281 | 2005 | Arthroconidia | BAL | H |
| B-111 | 07-864 | 2007 | Arthroconidia | Lung wash | H |
| B-112 | 07-865 | 2007 | Arthroconidia | BAL | H |
| B-113 | 07-866 | 2007 | Arthroconidia | BAL | H |
| B-114 | 07-1061 | 2007 | Sterile, crystal-encrusted hyphae | BAL | H |
| B-115 | 07-1076 | 2007 | Arthroconidia | BAL | H |
| B-116 | 07-1092 | 2007 | Arthroconidia, chlamydoconidia | Sputum | H |
| B-117 | 07-1095 | 2007 | Arthroconidia | BAL | H |
| B-118 | 06-2442 | 2006 | Arthroconidia | BAL | H |
| B-119 | 06-2441 | 2006 | Produces arthroconidia | BAL | H |
| B-120 | 06-2439 | 2006 | Arthroconidia, chlamydoconidia | BAL | H |
| B-121 | 06-2433 | 2006 | Sterile | Sputum | H |
| B-122 | 06-2432 | 2006 | Sterile, achanthohyphidia | BAL | H |
| B-123 | 06-2422 | 2006 | Sterile, chlamydoconidia, crystals | BAL | H |
| B-124 | 06-2420 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-125 | 06-2401 | 2006 | Sterile | Sputum | H |
| B-126 | 06-2362 | 2006 | Sterile, chlamydoconidia | Sputum | H |
| B-127 | 06-2358 | 2006 | Sterile | BAL | H |
| B-128 | 06-2354 | 2006 | Sterile | BAL | H |
| B-129 | 06-2341 | 2006 | Sterile | BAL | H |
| B-130 | 06-2304 | 2006 | Sterile | BAL | H |
| B-131 | 06-2581 | 2006 | Arthroconidia, crystal-encrusted hyphae | Right foot | H |
| B-132 | 06-2571 | 2006 | Arthroconidia | Sputum | H |
| B-133 | 06-2563 | 2006 | Sterile | BAL | H |
| B-134 | 06-2544 | 2006 | Sterile | BAL | H |
| B-135 | 06-2552 | 2006 | Sterile | BAL | H |
| B-136 | 06-2544 | 2006 | Sterile | Unknown | H |
| B-137 | 06-2536 | 2006 | Arthroconidia, chlamydoconidia, conidia | Lung | H |
| B-138 | 06-2486 | 2006 | Sterile | BAL | H |
| B-139 | 06-2736 | 2006 | Curved conidia | BAL | H |
| B-140 | 06-2734 | 2006 | Sterile | BAL | H |
| B-141 | 06-2729 | 2006 | Curved conidia | BAL | H |
| B-142 | 06-2725 | 2006 | Arthroconidia | BAL | H |
| B-143 | 06-2723 | 2006 | Sterile | BAL | H |
| B-144 | 06-2721 | 2006 | Sterile | BAL | H |
| B-145 | 06-2687 | 2006 | Sterile | BAL | H |
| B-146 | 06-2685 | 2006 | Sterile, clamp connections | BAL | H |
| B-147 | 06-2683 | 2006 | Arthroconidia | BAL | H |
| B-148 | 06-2670 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-149 | 06-2650 | 2006 | Arthroconidia | Cornea | H |
| B-150 | 06-2644 | 2006 | Curved conidia | BAL | H |
| B-151 | 06-2641 | 2006 | Sterile, clamp connections | BAL | H |
| B-152 | 06-2629 | 2006 | Sterile | Cornea | H |
| B-153 | 06-2624 | 2006 | Sterile | BAL | H |
| B-154 | 06-3057 | 2006 | Arthroconidia | BAL | H |
| B-155 | 06-3035 | 2006 | Sterile | BAL | H |
| B-156 | 06-3002 | 2006 | Sterile | BAL | H |
| B-157 | 06-3001 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-158 | 06-2997 | 2006 | Arthroconidia | BAL | H |
| B-159 | 06-2951 | 2006 | Arthroconidia | BAL | H |
| B-160 | 06-2949 | 2006 | Sterile, chlamydoconidia, orange | BAL | H |
| B-161 | 06-2947 | 2006 | Arthroconidia | BAL | H |
| B-162 | 06-2939 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-163 | 06-2860 | 2006 | Sterile, chlamydoconidia | BAL | H |
| B-164 | 06-2839 | 2006 | Arthroconidia | BAL | H |
| B-165 | 06-2833 | 2006 | Sterile, chlamydoconidia | Sinus fluid | H |
| B-166 | 06-2807 | 2006 | Arthroconidia | BAL | H |
| B-167 | 07-1060 | 2007 | Sterile | Sputum | H |
| B-168 | 07-1074 | 2007 | Sterile, spicules | Left sinus | H |
Year accessioned into the Fungus Testing Laboratory culture collection.
Determined by growth on potato flakes agar at 25°C.
H, human source; A, animal source; O, other source (e.g., the environment).
BAL, bronchoalveolar lavage.
DNA preparation.
The isolates were again subcultured onto PDA and were grown for 24 h at 30°C. DNA was isolated from the hyphae by use of the Prepman Ultra reagent (Applied Biosystems, Foster City, CA), in which a small amount of material (enough to fill a loop) from each isolate was suspended in 50 μl of Prepman Ultra reagent in a 0.5-ml microcentrifuge tube. The suspension was initially vortexed for 45 s to 60 s to disperse the hyphal material and was then heated for 15 min at 100°C. The suspension was vortexed briefly and was then pelleted for 5 min at a maximum speed of 16,000 × g in a microcentrifuge. The supernatant was transferred to a new tube and stored on ice until the PCRs could be set up (within 1 h).
PCR.
PCR was performed with a 50-μl volume, which contained the following: 3 μl of template DNA, 5 μl 10× PCR buffer, 5 μl of a 10 μM stock solution of each primer (ITS-1 forward primer [32] and NL-4 reverse primer [17, 19]), 1.5 μl of 10 mM deoxynucleoside triphosphates (Invitrogen, Carlsbad, CA), and 5.0 U of Triplemaster Taq DNA polymerase (Eppendorf, Westbury, NY). The PCRs were performed in an Eppendorf master thermocycler and were run with a temperature profile of 2 min at 94°C, followed by 35 cycles of 20 s at 94°C, 20 s at 60°C, and 1 min at 72°C. The 35 cycles were followed by 5 min at 72°C. A 5-μl aliquot of each PCR product and a negative no-DNA control were run on a 0.7% agarose gel, stained with ethidium bromide, and documented with a DC 290 imaging system (Eastman Kodak Co., Rochester, NY) to confirm that amplification took place. The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA), and both strands were sequenced through the original ITS-1 and NL-4 PCR primer sites. The sequences were obtained as overlapping runs of the two flanking primers (primers ITS-1 and NL-4), as well as runs of two internal primers (primers ITS-4 and NL-1) (9, 17, 32). Sequencing was performed at the UTHSCSA Advanced Nucleic Acids Core Facility, and data were obtained with Sequencing Analysis Software (version 5.3.1; Applied Biosystems).
Sequence analysis.
The sequence data were assembled and analyzed by the use of MacVector software (MacVector, Inc., Cary, NC) and were then searched by using the ITS-1 and ITS-4 primer sequences to delineate the ITS region, as well as the NL-1 and NL-4 sequences to delineate the D1/D2 region. Each sequence was parsed into both the ITS and the D1/D2 regions and was then separately used to perform individual nucleotide-nucleotide searches with the BLASTn algorithm at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). The outputs from the BLAST searches were sorted on the basis of the maximum identity and were recorded as they appeared without modification of genus or species names that may have been synonyms or teleomorphs of other genus or species names in other GenBank records. Sequence-based identities with a cutoff of 97% or greater were considered significant in this study, and the best hit was defined as the sequence with the highest maximum identity to the query sequence.
RESULTS
Morphological basidiomycete identification.
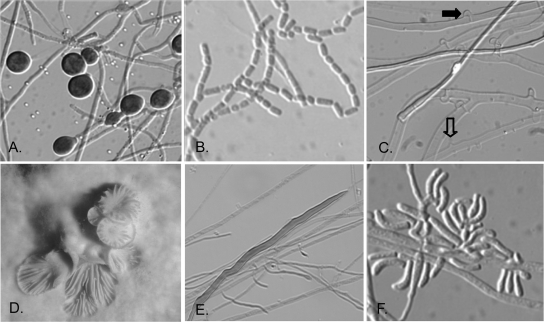
The isolates used in the study were identified as probable or presumptive basidiomycetes on the basis of their macroscopic, microscopic, and physiological features. Although a limited number of features of filamentous basidiomycetes are not diagnostic, they are suggestive for placement of the isolates in the phylum Basidiomycota. Growth is typically rapid, often up the side of the tube or plate; and colony colors are usually white, but they are sometimes cream to golden, orange, or slightly brownish on PDA. Microscopically, sterile basidiomycetes may display hyphae only or hyphae with chlamydoconidia (Fig. 1A). Some basidiomycetes do, however, produce conidia in culture. Most are arthroconidia, as seen in Fig. 1B, or compact clusters of arthroconidia, as seen for some Hormographiella species (anamorphs of some Coprinopsis [Coprinus] species). One of the more useful microscopic features for the identification of sterile isolates as basidiomycetes is the production of clamp connections, the defining characteristic for this phylum (Fig. 1C). Another important diagnostic feature of some sterile basidiomycetes is the production of spicules along the sides of hyphae (Fig. 1C, open arrow), with or without clamp connections (Fig. 1C, closed arrow), as in the case of Schizophyllum commune (Fig. 1C). Occasional dikaryons of this species may also produce basidiocarps (Fig. 1D). The recently reported species Inonotus (Phellinus) tropicalis (12, 15, 31), which is otherwise sterile in culture, may produce somewhat unusual hyphal elements known as setal hyphae (Fig. 1E); however, these types of hyphae may occur in other genera as well. Curved conidia, which are typical of Hormographiella species, may also be observed (Fig. 1F). The microscopic features of isolates included in this study are noted in Table 1. Finally, the ability of most basidiomycetes to grow on medium containing benomyl and their lack of growth on media containing cycloheximide further supported their probable identification.
FIG. 1.
Typical morphological features of basidiomycetes in culture. The morphological features that basidiomycetes may display in culture are shown. Microscopic features include chlamydoconidia (A), arthroconidia (B), spicules (open arrow) and clamp connections (solid arrow) of Schizophyllum commune (C), macroscopic basidiocarp of a dikaryotic Schizophyllum commune isolate (D), setal hyphae of Inonotus (Phellinus) tropicalis (E), and conidia of the Hormographiella anamorph of a Coprinus sp. (F). (A, B, C, E, F) Magnifications, ×880; (D) magnification, ×5.
A total of 168 filamentous isolates that had been identified as probable basidiomycetes by using the criteria cited above made up the study set that was sequenced.
Comparison of ITS and D1/D2 region BLAST results.
Comparison of the top hits from the GenBank database for the ITS and D1/D2 regions showed a number of isolates that returned the same species name for both the ITS region and the D1/D2 region (Table 2). However, when the number of disagreements was considered for the two regions, comparative ITS-D1/D2 sequencing for this set of isolates showed an overall striking lack of agreement. Although the BLASTn results for each isolate yielded a basidiomycete identification to the species level for 99.4% (167/168) of the isolates, the inconsistency of the outputs for the two regions made it impossible to assign a conclusive identification for 70.8% (119/168) of the isolates (Table 3). At the least-stringent level, in which agreement between the two sequences needed to consist only of the same genus name, regardless of the percent identity (i.e., the ITS sequence identified Phlebia tremellosa with 95% identity; the D1/D2 sequence identified Phlebia radiata with 96% identity), only 48.8% (82/168) of the results were in agreement (Table 4). For genus and species agreement, regardless of the percent identity (i.e., the ITS sequence identified Phlebia tremellosa with 97% identity; the D1/D2 sequence identified Phlebia tremellosa with 93% identity), the results for only 28.6% (48/168) of the specimens agreed. For genus and species agreement with a cutoff of ≥97% identity, the results for only 21.4% (36/168) of the specimens agreed. Further analysis showed that of the 168 sequences, the sequence of only a single isolate (0.6%) displayed matching ITS region- and D1/D2 region-based genus and species names with 100% identity.
TABLE 2.
Comparison of GenBank top hits for the ITS and D1/D2 regions which agreea
| Isolate | Organism ITS identified | % ITS identity | No. of ITS matches/no. identified in GenBank | Organism D1/D2 identified | % D1/D2 identity | No. of D1/D2 matches/no. identified in GenBank |
|---|---|---|---|---|---|---|
| 06-4444 | Bjerkandera adusta | 99 | 698/705 | Bjerkandera adusta | 100 | 624/624 |
| 06-3787 | Bjerkandera adusta | 99 | 692/695 | Bjerkandera adusta | 100 | 583/583 |
| 05-1243 | Bjerkandera adusta | 100 | 583/583 | Bjerkandera adusta | 99 | 889/895 |
| 05-3095 | Bjerkandera adusta | 99 | 598/605 | Bjerkandera adusta | 100 | 624/624 |
| 05-1853 | Ceriporiopsis subvermispora | 97 | 751/774 | Ceriporiopsis subvermispora | 95 | 619/645 |
| 05-2504 | Fomitopsis feei | 99 | 646/647 | Fomitopsis feei | 99 | 646/647 |
| 06-3335 | Fomitopsis rosea | 98 | 638/647 | Fomitopsis rosea | 98 | 638/647 |
| 06-3906 | Irpex lacteus | 99 | 660/663 | Irpex lacteus | 100 | 560/560 |
| 07-312 | Mycorrhizal basidiomycete | 99 | 622/626 | Mycorrhizal basidiomycete | 100 | 646/646 |
| 05-597 | Oxyporus corticola | 100 | 605/605 | Oxyporus corticola | 99 | 647/648 |
| 05-1822 | Oxyporus corticola | 98 | 389/395 | Oxyporus corticola | 94 | 844/894 |
| 06-3281 | Peniophora cinerea | 96 | 693/716 | Peniophora cinerea | 98 | 590/602 |
| 06-3093 | Peniophora cinerea | 95 | 605/641 | Peniophora cinerea | 98 | 591/602 |
| 07-1076 | Peniophora cinerea | 95 | 505/541 | Peniophora cinerea | 98 | 591/602 |
| 06-2439 | Peniophora cinerea | 97 | 625/641 | Peniophora cinerea | 99 | 591/602 |
| 06-2581 | Peniophora cinerea | 96 | 617/641 | Peniophora cinerea | 97 | 591/602 |
| 06-2670 | Peniophora cinerea | 98 | 735/741 | Peniophora cinerea | 97 | 591/602 |
| 06-3035 | Peniophora cinerea | 91 | 775/841 | Peniophora cinerea | 97 | 587/602 |
| 05-1560 | Phlebia acerina | 92 | 756/785 | Phlebia acerina | 99 | 640/646 |
| 06-3159 | Phlebia radiata | 93 | 711/767 | Phlebia radiata | 99 | 646/647 |
| 06-3082 | Phlebia radiata | 88 | 629/766 | Phlebia radiata | 97 | 632/650 |
| 07-56 | Phlebia tremellosa | 97 | 646/664 | Phlebia tremellosa | 99 | 620/623 |
| 07-31 | Phlebia tremellosa | 99 | 620/623 | Phlebia tremellosa | 99 | 634/640 |
| 06-4410 | Phlebia tremellosa | 100 | 498/498 | Phlebia tremellosa | 100 | 603/603 |
| 06-4285 | Phlebia tremellosa | 98 | 644/651 | Phlebia tremellosa | 100 | 603/603 |
| 07-315 | Phlebia tremellosa | 98 | 585/592 | Phlebia tremellosa | 100 | 633/633 |
| 05-738 | Phlebia tremellosa | 98 | 585/592 | Phlebia tremellosa | 100 | 633/633 |
| 06-2422 | Phlebia tremellosa | 98 | 646/664 | Phlebia tremellosa | 99 | 620/623 |
| 06-2486 | Phlebia tremellosa | 96 | 646/664 | Phlebia tremellosa | 99 | 620/623 |
| 06-2644 | Phlebia tremellosa | 97 | 646/664 | Phlebia tremellosa | 93 | 575/623 |
| 06-3806 | Polyporus tricholoma | 98 | 647/655 | Polyporus tricholoma | 100 | 611/611 |
| 06-2860 | Psathyrella cf. gracilis | 98 | 694/706 | Psathyrella cf. gracilis | 99 | 644/646 |
| 06-3176 | Schizophyllum radiatum | 100 | 616/616 | Schizophyllum radiatum | 99 | 909/912 |
| 06-4124 | Termitomyces albuminosus | 99 | 638/639 | Termitomyces albuminosus | 99 | 622/623 |
| 05-1553 | Termitomyces albuminosus | 99 | 700/702 | Termitomyces albuminosus | 99 | 622/623 |
| 05-2641 | Termitomyces albuminosus | 99 | 700/702 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3310 | Termitomyces albuminosus | 99 | 745/746 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3259 | Termitomyces albuminosus | 99 | 745/746 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3212 | Termitomyces albuminosus | 99 | 705/706 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3194 | Termitomyces albuminosus | 99 | 732/733 | Termitomyces albuminosus | 99 | 621/623 |
| 06-3183 | Termitomyces albuminosus | 99 | 926/928 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3080 | Termitomyces albuminosus | 99 | 705/706 | Termitomyces albuminosus | 99 | 622/623 |
| 05-3255 | Termitomyces albuminosus | 99 | 705/706 | Termitomyces albuminosus | 99 | 622/623 |
| 05-2677 | Termitomyces albuminosus | 99 | 745/746 | Termitomyces albuminosus | 99 | 622/623 |
| 06-2433 | Termitomyces albuminosus | 99 | 645/646 | Termitomyces albuminosus | 99 | 622/623 |
| 06-2571 | Termitomyces albuminosus | 99 | 745/746 | Termitomyces albuminosus | 98 | 622/623 |
| 06-2650 | Termitomyces albuminosus | 95 | 712/746 | Termitomyces albuminosus | 98 | 622/623 |
| 07-1074 | Termitomyces albuminosus | 98 | 745/746 | Termitomyces albuminosus | 97 | 615/623 |
| 05-2354 | Trametes versicolor | 99 | 759/763 | Trametes versicolor | 99 | 645-646 |
Differences in sequence matches between multiple isolates of the same species and what was returned by BLAST reflect the different percent identities of multiple GenBank records for the same species, one of which had the closest identity to our sequence but which could differ with each search. The table was sorted alphabetically.
TABLE 3.
Comparison of GenBank top hits for the ITS and D1/D2 regions which disagreea
| Isolate | Organism ITS identified | % ITS identity | No. of ITS matches/no. identified in GenBank | Organism D1/D2 identified | % D1/D2 identity | No. of D1/D2 matches/no. identified in GenBank |
|---|---|---|---|---|---|---|
| 07-551 | Antrodia albida | 99 | 443/446 | Fomes fomentarius | 98 | 636/646 |
| 06-3321 | Antrodia malicola | 96 | 348/360 | Bjerkandera adusta | 95 | 1136/1194 |
| 06-2304 | Antrodia malicola | 96 | 448/460 | Bjerhandera adusta | 95 | 736/794 |
| 06-2544 | Antrodia malicola | 96 | 448/460 | Bjerhandera adusta | 97 | 745/794 |
| 06-4454 | Bjerkandera adusta | 98 | 694/706 | Antrodia malicola | 99 | 644/646 |
| 06-4450 | Bjerkandera adusta | 96 | 604/627 | Antrodia malicola | 100 | 644/644 |
| 06-4161 | Bjerkandera adusta | 98 | 675/685 | Antrodia malicola | 100 | 646/646 |
| 06-3795 | Bjerkandera adusta | 100 | 583/583 | Thanatephorus cucumeris | 99 | 591/596 |
| 06-3769 | Bjerkandera adusta | 92 | 630/668 | Oudemansiella canarii | 92 | 630/670 |
| 06-2441 | Bjerkandera adusta | 98 | 894/906 | Antrodia malicola | 99 | 644/646 |
| 06-2552 | Bjerkandera adusta | 98 | 690/706 | Antrodia malicola | 99 | 644/646 |
| 06-2725 | Bjerkandera adusta | 97 | 690/706 | Antrodia malicola | 99 | 644/646 |
| 06-2683 | Bjerkandera adusta | 100 | 583/583 | Thanatephorus cucumeris | 98 | 591/596 |
| 06-3002 | Bjerkandera adusta | 97 | 604/627 | Antrodia malicola | 100 | 644/644 |
| 06-3001 | Bjerkandera adusta | 100 | 583/583 | Thanatephorus cucumeris | 99 | 591/596 |
| 06-2939 | Bjerkandera adusta | 96 | 604/627 | Antrodia malicola | 100 | 644/644 |
| 05-2954 | Coprinopsis cinera | 100 | 626/626 | Coprinopsis domesticus | 99 | 581/583 |
| 07-865 | Coprinopsis cinera | 100 | 626/626 | Coprinopsis domesticus | 99 | 581/583 |
| 05-567 | Coprinopsis cinerea | 100 | 639/639 | Coprinus trisporus | 99 | 613/614 |
| 06-3970 | Coprinus echinosporus | 93 | 473/504 | Coprinus trisporus | 100 | 612/612 |
| 06-2354 | Coprinus echinosporus | 97 | 473/504 | Coprinus trisporus | 100 | 612/612 |
| 06-2687 | Coprinus echinosporus | 95 | 473/504 | Coprinus trisporus | 100 | 612/612 |
| 06-2949 | Coprinus echinosporus | 93 | 473/504 | Coprinus trisporus | 100 | 612/612 |
| 06-3497 | Coprinus radians | 100 | 658/658 | Phlebia chrysocreas | 95 | 582/608 |
| 06-3341 | Coprinus radians | 100 | 658/658 | Phlebia chrysocreas | 95 | 582/608 |
| 05-1063 | Coprinus radians | 100 | 637/637 | Ceriporiopsis subvermispora | 99 | 622/624 |
| 05-1575 | Coriolopsis caperata | 96 | 651/673 | Microporus affinis | 98 | 638/648 |
| 05-459 | Fomes fomentarius | 97 | 751/774 | Microporus affinis | 98 | 636/646 |
| 06-2401 | Fomes fomentarius | 97 | 751/774 | Microporus affinis | 98 | 636/646 |
| 06-2563 | Fomes fomentarius | 96 | 751/774 | Microporus affinis | 98 | 636/646 |
| 06-3768 | Fomitopsis pinicola | 95 | 736/794 | Fomitopsis feei | 99 | 589/590 |
| 07-495 | Hymenochaete spreta | 98 | 699/707 | Hydnochaete olivacea | 99 | 654/657 |
| 06-3994 | Irpex lacteus | 99 | 660/663 | Phlebia tremellosa | 98 | 630/644 |
| 06-3888 | Irpex lacteus | 99 | 622/625 | Antrodia malicola | 99 | 633/635 |
| 06-3788 | Irpex lacteus | 93 | 450/468 | Trametes maxima | 93 | 450/468 |
| 06-3536 | Oudemansiella canarii | 97 | 429/437 | Polyporus brumalis | 100 | 620/620 |
| 07-1092 | Oudemansiella canarii | 97 | 329/337 | Polyporus brumalis | 100 | 620/620 |
| 06-2341 | Oudemansiella canarii | 98 | 431/437 | Polyporus brumalis | 100 | 620/620 |
| 06-2721 | Oudemansiella canarii | 97 | 329/337 | Polyporus brumalis | 100 | 620/620 |
| 05-2369 | Oxyporus corticola | 93 | 819/874 | Panus strigellus | 97 | 751/775 |
| 06-2629 | Peniophora cinerea | 93 | 701/779 | Phlebia tremellosa | 98 | 591/596 |
| 06-3869 | Phanerochaete carnosa | 93 | 450/468 | Phanerochaete velutina | 92 | 557/603 |
| 06-3499 | Phanerochaete velutina | 98 | 636/646 | Phanerochaete sordida | 98 | 735/747 |
| 05-2219 | Phlebia acerina | 93 | 719/776 | Phlebia radiata | 96 | 625/647 |
| 05-2777 | Phlebia acerina | 96 | 503/526 | Phlebia radiata | 95 | 620/648 |
| 05-2582 | Phlebia acerina | 96 | 503/526 | Phlebia radiata | 95 | 620/648 |
| 05-2742 | Phlebia lilascens | 94 | 585/616 | Coprinellus disseminatus | 100 | 621/621 |
| 05-3058 | Phlebia lilascens | 99 | 667/673 | Coprinopsis domesticus | 100 | 621/621 |
| 05-2353 | Phlebia radiata | 92 | 770/829 | Schizophyllum radiatum | 96 | 626/646 |
| 05-2587 | Phlebia radiata | 94 | 844/894 | Hymenochaete spreta | 95 | 850/894 |
| 07-797 | Phlebia radiata | 93 | 711/767 | Phlebia uda | 99 | 646/647 |
| 07-864 | Phlebia radiata | 93 | 511/567 | Phlebia uda | 99 | 646/647 |
| 06-2723 | Phlebia radiata | 93 | 711/767 | Polyporus brumalis | 99 | 646/647 |
| 06-2997 | Phlebia radiata | 93 | 811/867 | Polyporus brumalis | 99 | 646/647 |
| 06-3460 | Phlebia subserialis | 99 | 578/579 | Phlebia chrysocreas | 95 | 582/608 |
| 05-2308 | Phlebia subserialis | 99 | 554/556 | Phlebia chrysocreas | 95 | 619/645 |
| 05-2474 | Phlebia subserialis | 99 | 563/564 | Phlebia chrysocreas | 95 | 619/645 |
| 06-3223 | Phlebia uda | 93 | 662/702 | Schizophyllum commune | 100 | 613/613 |
| 05-2738 | Phlebia uda | 93 | 662/702 | Phlebia subochracea | 94 | 699/751 |
| 07-729 | Phlebia uda | 93 | 662/702 | Termitomyces albuminosus | 99 | 622/623 |
| 07-1095 | Phlebia uda | 93 | 662/702 | Phanerochaete velutina | 99 | 635/636 |
| 06-2358 | Phlebia uda | 93 | 662/702 | Bjerkandera adusta | 99 | 642/646 |
| 06-2624 | Phlebia uda | 93 | 662/702 | Coprinus quadrifidus | 98 | 636/647 |
| 06-4057 | Polyporus brumalis | 98 | 633/640 | Termitomyces albuminosus | 99 | 622/623 |
| 06-3349 | Polyporus brumalis | 97 | 627/651 | Lentinus bertieri | 99 | 611/612 |
| 05-2588 | Polyporus brumalis | 97 | 647/664 | Lentinus bertieri | 99 | 611/613 |
| 05-1932 | Rhizochaete filamentosa | 93 | 617/679 | Phlebiopsis gigantea | 96 | 625/648 |
| 05-2586 | Rhizochaete filamentosa | 94 | 693/711 | Phlebiopsis gigantea | 96 | 628/650 |
| 06-3298 | Rhizochaete filamentosa | 93 | 592/652 | Phlebiopsis gigantea | 96 | 628/650 |
| 06-3297 | Rhizochaete filamentosa | 92 | 677/749 | Phanerochaete velutina | 99 | 639/645 |
| 06-3094 | Rhizochaete filamentosa | 93 | 692/752 | Phlebiopsis gigantea | 96 | 628/650 |
| 05-2061 | Rhizochaete fouquieriae | 95 | 701/779 | Trametes versicolor | 96 | 625/648 |
| 05-2164 | Rhizochaete fouquieriae | 98 | 389/395 | Panus strigellus | 99 | 611/612 |
| 05-1416 | Schizophyllum commune | 94 | 644/694 | Hymenochaete spreta | 96 | 850/894 |
| 05-2239 | Schizophyllum commune | 99 | 634/637 | Schizophyllum radiatum | 100 | 613/613 |
| 05-1442 | Schizophyllum commune | 93 | 917/979 | Polyporus brumalis | 97 | 751/774 |
| 06-2442 | Schizophyllum commune | 94 | 544/594 | Hymenochaete spreta | 96 | 850/894 |
| 06-2432 | Schizophyllum commune | 95 | 501/579 | Trametes versicolor | 96 | 625/648 |
| 06-2420 | Schizophyllum commune | 94 | 644/694 | Hymenochaete spreta | 96 | 850/894 |
| 06-2729 | Schizophyllum commune | 95 | 701/779 | Trametes versicolor | 97 | 625/648 |
| 06-2641 | Schizophyllum commune | 95 | 731/779 | Trametes versicolor | 96 | 625/648 |
| 06-2807 | Schizophyllum commune | 94 | 644/694 | Hymenochaete spreta | 96 | 850/894 |
| 06-3190 | Schizophyllum radiatum | 99 | 625/626 | Termitomyces albuminosus | 99 | 625/626 |
| 06-3182 | Schizophyllum radiatum | 92 | 892/960 | Schizophyllum commune | 91 | 711/899 |
| 07-1061 | Schizophyllum radiatum | 99 | 625/626 | Termitomyces albuminosus | 99 | 625/626 |
| 06-2544 | Schizophyllum radiatum | 99 | 625/626 | Termitomyces albuminosus | 99 | 625/626 |
| 07-1060 | Schizophyllum radiatum | 99 | 625/626 | Termitomyces albuminosus | 99 | 625/626 |
| 06-4137 | Termitomyces albuminosus | 99 | 609/610 | Phlebia tremellosa | 99 | 634/640 |
| 06-3466 | Termitomyces albuminosus | 99 | 700/702 | Phanerochaete sordida | 98 | 735/747 |
| 05-1037 | Termitomyces albuminosus | 97 | 751/774 | Ceriporiopsis subvermispora | 98 | 616/623 |
| 05-2112 | Termitomyces albuminosus | 99 | 609/614 | Trametes versicolor | 99 | 642/645 |
| 05-2269 | Termitomyces albuminosus | 96 | 765/789 | Ceriporiopsis subvermispora | 99 | 616/623 |
| 06-2736 | Termitomyces albuminosus | 99 | 600/602 | Phanerochaete sordida | 97 | 735/747 |
| 06-3057 | Termitomyces albuminosus | 99 | 700/702 | Phanerochaete sordida | 98 | 735/747 |
| 06-3924 | Thanatephorus cucumeris | 99 | 624/626 | Antrodia malicola | 99 | 644/646 |
| 06-3821 | Thanatephorus cucumeris | 99 | 624/626 | Bjerkandera adusta | 100 | 583/583 |
| 06-4341 | Trametes maxima | 96 | 781/813 | Donkioporia expansa | 97 | 628/645 |
| 06-4103 | Trametes maxima | 95 | 638/682 | Polyporus brumalis | 100 | 620/620 |
| 06-3621 | Trametes maxima | 94 | 517/575 | Polyporus brumalis | 100 | 620/620 |
| 06-3308 | Trametes maxima | 94 | 635/650 | Polyporus tricholoma | 97 | 621/622 |
| 05-3281 | Trametes maxima | 95 | 638/682 | Polyporus brumalis | 100 | 620/620 |
| 07-866 | Trametes maxima | 95 | 638/682 | Polyporus brumalis | 100 | 620/620 |
| 06-2734 | Trametes maxima | 95 | 638/682 | Polyporus brumalis | 100 | 620/620 |
| 06-2951 | Trametes maxima | 95 | 781/813 | Donkioporia expansa | 98 | 628/645 |
| 06-2947 | Trametes maxima | 95 | 838/882 | Polyporus brumalis | 100 | 620/620 |
| 06-2833 | Trametes maxima | 96 | 781/813 | Donkioporia expansa | 97 | 628/645 |
| 05-2034 | Trametes ochracea | 97 | 615/636 | Trametes hirsuta | 99 | 644/646 |
| 05-2341 | Trametes ochracea | 97 | 751/774 | Trametes versicolor | 96 | 625/648 |
| 06-3320 | Trametes versicolor | 94 | 554/569 | Bjerkandera adusta | 100 | 583/583 |
| 05-679 | Trametes versicolor | 94 | 844/894 | Trametes lactinea | 100 | 613/613 |
| 05-3313 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 05-3368 | Trametes versicolor | 94 | 630/681 | Trametes lactinea | 100 | 611/611 |
| 05-2585 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 07-793 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 05-2661 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 06-2362 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 06-2536 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 06-2685 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
| 06-2839 | Trametes versicolor | 93 | 750/798 | Trametes lactinea | 100 | 613/613 |
Differences in sequence matches between multiple isolates of the same species and what was returned by BLAST reflect the different percent identities of multiple GenBank records for the same species, one of which had the closest identity to our sequence but which could differ with each search. The table was sorted alphabetically on the basis of the ITS name.
TABLE 4.
ITS-D1/D2 BLAST output comparison
| % Identity cutoff | No.a | % Agreement |
|---|---|---|
| Genus only agreementb | 82 | 48.8 |
| Genus + species agreement,c any % identity | 48 | 28.6 |
| Genus + species agreement, one ≥97% identity | 48 | 28.6 |
| Genus + species agreement, both ≥97% identity | 36 | 21.4 |
| Genus + species agreement, one ≥98% identity | 43 | 25.6 |
| Genus + species agreement, both ≥98% identity | 32 | 19 |
| Genus + species agreement, one ≥99% identity | 36 | 21.4 |
| Genus + species agreement, both ≥99% identity | 24 | 14.3 |
| Genus + species agreement, one 100% identity | 12 | 7.1 |
| Genus + species agreement, both 100% identity | 1 | 0.6 |
Number of isolates of 168 isolates tested with given identity.
Any percent identity.
Genus plus species agreement represents BLAST outputs in which the ITS genus and species name matched the D1/D2 genus and species name.
Comparison of ITS and D1/D2 GenBank deposits.
In order to investigate possible causes for the low frequency of agreement for the ITS and D1/D2 regions, the GenBank database was searched for the presence of sequence deposits that corresponded to these two sequences for each species. This analysis revealed that there were no entries in GenBank for 14% of the top ITS hits (7/50) and 16% (8/50) of the top D1/D2 hits for the isolates on our list (Table 5). Therefore, 30% of the species that were identified in this study had either an ITS or a D1/D2 sequence that matched a deposit in GenBank, but not both.
TABLE 5.
Presence of species-specific GenBank ITS and D1/D2 deposits
| Species | GenBank recorda |
|
|---|---|---|
| ITS region | D1/D2 region | |
| Antrodia albida | X (complete) | X (partial) |
| Antrodia malicola | X (complete) | X (partial) |
| Bjerkandera adusta | X (partial) | X (partial) |
| Ceriporiopsis subvermispora | X (partial) | X (partial) |
| Coprinellus disseminatus | X (partial) | X (partial) |
| Coprinopsis cinera | X (partial) | No deposit |
| Coprinopsis domesticus | No deposit | X (partial) |
| Coprinus echinosporus | X (partial) | No deposit |
| Coprinus quadrifidus | X (partial) | X (partial) |
| Corpinus radians | X (partial) | X (partial) |
| Corpinus trisporus | No deposit | X (partial) |
| Coriolopsis caperata | X (partial) | X (partial) |
| Donkiopora expansa | X (partial) | X (partial) |
| Fomes fomentarius | X (partial) | X (partial) |
| Fomitopsis feei | X (partial) | X (partial) |
| Fomitopsis pinicola | X (partial) | X (partial) |
| Fomitopsis rosea | X (complete) | X (partial) |
| Hydnochaete olivacea | X (partial) | No deposit |
| Hymenochaete spreta | X (partial) | X (partial) |
| Irpex lacteus | X (partial) | X (partial) |
| Lentinus bertieri | No deposit | X (partial) |
| Microporus affinis | No deposit | X (partial) |
| Oudemansiella canarii | X (partial) | No deposit |
| Oxyporus corticola | X (partial) | X (partial) |
| Panus strigellus | No deposit | X (partial) |
| Peniophora cinerea | X (partial) | X (partial) |
| Phanerochaete carnosa | X (partial) | No deposit |
| Phanerochaete sordida | X (partial) | X (partial) |
| Phanerochaete velutina | X (partial) | X (partial) |
| Phlebia acerina | X (partial) | X (partial) |
| Phlebia chrysocreas | No deposit | X (complete) |
| Phlebia lilascens | No deposit | X (partial) |
| Phlebia radiata | X (partial) | X (partial) |
| Phlebia subochracea | X (partial) | X (partial) |
| Phlebia subserialis | X (partial) | X (partial) |
| Phlebia tremellosa | X (partial) | X (partial) |
| Phlebia uda | X (partial) | X (complete) |
| Phlebiopsis gigantea | X (partial) | X (partial) |
| Polyporus brumalis | X (partial) | X (complete) |
| Polyporus tricholoma | X (partial) | X (partial) |
| Rhizochaete filamentosa | X (partial) | X (complete) |
| Rhizochaete fouquieriae | X (partial) | No deposit |
| Schizophyllum commune | X (partial) | X (partial) |
| Schizophyllum radiatum | X (partial) | X (complete) |
| Termitomyces albuminosus | X (complete) | No deposit |
| Thanatephorus cucumeris | X (partial) | No deposit |
| Trametes lactinea | X (partial) | X (partial) |
| Trametes maxima | X (partial) | X (partial) |
| Trametes ochracea | X (partial) | X (partial) |
| Trametes versicolor | X (partial) | X (partial) |
X, a sequence deposit was made for this species. No deposit, no GenBank record could be found for the corresponding sequence; partial and complete, the sequence length between the ITS-1 and ITS-4 primers (ITS region) or the NL-1 and NL-4 primers (D1/D2 region).
Analysis of ITS and D1/D2 GenBank deposit sequence lengths.
The top hits for each BLAST query for both the ITS and the D1/D2 regions with an identity of 97% or greater were evaluated for their completeness, which was defined as a sequence whose length matched the length of our query sequence, excluding internal deletions or insertions. Comparison of GenBank deposit sequence lengths of both the ITS and the D1/D2 regions for each isolate showed that the deposit entries for each region were largely truncated compared to the lengths of the regions that we sequenced. Of the 50 species represented, only 8% of the ITS regions and 10% of the D1/D2 regions had complete sequence data in GenBank (Table 5).
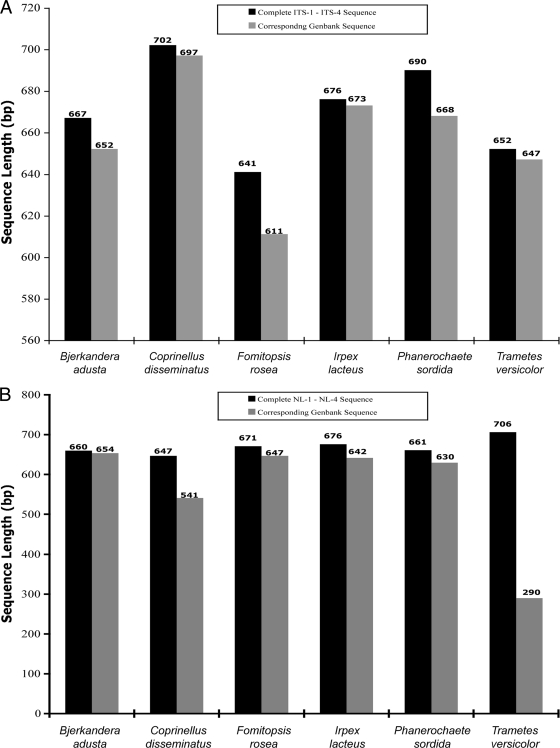
Since most of the species that we identified are rare and in many cases had only a single GenBank deposit, we selected six species that were the most redundant from the BLAST output list (Table 2) and obtained the sequence lengths from each record with the highest percent identity. Sequences were recovered for Bjerkandera adusta (GenBank accession number EU918694), Coprinellus disseminatus (GenBank accession number FN386275), Fomitopsis rosea (GenBank accession number DQ491412), Irpex lacteus (GenBank accession number FJ462768.1), Phanerochaete sordida (GenBank accession number EU118653.1), and Trametes versicolor (GenBank accession number FJ810146). The ITS sequence analysis (Fig. 2A) showed that regardless of the species, there were no complete sequences compared to our ITS sequences. The most complete sequence of Coprinellus disseminatus found in GenBank was 697 bp (GenBank accession number FN386275), whereas the ITS sequence that we obtained was 702 bp. The other GenBank ITS sequences varied in length and were found to be incomplete as well. The ITS sequence lengths obtained from GenBank ranged from 95% to 99% compared to the complete ITS sequences that we derived by sequencing with primers ITS-1 and ITS-4. The D1/D2 sequence length data (Fig. 2B) also proved to be largely incomplete. The most complete sequence of Bjerkandera adusta found in the GenBank database was 654 bp (GenBank accession number AB096738), whereas the D1/D2 sequence that we obtained was 660 bp. The D1/D2 sequence lengths obtained from GenBank ranged from 41% to 99% complete compared to the complete D1/D2 sequences that we obtained by sequencing with primers NL-1 and NL-4. These data indicate that many of the current GenBank sequences for basidiomycetes have incomplete sequence data for the regions that we used for identification. Importantly, these results were obtained for the most redundant species recovered from our BLAST searches and therefore would be expected to have a higher likelihood for a complete deposit due to the presence of multiple records.
FIG. 2.
Comparison of ITS and D1/D2 sequence lengths. (A) Comparison of ITS lengths to ITS lengths in GenBank. The sequence lengths of the ITS regions of our isolates were compared to those found in GenBank. The six species represented here were chosen on the basis of being the most redundant among the results from the BLASTn search. (B) Comparison of D1/D2 lengths to D1/D2 lengths in GenBank. The sequence lengths of the D1/D2 regions of our isolates were compared to those found in GenBank.
DISCUSSION
The frequency of human mycoses due to filamentous fungi is steadily increasing, and mycoses mostly affect defined risk groups, such as immunocompromised or severely ill patients. In addition to the well-known opportunistic basidiomycetous pathogen Cryptococcus neoformans, other basidiomycetous yeasts such as Malassezia spp. and Rhodotorula spp. are now considered emergent opportunistic pathogens and are recovered at increasing frequencies (6, 14, 20, 22). Basidiomycetous molds, with few exceptions, are rarely recovered as human pathogens because of the difficulty identifying these fungi or the difficulty distinguishing colonizers from invasive isolates in patient specimens. Sterile and/or arthroconidium-forming basidiomycetes are a subset of this class and cannot be conclusively identified by standard phenotypic methods because they do not produce distinguishing structures. Although these mostly sterile isolates may be morphologically identified as basidiomycetes when clamp connections are present, many genera of basidiomycetes do not produce clamp connections in culture. Consequently, they may simply be described as nonsporulating molds with unknown clinical significance.
In a study by Pounder et al., which also used a sequencing strategy for identification, 31 of the 48 (65%) isolates were classified as basidiomycetes (23) by use of a sequence derived from the ITS region. Under the cutoff criteria of a sequence length of at least 400 bp, ≥99% identity for a species-level identification, and ≥93% identity for a genus-level identification, 92% of the isolates were identified to the genus level and 79% were identified to the species level. Because of the relatively high identification rate, we decided to use a similar strategy to identify our isolates. A large number of the initial ITS sequences that we obtained did not meet the 97% cutoff criterion that we established for identity. Therefore, we decided to add the D1/D2 region as a second locus, under the assumption that the results from D1/D2 searches would yield more identities higher than 97%, thereby allowing an identification. However, we were surprised to find that while in many cases we obtained a D1/D2 identity of ≥97%, we observed a striking amount of disagreement between the best hit (the highest level of identity) for the ITS search and the best hit for the D1/D2 search. The agreement between the two sequences for the same isolate was only 28.6% at any level of identity, whereas with a more stringent cutoff of ≥97% identity, agreement occurred only 21.4% of the time. We suspect that this low level of agreement would likely be the same for any mold that is rarely studied at the molecular level, whether it is sterile or not, due to the absence of searchable data in GenBank.
The low level of ITS-D1/D2 agreement led us to investigate why the results were so disparate. Of the 50 species that we identified, almost a third did not have a GenBank deposit for the ITS region or the D1/D2 region. When all significant hits (≥97% identity) were considered for each search output, two-thirds (66%) of the records had either an ITS deposit or a D1/D2 deposit, but not both. As a result of this discrepancy, error can be introduced during the BLAST search output when the next-highest identity, which will be a different species, becomes the top hit in the search. We also found that the sequences in GenBank were largely incomplete compared to our query sequences. It is not clear how much sequence would need to be truncated from either or both ends before a significant impact on identity occurs; however, sequence alignments demonstrate that sequence variation can occur very close (within a few bases) to the primer sites that we used (data not shown). These variable regions may not be present in the sequence if the sequence is truncated due to single-stranded sequencing, if the sequence is derived from a different primer combination or a partially overlapping region, or if the sequencing run terminates and does not proceed through the primers. These observations, combined with known GenBank issues such as nomenclature errors (5) and poor-quality deposits (18, 23), can complicate sequence-based identification. In fact, fungal GenBank deposits may be more adversely affected by issues involving nomenclature than GenBank deposits for other microbial organisms. Few investigators working with fungi outside of classical mycology are well versed in the rules governing how and when the anamorph and teleomorph nomenclature is properly used. Similarly, isolates may be identified by their obsolete or synonymous names, and selection of the currently accepted name is difficult even for classical mycologists, since names are often changed on the basis of basic research, including some of the molecular techniques used in this study, and may not be widely reported or even accepted. The sequences of basidiomycetes in the GenBank database, with the possible exception of those of C. neoformans, may be more adversely affected by these issues, since taxonomic studies of basidiomycete species pathogenic for humans may be lagging similar studies of other fungi, such as the aspergilli or the fusaria, for which detailed analysis has resulted in revised classifications (2, 3). These issues were not addressed in the data analysis, since the study focused on the actual GenBank outputs; consequently, it is possible that the levels of agreement would improve slightly due to a correct agreement being masked by the erroneous or inconsistent naming of the deposit.
Our results and the results of Pounder et al. (23) suggest that sterile molds recovered from human clinical specimens may comprise a substantial number of basidiomycetes. In fact, our study utilized a subset of sterile and/or arthroconidium-producing isolates from human clinical specimens phenotypically identified as probable basidiomycetes (on the basis of the morphological criteria that we used for our study) that had been sent to the Fungus Testing Laboratory. Both studies had six species in common, including Polyporus tricholoma, Irpex lacteus, Schizophyllum commune, Phlebia subserialis, Trametes versicolor, and Thanatephorus cucumeris. While it is highly likely that most filamentous basidiomycetes identified from clinical specimens are clinically insignificant because they are noncolonizers abundant in ambient air, a number of our specimens were from sites other than the respiratory tract that are normally sterile (i.e., cerebrospinal fluid). The host status, the route of infection, and the shear number and variety of fungal elements that a patient is exposed to likely determine whether a basidiomycosis can occur.
While this study has highlighted issues that need to be carefully considered when sequence-based identification is employed, sequence-based identification has some major diagnostic strengths and continues to be extremely useful to our group for fungal identification. It clearly has great diagnostic value for common fungi and/or fungi that have numerous GenBank deposits. The sequence data in GenBank are also useful if they are combined with additional nonsequence data, even if the sequencing results are somewhat ambiguous. In fact, our sequencing results for the 168 isolates were in complete agreement with the preliminary morphological results, in that all BLAST results were consistent with the organism being a basidiomycete. However, as a general rule, on the basis of the results of this study, we now utilize both the ITS and D1/D2 regions when we make sequence-based identifications for any fungus and double check if there is disagreement to make sure there is a GenBank deposit for both sequences. While this strategy does not guarantee that the sequence results will be 100% accurate, it can rapidly reveal whether there are enough data in GenBank for sequencing to even be used in the identification process. In our specific study, unfortunately, there does not seem to be enough data in GenBank to identify any unknown sterile basidiomycete with a high degree of confidence by ITS and/or D1/D2 sequencing.
As sequencing moves toward broader acceptance in the clinical laboratory, an important challenge to be overcome will be the development of a process that can provide a platform that certification bodies (the Clinical Laboratory Improvement Amendments [CLIA], the College of American Pathologists [CAP]) can standardize. In fact, guidelines are now being established to facilitate standardization (11). Evaluation of the data source should clearly be included in this platform, a major part of which should be a determination of how databases, whether they are public, such as GenBank, or private, could fit into the process. Unfortunately, the choice of database is not going to be a trivial issue. Despite the known errors with GenBank records, the depth of the sequences with regard to the number of potential species included in the database cannot be matched. Even imprecise GenBank records can be informative in some cases, since some taxonomic information may be identifiable, despite incorrect genus or species names. Conversely, private or closed databases may be more accurate due to the confirmation of each entry and the deposit of high-quality sequences. However, these databases will likely sacrifice species diversity and redundancy due to the smaller number of entries. Despite this shortcoming, a closed database may be more amenable to standardization, particularly if sequences are generated specifically for the database (versus downloading from another source), since primers, completeness, and identities can be standardized and confirmed.
In summary, this study has shown that in addition to the well-known concerns with the use of the sequences in a public database for sequence-based identification, missing data can also contribute to erroneous conclusions during searches. These errors may be caught for fungi for which substantial phenotypic data are available for comparison to the sequencing results; however, when there are few phenotypic data, such as for sterile basidiomycetes or other molds, erroneous conclusions could be quite common.
Acknowledgments
A.M.R. was supported by NIDCR grant DE14318 (CO STAR). B.L.W. was supported by grant PR054228 from the U.S. Army Medical Research and Materiel Command, Office of Congressionally Directed Medical Research Programs.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Atkins, S. D., and I. M. Clark. 2004. Fungal molecular diagnostics: a mini review. J. Appl. Genet. 1:3-15. [PubMed] [Google Scholar]
- 2.Balajee, S. A., A. M. Borman, M. E. Brandt, J. Cano, M. Cuenca-Estrella, E. Dannaoui, J. Guarro, G. Haase, C. C. Kibbler, W. Meyer, K. O'Donnell, A. Petti, J. L. Rodriguez, D. Sutton, A. Velegraki, and B. L. Wickes. 2009. Sequence based identification of Aspergillus, Fusarium, and Mucorlaes species in the clinical mycology laboratory: where should we go from here? J. Clin. Microbiol. 47:877-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A. 2007. Aspergillus species identification in the clinical setting. Stud. Mycol. 1:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balajee, S. A., L. Sigler, and M. E. Brandt. 2007. DNA and the classical way: identification of medically important molds in the 21st century. J. Med. Mycol. 45:475-490. [DOI] [PubMed] [Google Scholar]
- 5.Bidartondo, M. I. 2008. Preserving accuracy in GenBank. Science 319:1616. [DOI] [PubMed] [Google Scholar]
- 6.Borman, A. M., C. J. Linton, S. J. Miles, and E. M. Johnson. 2008. Molecular identification of pathogenic fungi. J. Antimicrob. Chemother. 61:7-12. [DOI] [PubMed] [Google Scholar]
- 7.Bruns, T. D., T. J. White, and J. W. Taylor. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Syst. 22:525-564. [Google Scholar]
- 8.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirado, D. E., G. Schar, M. Altwegg, E. C. Bottger, and P. P. Bosshard. 2007. Identification of moulds in the diagnostic laboratory—an algorithm implementing molecular and phenotypic methods. Diagn. Microbiol. Infect. Dis. 59:49-60. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA sequencing; approved guideline. CLSI MM18-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Davis, C. M., L. M. Noroski, M. K. Dishop, D. A. Sutton, R. M. Braverman, M. E. Paul, and H. M. Rosenblatt. 2007. Basidiomycetous fungal Inonotus tropicalis sacral osteomyelitis in X-linked chronic granulomatous disease. Pediatr. Infect. Dis. J. 26:655-656. [DOI] [PubMed] [Google Scholar]
- 13.Erjavec, Z., and P. E. Verweij. 2002. Recent progress in the diagnosis of fungal infections in the immunocompromised host. Drug Resist. Updat. 5:3-10. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, A., R. Sierra, R. E. Cardenas, A. Grajales, S. Restrepo, M. C. Cepero de Garcia, and A. Celis. 2009. Physiological and molecular characterization of atypical isolates of Malassezia furfur. J. Clin. Microbiol. 47:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, G. M., D. A. Sutton, E. Thompson, E. Tijerina, and M. G. Rinaldi. 2001. In vitro activities of approved and investigational antifungal agents against 44 clinical isolates of basidiomycetous fungi. Antimicrob. Agents Chemother. 45:633-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2004. Diagnostic PCR: making internal amplification control mandatory. Lett. Appl. Microbiol. 38:79-80. [DOI] [PubMed] [Google Scholar]
- 17.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. J. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 18.Janda, J. M., and S. L. Abbott. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirza, S. A., M. Phelan, D. Rimland, E. Graviss, R. Hamill, M. E. Brandt, T. Gardner, M. Sattah, G. Ponce de Leon, W. Baughman, and R. A. Hajjeh. 2003. The changing epidemiology of cryptococcosis: an update from population-based active surveillance in 2 large metropolitan areas, 1992-2000. Clin. Infect. Dis. 36:789-794. [DOI] [PubMed] [Google Scholar]
- 21.Pfaller, M., and R. Wenzel. 1992. Impact of the changing epidemiology of fungal infections in the 1990s. Eur. J. Clin. Microbiol. Infect. Dis. 11:287-291. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller, M. A., and D. J. Diekema. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pounder, J. I., K. E. Simmon, C. A. Barton, S. L. Hohmann, M. E. Brandt, and C. A. Petti. 2007. Discovering potential pathogens among fungi identified as nonsporulating molds. J. Clin. Microbiol. 45:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakeman, J. L., U. Bui, K. LaFe, Y. Chen, R. J. Honeycutt, and B. T. Cookson. 2005. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J. Clin. Microbiol. 43:3324-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinaldi, M. G. 1982. Use of potato flakes agar in clinical mycology. J. Clin. Microbiol. 15:1159-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorr, A. F., R. Lazarus, J. H. Sherner, W. L. Jackson, M. Morrel, V. J. Frasier, and M. H. Kollef. 2007. Do clinical features allow for accurate prediction of fungal pathogenesis in bloodstream infections? Potential implications of the increasing prevalence of non-albicans candidemia. J. Soc. Crit. Care Med. 35:1077-1083. [DOI] [PubMed] [Google Scholar]
- 27.Sigler, L., and S. L. Abbott. 1997. Characterizing and conserving diversity of filamentous basidiomycetes from human sources. Microb. Cult. Collect. 13:21-27. [Google Scholar]
- 28.Steinbach, W. J., T. G. Mitchell, W. A. Schell, A. Espinel-Ingroff, R. F. Coico, T. J. Walsh, and J. R. Perfect. 2003. Status of medical mycology education J. Med. Mycol. 41:457-467. [DOI] [PubMed] [Google Scholar]
- 29.Stevens, D. A. 2002. Diagnosis of fungal infections: current status J. Antimicrob. Chemother. 49:11-19. [DOI] [PubMed] [Google Scholar]
- 30.Summerbell, R. C. 1993. The benomyl test as a fundamental diagnostic method for medical mycology. J. Clin. Microbiol. 31:572-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutton, D. A., E. H. Thompson, M. G. Rinaldi, P. C. Iwen, K. K. Nakasone, H. S. Jung, H. M. Rosenblatt, and M. E. Paul. 2005. Identification and first report of Inonotus (Phellinus) tropicalis as an etiologic agent in a patient with chronic granulomatous disease. J. Clin. Microbiol. 43:982-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, T. J., T. D. Bruns, S. B. Lee, and J. W. Taylor. 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In PCR protocols and applications: a laboratory manual. Academic Press, New York, NY.