Abstract
We applied multiplex-tandem PCR (MT-PCR) to 255 EDTA whole-blood specimens, 29 serum specimens, and 24 plasma specimens from 109 patients with Candida bloodstream infection (candidemia) to determine whether a diagnosis could be expedited in comparison with the time to diagnosis by the use of standard blood culture. Overall, the MT-PCR performed better than blood culture with DNA extracted from whole blood from 52/74 (70%) patients, accelerating the time to detection (blood culture flagging) and determination of the pathogenic species (by use of the API 32C system [bioMérieux, Marcy l'Etoile, France]) by up to 4 days (mean, 2.2 days; range, 0.5 to 8 days). Candida DNA was detected more often in serum (71%) and plasma (75%) than in whole blood (54%), although relatively small numbers of serum and plasma specimens were tested. The sensitivity, specificity, positive predictive value, and negative predictive value of the assay with whole blood were 75%, 97%, 95%, and 85%, respectively. Fungal DNA was not detected by MT-PCR in 6/24 (25%) whole-blood samples drawn simultaneously with the positive blood culture sample. MT-PCR performed better with whole-blood specimens stored at −20°C (75%) and when DNA was extracted within 1 week of sampling (66%). The molecular and culture identification results correlated for 61 of 66 patients (92%); one discrepant result was due to misidentification by culture. All but one sample from 53 patients who were at high risk of candidemia but did not have proven disease were negative by MT-PCR. The results demonstrate the good potential of MT-PCR to detect candidemia, to provide Candida species identification prior to blood culture positivity, and to provide improved sensitivity when applied to with serum and plasma specimens.
Fungal bloodstream infections (BSIs) are major causes of morbidity and mortality in hospitalized patients, including critically ill individuals. Candida spp. rank as the third most common cause of nosocomial BSIs in the United States (39) and the eighth most common cause of all BSIs in Australia and Europe (3, 5). The outcomes of candidemia are poor, with the crude mortality rate being 40% (28) and the attributable mortality rate being 25 to 49% (10). Other consequences include excess hospital stay and costs estimated to be U.S. $40,000 per episode (7, 25).
The early initiation of targeted antifungal therapy is essential for improving patient outcomes (8, 26). However, the diagnosis of fungemia by blood culture, the current “gold standard,” is slow and insensitive, as yeasts are not detected in ∼50% of patients with candidemia and molds are rarely recovered (15). In addition, 1 to 2 days is required for isolation and phenotype-based identification from cultures. A number of alternative techniques, including peptide nucleic acid fluorescence in situ hybridization (PNA FISH; AdvanDx, Woburn, MA) (9, 30), direct germ tube analysis (31, 36), and nucleic acid detection tests (18, 24), have been developed to enable the more rapid identification of fungi directly from blood cultures that have flagged positive. However, these remain reliant on the sensitivity of the blood culture system.
Efforts to facilitate the earlier diagnosis of fungemia by direct testing of whole blood, serum, and plasma specimens have included the application of DNA microarray chips, the Luminex technology, and the commercial LightCycler SeptiFast system (Roche Diagnostics, Mannheim, Germany) (16, 17, 22, 34, 37). Although these tests are highly sensitive, the clinical utility of these platforms is limited by the lack of standardization and high setup costs. Furthermore, a significant proportion of assays target only a small number of pathogens (Candida albicans, Candida glabrata, Candida krusei, Candida parapsilosis, and Candida tropicalis) due to the limited multiplexing capacities of most real-time PCR instruments. As a result, other less common but nonetheless important causes of fungemia, such as other Candida species, Saccharomyces cerevisiae, and Fusarium and Scedosporium species are overlooked (12, 29, 32).
We recently demonstrated the clinical value of a novel, multiplex-tandem PCR (MT-PCR) platform for the accurate, simultaneous identification of up to 16 fungal pathogens from blood cultures and fungal culture plates (18, 19). The MT-PCR assay provided rapid (<2 h) identification compared with the time to identification by conventional phenotypic methods. In the study described here, we sought to determine whether the time to the detection of candidemia could be expedited by applying MT-PCR to DNA extracted from whole blood, serum, and plasma specimens collected up to 5.5 days prior to and/or up to 14 days subsequent to the time of collection of the Candida-positive blood sample. The sensitivity of the MT-PCR with different blood fractions and the impact of technical variables on DNA isolation and detection were also assessed. The specificity of the assay was further examined with whole-blood samples collected from a control cohort of patients considered to be at high risk of candidemia.
(This work was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy/Infectious Diseases Society of America 46th Annual Meeting, Washington, DC, 25 to 28 October 2008.)
MATERIALS AND METHODS
Patients. (i) Test population.
Two hundred fifty-five whole-blood specimens collected in 5-ml EDTA-coated tubes were obtained from 109 candidemic patients. Twenty-nine serum samples were collected from 14 of the candidemic patients, and 24 plasma specimens were collected from 5 of the candidemic patients. All specimens were obtained from the Clinical Haematology Laboratories of two Australian hospitals (Westmead Hospital, Westmead, New South Wales, and Princess Alexandra Hospital, Brisbane, Queensland) and from samples collected as part of the Australian Candidemia Study (3) from August 2001 to July 2009. Approval for the study was obtained from the institutions' human research ethics committees.
Specimens retrieved following the diagnosis of candidemia were categorized into four time (T) points: (i) T < 0, blood drawn prior to collection of the positive blood culture sample; (ii) T = 0, blood drawn simultaneously with the collection of the positive blood culture sample; (iii) T > 0, blood drawn after collection of the positive blood culture sample; and (iv) T > 0 flag+, blood drawn after the blood culture had flagged positive. Seven of 39 (18%) patients at T < 0, 9 of 43 (21%) patients at T > 0, and 39 of 53 (74%) patients at T > 0 flag+ were receiving antifungal therapy at the time of sampling. Specimens obtained between 2001 and 2006 were stored at −20°C prior to use. The remaining samples were stored at 4°C.
(ii) Control population.
Seventy-seven whole-blood specimens collected in EDTA-coated tubes were obtained from 53 medical and surgical intensive care unit (ICU) patients considered to be at high risk for candidemia (27).
Total nucleic acid extraction.
Nucleic acid was extracted in a class II laminar-flow cabinet, as follows.
(i) Whole blood in EDTA-coated tubes.
A High Pure PCR template preparation kit (Roche Diagnostics, Castle Hill, New South Wales, Australia) was used to isolate DNA from 3-ml blood samples. Briefly, specimens were divided into three 1-ml aliquots and washed twice in erythrocyte lysis buffer (0.155 M NH4Cl, 0.01 M NH4HCO3, 0.1 mM EDTA [pH 7.4]) (33) for 10 min at −20°C. A leukocyte pellet was collected following centrifugation at 4,000 × g for 10 min. Spheroplasts were resuspended in 1 ml molecular biology-grade water (Sigma-Aldrich, Castle Hill, New South Wales, Australia), vortexed thoroughly, and centrifuged at 5,400 × g for 5 min. The supernatant was removed, and the pellet was incubated in 300 μl tissue lysis buffer and 60 μl proteinase K (Roche Diagnostics) at 55°C for 1 h to overnight. DNA was extracted according to the manufacturer's instructions with a final elution volume of 60 μl.
(ii) Serum and plasma.
Plain and EDTA-coated blood collection tubes were centrifuged at 1,258 × g for 10 min to gather cells; the serum and plasma, respectively, were then removed. DNA was extracted from a 1-ml sample by using the protocol for whole blood on a nucliSENS easyMAG instrument (bioMérieux, Baulkham Hills, New South Wales, Australia) with a final elution volume of 60 μl.
MT-PCR amplification and detection.
Master mix reagents and 72-well gene discs containing lyophilized primers were prepared by AusDiagnostics Pty. Ltd. (Alexandria, New South Wales, Australia). The fungal ribosomal DNA (rDNA) internal transcribed spacer 1 (ITS1) and ITS2 regions, elongation factor 1-alpha (EF1-α), and β-tubulin gene loci were used to design primers specific for the 10 most frequently encountered Candida species in Australia (3), C. albicans, C. dubliniensis, C. famata, C. glabrata, C. guilliermondii, C. kefyr, C. krusei, C. lusitaniae, C. parapsilosis complex (C. parapsilosis, C. metapsilosis, and C. orthopsilosis), and C. tropicalis, as well as Yarrowia lipolytica, Cryptococcus neoformans complex (C. neoformans var. grubii, C. neoformans var. neoformans, and Cryptococcus gattii), Saccharomyces cerevisiae, Fusarium solani, Fusarium spp., and Scedosporium prolificans. The primer sequences are not shown due to commercial confidentiality agreements with AusDiagnostics. Product details are available from AusDiagnostics.
(i) First-round multiplexed preamplification.
PCRs were performed in a 20-μl volume consisting of 8 μl Step 1 master mix (the composition is commercial and confidential), 1 U MangoTaq DNA polymerase (Bioline, Alexandria, New South Wales, Australia) containing 20 U Moloney murine leukemia virus reverse transcriptase (Invitrogen, Mt. Waverly, Victoria, Australia), and 11 μl DNA template. Amplification was performed on a RotorGene thermal cycler (RG6000; Qiagen, Doncaster, Victoria, Australia). The thermal cycling conditions were 55°C for 2 min and 95°C for 5 min, followed by 15 cycles of 95°C for 10 s, 60°C for 20 s, and 72°C for 20 s. The PCRs were monitored for inhibition by the inclusion of an internal positive control (consisting of artificial DNA) with each specimen and were monitored for contamination by the inclusion of a negative water control in each test run.
(ii) Second-round quantification amplification.
A total of 5.3 μl of each multiplexed amplification product was diluted into 200 μl Step 2 master mix (the composition is commercial and confidential), 184.5 μl molecular biology-grade water, and 10.1 U MangoTaq DNA polymerase. A 20-μl aliquot was then added to the corresponding 17 positions on the gene disc containing the lyophilized inner primers. Amplification was performed on the RG6000 thermal cycler. The cycling conditions were 55°C for 2 min, followed by 30 cycles of 95°C for 1 s, 60°C for 10 s, and 72°C for 10 s. The fluorescence was measured at the end of each 72°C extension step. Following cycling, a melting curve was generated from 72°C to 95°C at 0.5°C intervals. The presence or the absence of an organism was determined by the use of analysis software (AusDiagnostics), which compared the given melting temperature to the expected melting temperature.
The analytical sensitivity and specificity of the assay were determined previously (18). The detection limits were 10 CFU/ml of blood for C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis and 100 CFU/ml of blood for C. krusei. All primers hybridized specifically with their respective target templates. No cross-reactions with other clinically relevant organisms were observed (18).
RESULTS
A total of 255 whole-blood specimens in EDTA-coated tubes, 29 serum specimens, and 24 plasma specimens from 109 patients with candidemia were tested by MT-PCR.
Assay performance compared with that of blood culture.
The performance of the MT-PCR assay was evaluated by using 61 whole-blood samples from 49 patients from whom a blood sample for culture was drawn simultaneously. For 19 samples with a positive result by MT-PCR, 18 were positive and 1 was negative by culture. For 42 samples with a negative result by MT-PCR, 6 were positive and 36 were negative by culture. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the MT-PCR assay were 75%, 97%, 95%, and 85%, respectively. There was no evidence of PCR inhibition, as the internal positive control in the MT-PCR assay for each sample always tested positive.
MT-PCR analysis of whole blood in EDTA-coated tubes, serum, and plasma samples from patients with documented candidemia.
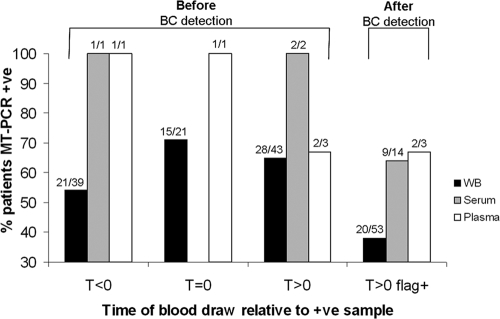
The results of MT-PCR with whole-blood, serum, and plasma samples relative to the time of collection of the first blood sample that flagged positive by culture are illustrated in Fig. 1. Fungal DNA was detected an average of 2.2 days (range, 0.5 to 8 days) before the blood cultures signaled positive for 52/74 (70%) patients, enabling the detection of fungemia as well as Candida species identification to be expedited by up to 4 days (the time required for the blood culture to signal positive and the time for species identification by use of the API 32C system [bioMérieux]). For 21/39 patients (54%), Candida DNA was detected an average of 1.5 days (range, 0.5 to 4.5 days) prior to the time of collection of the first positive blood culture sample (T < 0). Although the sample numbers were small, the MT-PCR assay performed better with serum (71% positivity rate) and plasma (75%) than with whole blood (54%). The sensitivities of the MT-PCR with serum, plasma, and whole-blood samples collected after blood culture positivity (T > 0 flag+) and up to 12 days after the blood sample positive by culture was drawn were 64%, 67%, and 38%, respectively.
FIG. 1.
Results of MT-PCR detection for 109 candidemic patients which have been stratified for time of collection relative to the time of collection of the first blood sample that flagged positive by culture (T = 0); i.e., those obtained before the time of collection of the blood sample that flagged positive by blood culture (BC) (T < 0), simultaneously with the time of collection of the blood sample that flagged positive by blood culture (T = 0), or after collection of the blood sample that flagged positive by blood culture (T > 0). T > 0 flag+, blood samples drawn after the blood culture had flagged positive. +ve, positive; WB, whole blood. Note that samples were collected from some patients at multiple time points.
Effect of time from T = 0 on MT-PCR detection.
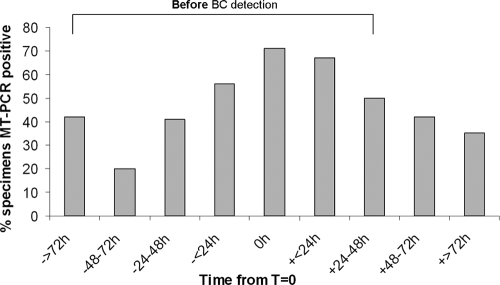
Whole-blood samples were collected up to 5.5 days prior to and up to 14 days after T = 0. Over this period, the detection of fungal DNA was the most sensitive with samples collected at T = 0 (71%) or within ±24 h of T = 0 (56 to 67%). There was a linear drop in sensitivity as the time increased from T = 0 (Fig. 2).
FIG. 2.
Percentage of MT-PCR-positive whole-blood samples stratified by collection times relative to the time that the first positive blood culture (BC) sample was drawn (T = 0) for 255 specimens collected from 109 candidemic patients.
Fungal species.
Of the 109 patients with candidemia, at least one blood specimen from 66 patients had positive MT-PCR results. C. albicans was isolated the most frequently (38%), followed by C. parapsilosis (30%); C. glabrata (14%); C. tropicalis (6%); and 1.5% each of C. krusei, C. kefyr, C. neoformans complex, and S. cerevisiae. Four patients had candidemia due to two Candida species: two patients had mixed C. albicans and C. glabrata infections and one patient each had mixed C. dubliniensis and C. parapsilosis infections and C. glabrata and C. tropicalis infections. The results of molecular- and culture-based species identification were concordant for 61 of 66 (92%) patients. In one case, C. albicans was phenotypically identified incorrectly as C. dubliniensis: MT-PCR confirmed that the original culture isolate was C. albicans. Discrepant identification (phenotypic versus MT-PCR identifications) for the remaining four cases was noted with C. albicans and C. tropicalis, Candida nivariensis and C. albicans, C. glabrata and C. parapsilosis, and C. parapsilosis and C. albicans. Phenotypic identification was confirmed by MT-PCR with the original culture isolate. With culture as the gold standard, MT-PCR yielded a single incorrect result, which was consistent on repeat MT-PCR analysis of the positive specimen.
MT-PCR results for paired blood and serum samples and paired blood and plasma samples.
Paired whole-blood and serum samples (n = 22 samples from 14 patients) and whole-blood and plasma samples (n = 14 samples from 5 patients) were tested by MT-PCR. Concordant positive or negative MT-PCR results were obtained for 50% of the blood-serum sample pairs and 79% of the blood-plasma sample pairs. For whole-blood negative cases, MT-PCR performed better with serum specimens (50%) than with plasma specimens (7%), although the results were not statistically significant, given the small number of samples tested (Table 1). On a patient basis, the assay was positive with sera obtained from five of six (83%) patients with positive whole-blood samples and seven of eight (88%) patients with negative whole-blood samples. The assay was also positive for concurrently drawn whole-blood and plasma samples obtained from four of five (80%) patients. Eight of nine (89%) patients not receiving antifungal therapy had MT-PCR-positive sera, while three of four (75%) patients not receiving antifungal therapy had positive plasma samples. For one patient with a mixed infection, both C. glabrata and C. tropicalis were detected in the patient's plasma, whereas only C. glabrata was detected in the patient's serum and whole blood.
TABLE 1.
MT-PCR results for paired whole-blood and serum samples and paired whole-blood and plasma samples
| Samples | No. (%) of paired samples with the following MT-PCR results: |
|||
|---|---|---|---|---|
| WBa and serum or plasma positive | WB positive and serum or plasma negative | WB negative and serum or plasma positive | WB and serum or plasma negative | |
| WB and serum (n = 22, 14 patients) | 8 (36) | 0 (0) | 11 (50) | 3 (14) |
| WB and plasma (n = 14, 5 patients) | 8 (57) | 2 (14) | 1 (7) | 3 (22) |
WB, whole blood.
MT-PCR of whole-blood samples from high-risk control patients.
Only 1 of 77 whole-blood samples collected from the control group of patients at high risk of developing an invasive fungal infection tested positive by MT-PCR. Repeat testing of the same specimen confirmed the presence of C. kefyr DNA; all other samples were negative by MT-PCR. All control patients remained without documented candidemia during the relevant episode of hospitalization.
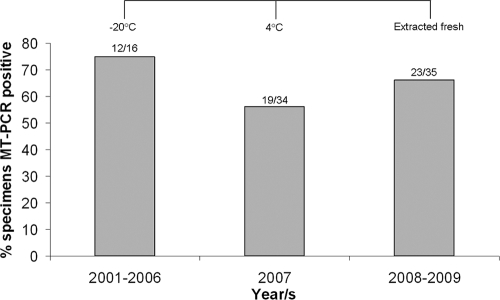
Effect of specimen storage on MT-PCR performance.
Specimens collected between 2001 and 2006 were stored at −20°C prior to DNA extraction, and those collected in 2007 were kept at 4°C prior to DNA extraction. Samples obtained in 2008 and 2009 were also stored at 4°C, but DNA was extracted within 1 week of specimen acquisition. Figure 3 shows the MT-PCR data for 85 specimens (from 62 patients) that were collected within ±24 h of the time that the blood sample positive by culture was drawn. Overall, the detection rate by MT-PCR was 64%. The assay performed best with whole-blood samples that were stored frozen (75%) rather than those that were stored at 4°C (56%) for extended periods (6 to 9 months). The rate of detection improved by 10% when DNA was extracted from fresh samples (<1 week old).
FIG. 3.
Effect of storage on MT-PCR detection of fungi from 85 whole-blood specimens from 62 candidemic patients.
DISCUSSION
The limitations of conventional blood culture systems for the detection of fungemia have prompted the need to develop alternative rapid, sensitive, and accurate methods. We have previously demonstrated the value of the fungal MT-PCR assay for the rapid identification of Candida spp. from blood cultures that had signaled positive and demonstrated yeasts on Gram stain (18). MT-PCR performed better than blood culture with DNA extracted from whole blood for 52/74 (70%) patients, accelerating the time to detection and identification of the pathogenic species by up to 4 days.
The performance of the MT-PCR assay was first assessed with whole-blood samples drawn simultaneously with the positive blood culture sample. Fungal cells are estimated to circulate in the blood at levels of <10 CFU/ml, with 25% of cases having <1 CFU/ml (38). A low fungal load at the time of sampling, the intermittent hepatic clearance of fungal cells from blood, and/or the short life of circulating DNA (2, 4) may have contributed to the lower sensitivity (75%) and NPV (85%) reported in this study. The single MT-PCR-positive/culture-negative result for C. kefyr (97% specificity) is not necessarily a false-positive result since nonviable cells or insufficient numbers of cells may have been present in the blood inoculated for culture. Furthermore, the sensitivities of blood culture systems are known to be poor even when viable cells are inoculated into the bottle (11).
The rate of concordance between MT-PCR and culture identification was high (92%; n = 66 samples). Concurrent infections with two species of Candida were also identified by MT-PCR of blood from 4 patients, and the fungal species in 11 patients with combined bacteremia and fungemia were correctly identified. Discrepant results were obtained for a single sample from each of five patients. In one case, the phenotypic misidentification of C. albicans was made (C. dubliniensis by culture), highlighting the difficulty of differentiating these two closely related species by standard methods (35). The remaining discrepancies are unlikely to have been due to cross-reactions of primer sequences since none were observed when the primers were tested with >500 culture isolates and blood culture specimens (18, 19). Contamination of the sample (e.g., during routine hematological examination) prior to DNA extraction is a plausible explanation.
There is currently no consensus on the optimal blood fraction from which to isolate fungal DNA (21). Although small numbers of paired samples were tested, we detected Candida DNA more often in serum (71%) and plasma (75%) than in whole blood (56%) (Fig. 1), as has been observed in other studies (1, 14, 23). It has been demonstrated in rabbit models that free DNA (in the serum and plasma fractions) is cleared more slowly than intracellular DNA, and the efficiency of extraction of fungal DNA from plasma or serum is higher than that from whole blood (1, 14). Although whole blood contains both free and cell-associated fungal DNA, all fungal DNA except that associated with leukocytes and/or whole fungal cells is removed during the process of red blood cell lysis and the removal of PCR inhibitors. In one study, a 50% loss of C. albicans DNA was noted following the centrifugation and removal of erythrocytes (13). Notably, the time needed to extract DNA from serum and plasma (1 h) was significantly shorter and less laborious than that needed to extract DNA from whole blood (at least 4 h), since cell lysis was not required. The optimal blood fraction for the isolation of DNA of pathogenic molds is less well studied but can be expected to differ from that of yeasts due to differences in cell wall composition/structure and DNA kinetics (20, 40).
The sensitivity of DNA detection may also vary with the time of blood sampling during the course of the infection (23). MT-PCR was more likely to be positive when it was performed with whole blood collected within ±24 h of T = 0 (Fig. 2). One possible explanation for the lower yield at T < 0 is the absence of infection, since candidemia is usually acute in onset. We considered whether the reduced sensitivity of the MT-PCR assay with whole-blood specimens at T > 0 was due to antifungal therapy, which can be expected to release DNA from damaged fungal cells, thereby favoring the detection of DNA in serum and plasma samples. Our results did not support this hypothesis since the serum and plasma samples from 89% and 75% of patients, respectively, who had not received antifungal agents at or before the time of sampling were MT-PCR positive. Since DNA from both viable and nonviable cells is detected by MT-PCR, the results used to make clinical decisions should always be interpreted in combination with other microbiological data.
Our results also indicate that the temperature and duration of specimen storage can affect the sensitivity of MT-PCR (Fig. 3). The MT-PCR assay performed the best (75% sensitive) with whole-blood samples stored at −20°C. Freeze-thaw cycles may help disrupt the integrity of the fungal cell wall, leading to the better recovery of DNA. On the basis of these results, we propose that samples be stored at −20°C until they are processed to optimize the sensitivity of the assay.
The MT-PCR platform employed in the present study has a major advantage over other real-time multiplexing platforms (e.g., microarrays and the Luminex technology), in that it does not require expensive or specialized equipment (e.g., a flow cytometer), as it utilizes Evagreen dye and melting curve analysis for species identification. Flexibility in the number of detection targets also enables the number of samples that can be tested in a single run to be varied, and the platform can be automated for use by laboratory staff with little experience with molecular processing and analysis. The costs are similar to those of current laboratory tests at $14 per sample and 30 min of labor cost per test run. The assay had a detection limit of 10 CFU/ml for the major Candida species (except C. krusei, for which the detection limit is 100 CFU/ml), which compares favorably to that of the LightCycler SeptiFast system (30 CFU/ml; Roche Diagnostics).
The results demonstrate that MT-PCR is capable of providing results within a single working day (including DNA extraction, PCR amplification, and analysis). Its clinical utility, however, is dependent in part on the frequency of the test runs. We envisage this assay to be performed thrice weekly in our laboratory. Taking into consideration the potential delays in specimen transport and handling, we anticipate that the diagnosis of candidemia would realistically be expedited by 1 to 2 days (compared with the times for blood culture detection and species identification by use of the API 32C system [bioMérieux]). Even so, this compares favorably or is similar to the turnaround times of other rapid identification methods, such as PNA FISH techniques or rapid trehalose tests (RTTs), which require either the flagging of a blood culture or a pure culture of Candida. Furthermore, the MT-PCR assay has the advantage of enabling the species identification of a large range of species (n = 16). The Yeast Traffic Light PNA FISH protocol enables group identification of C. albicans/C. parapsilosis, C. glabrata/C. krusei, and C. tropicalis but does not select for each species (www.advandx.com), while the RTT selectively identifies only C. glabrata (6).
A limitation of this assay was that it was unable to detect fungal DNA in six (25%) whole-blood specimens drawn simultaneously with the positive blood culture sample. We therefore propose that the platform be used to complement blood culture until large-scale clinical evaluations are performed to position the role of MT-PCR in the diagnostic laboratory. Nonetheless, the diagnosis of candidemia and the identity of the pathogen were available earlier for the remaining 75% of the patients. Another limitation was the nonsystematic collection and storage of samples, given the retrospective nature of this study. This methodology was used since it was the most efficient way to assess the potential utility of the MT-PCR assay for the early diagnosis of candidemia. Whole-blood samples were primarily collected for routine hematological examination. As such, blood samples for culture were simultaneously drawn from few patients at the earlier time points, presumably because there was no clinical evidence of sepsis. It is therefore possible that the higher rate of MT-PCR positivity was driven by the more frequent sampling of whole-blood specimens. Despite these limitations, a proof of principle was established and important data on technical variables (e.g., specimen type and storage conditions) were ascertained.
In conclusion, the MT-PCR assay presents a promising platform for the real-time screening and potentially earlier diagnosis of candidemia. Our results additionally suggest that serum and plasma samples are preferable to whole blood, although a greater number of paired specimens need to be tested. Pending further interlaboratory validation and studies with samples from a larger patient cohort, the platform should be used in conjunction with blood culture. Although molds were not tested in the present study, the MT-PCR assay offers the potential for the early detection of BSIs with molds such as Fusarium spp. and S. prolificans. Further prospective studies are warranted to determine the extent to which the early detection of candidemia affects clinical practice.
Acknowledgments
We thank the Australian Candidemia Study for access to 38 blood samples and for providing data on antifungal therapy. We also thank the staff of the Clinical Haematology Laboratories at Westmead Hospital and Princess Alexandra Hospital for assistance with the provision of whole-blood specimens in EDTA-coated tubes and Dani Goh and Martin Cullen for help in identifying appropriate control patients.
This work was supported in part by a Centre for Clinical Research Excellence grant (grant no. 264625) from the National Health and Medical Research Council of Australia. A.L. is supported by an Australian Postgraduate Award.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Bougnoux, M., C. Dupont, J. Mateo, P. Saulnier, V. Faivre, D. Payen, and M. Nicolas-Chanoine. 1999. Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR. J. Clin. Microbiol. 37:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bretagne, S., J. M. Costa, E. Bart-Delabesse, N. Dhedin, C. Rieux, and C. Cordonnier. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S., M. Slavin, Q. Nguyen, D. Marriott, E. G. Playford, D. Ellis, T. Sorrell, and Australian Candidemia Study. 2006. Active surveillance for candidemia, Australia. Emerg. Infect. Dis. 12:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Repentigny, L. 1992. Serodiagnosis of candidiasis, aspergillosis, and cryptococcosis. Clin. Infect. Dis. 14(Suppl. 1):S11-S22. [DOI] [PubMed] [Google Scholar]
- 5.Fluit, A. C., M. E. Jones, F. J. Schmitz, J. Acar, R. Gupta, and J. Verhoef. 2000. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin. Infect. Dis. 30:454-460. [DOI] [PubMed] [Google Scholar]
- 6.Freydiere, A. M., J. D. Perry, O. Faure, B. Willinger, A. M. Tortorano, A. Nicholson, J. Peman, and P. E. Verweij. 2004. Routine use of a commercial test, GLABRATA RTT, for rapid identification of Candida glabrata in six laboratories. J. Clin. Microbiol. 42:4870-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fridkin, S. K. 2005. The changing face of fungal infections in health care settings. Clin. Infect. Dis. 41:1455-1460. [DOI] [PubMed] [Google Scholar]
- 8.Garey, K. W., M. Rege, M. P. Pai, D. E. Mingo, K. J. Suda, R. S. Turpin, and D. T. Bearden. 2006. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin. Infect. Dis. 43:25-31. [DOI] [PubMed] [Google Scholar]
- 9.Gherna, M., and W. G. Merz. 2009. Identification of Candida albicans and Candida glabrata within 1.5 hours directly from positive blood culture bottles with a shortened peptide nucleic acid fluorescence in situ hybridization protocol. J. Clin. Microbiol. 47:247-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudlaugsson, O., S. Gillespie, K. Lee, J. Vande Berg, J. Hu, S. Messer, L. Herwaldt, M. Pfaller, and D. Diekema. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172-1177. [DOI] [PubMed] [Google Scholar]
- 11.Horvath, L. L., D. R. Hospenthal, C. K. Murray, and D. P. Dooley. 2003. Detection of simulated candidemia by the BACTEC 9240 system with plus aerobic/F and anaerobic/F blood culture bottles. J. Clin. Microbiol. 41:4714-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, T. G., B. Gahrn-Hansen, M. Arendrup, and B. Bruun. 2004. Fusarium fungaemia in immunocompromised patients. Clin. Microbiol. Infect. 10:499-501. [DOI] [PubMed] [Google Scholar]
- 13.Jordan, J. A. 1994. PCR identification of four medically important Candida species by using a single primer pair. J. Clin. Microbiol. 32:2962-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasai, M., A. Francesconi, R. Petraitiene, V. Petraitis, A. M. Kelaher, H. S. Kim, J. Meletiadis, T. Sein, J. Bacher, and T. J. Walsh. 2006. Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive candidiasis. J. Clin. Microbiol. 44:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontoyiannis, D., V. Wessel, G. Bodey, et al. 2000. Zygomycosis in the 1990s in a tertiary care centre. Clin. Infect. Dis. 30:851-856. [DOI] [PubMed] [Google Scholar]
- 16.Landlinger, C., S. Preuner, B. Willinger, B. Haberpursch, Z. Racil, J. Mayer, and T. Lion. 2009. Species-specific identification of a wide range of clinically relevant fungal pathogens by use of Luminex xMAP technology. J. Clin. Microbiol. 47:1063-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau, A., S. Chen, S. Sleiman, and T. Sorrell. 2009. Current status and future perspectives of molecular and serological methods in diagnostic mycology. Future Microbiol. 4:1185-1222. [DOI] [PubMed] [Google Scholar]
- 18.Lau, A., T. C. Sorrell, S. Chen, K. Stanley, J. Iredell, and C. Halliday. 2008. Multiplex tandem PCR: a novel platform for rapid detection and identification of fungal pathogens from blood culture specimens. J. Clin. Microbiol. 46:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau, A., T. C. Sorrell, O. Lee, K. Stanley, and C. Halliday. 2008. Colony multiplex-tandem PCR for rapid, accurate identification of fungal cultures. J. Clin. Microbiol. 46:4058-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeffler, J., H. Hebart, U. Brauchle, U. Schumacher, and H. Einsele. 2000. Comparison between plasma and whole blood specimens for detection of Aspergillus DNA by PCR. J. Clin. Microbiol. 38:3830-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maaroufi, Y., N. Ahariz, M. Husson, and F. Crokaert. 2004. Comparison of different methods of isolation of DNA of commonly encountered Candida species and its quantitation by using a real-time PCR-based assay. J. Clin. Microbiol. 42:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMullan, R., L. Metwally, P. V. Coyle, S. Hedderwick, B. McCloskey, H. J. O'Neill, C. C. Patterson, G. Thompson, C. H. Webb, and R. J. Hay. 2008. A prospective clinical trial of a real-time polymerase chain reaction assay for the diagnosis of candidemia in nonneutropenic, critically ill adults. Clin. Infect. Dis. 46:890-896. [DOI] [PubMed] [Google Scholar]
- 23.Metwally, L., D. J. Fairley, P. V. Coyle, R. J. Hay, S. Hedderwick, B. McCloskey, H. J. O'Neill, C. H. Webb, and R. McMullan. 2008. Comparison of serum and whole-blood specimens for the detection of Candida DNA in critically ill, non-neutropenic patients. J. Med. Microbiol. 57:1269-1272. [DOI] [PubMed] [Google Scholar]
- 24.Metwally, L., G. Hogg, P. V. Coyle, R. J. Hay, S. Hedderwick, B. McCloskey, H. J. O'Neill, G. M. Ong, G. Thompson, C. H. Webb, and R. McMullan. 2007. Rapid differentiation between fluconazole-sensitive and -resistant species of Candida directly from positive blood-culture bottles by real-time PCR. J. Med. Microbiol. 56:964-970. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, J., M. I. Meltzer, B. D. Plikaytis, A. N. Sofair, S. Huie-White, S. Wilcox, L. H. Harrison, E. C. Seaberg, R. A. Hajjeh, and S. M. Teutsch. 2005. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect. Control Hosp. Epidemiol. 26:540-547. [DOI] [PubMed] [Google Scholar]
- 26.Morrell, M., V. J. Fraser, and M. H. Kollef. 2005. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 49:3640-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrosky-Zeichner, L., C. Sable, J. Sobel, B. D. Alexander, G. Donowitz, V. Kan, C. A. Kauffman, D. Kett, R. A. Larsen, V. Morrison, M. Nucci, P. G. Pappas, M. E. Bradley, S. Major, L. Zimmer, D. Wallace, W. E. Dismukes, and J. H. Rex. 2007. Multicenter retrospective development and validation of a clinical prediction rule for nosocomial invasive candidiasis in the intensive care setting. Eur. J. Clin. Microbiol. Infect. Dis. 26:271-276. [DOI] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietrucha-Dilanchian, P., R. E. Lewis, H. Ahmad, and A. E. Lechin. 2001. Candida lusitaniae catheter-related sepsis. Ann. Pharmacother. 35:1570-1574. [DOI] [PubMed] [Google Scholar]
- 30.Shepard, J. R., R. M. Addison, B. D. Alexander, P. Della-Latta, M. Gherna, G. Haase, G. Hall, J. K. Johnson, W. G. Merz, H. Peltroche-Llacsahuanga, H. Stender, R. A. Venezia, D. Wilson, G. W. Procop, F. Wu, and M. J. Fiandaca. 2008. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J. Clin. Microbiol. 46:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheppard, D. C., M. C. Locas, C. Restieri, and M. Laverdiere. 2008. Utility of the germ tube test for direct identification of Candida albicans from positive blood culture bottles. J. Clin. Microbiol. 46:3508-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simarro, E., F. Marin, A. Morales, E. Sanz, J. Perez, and J. Ruiz. 2001. Fungemia due to Scedosporium prolificans: a description of two cases with fatal outcome. Clin. Microbiol. Infect. 7:645-647. [DOI] [PubMed] [Google Scholar]
- 33.Skladny, H., D. Buchheidt, C. Baust, F. Krieg-Schneider, W. Seifarth, C. Leib-Mosch, and R. Hehlmann. 1999. Specific detection of Aspergillus species in blood and bronchoalveolar lavage samples of immunocompromised patients by two-step PCR. J. Clin. Microbiol. 37:3865-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiess, B., W. Seifarth, M. Hummel, O. Frank, A. Fabarius, C. Zheng, H. Morz, R. Hehlmann, and D. Buchheidt. 2007. DNA microarray-based detection and identification of fungal pathogens in clinical samples from neutropenic patients. J. Clin. Microbiol. 45:3743-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, D., and D. Coleman. 1998. Candida dubliniensis: characteristics and identification. J. Clin. Microbiol. 36:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terlecka, J. A., P. A. du Cros, C. Orla Morrissey, and D. Spelman. 2007. Rapid differentiation of Candida albicans from non-albicans species by germ tube test directly from BacTAlert blood culture bottles. Mycoses 50:48-51. [DOI] [PubMed] [Google Scholar]
- 37.Westh, H., G. Lisby, F. Breysse, B. Boddinghaus, M. Chomarat, V. Gant, A. Goglio, A. Raglio, H. Schuster, F. Stuber, H. Wissing, and A. Hoeft. 2009. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect. 15:544-551. [DOI] [PubMed] [Google Scholar]
- 38.White, P. L., A. Shetty, and R. A. Barnes. 2003. Detection of seven Candida species using the Light-Cycler system. J. Med. Microbiol. 52:229-238. [DOI] [PubMed] [Google Scholar]
- 39.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 40.Yamakami, Y., A. Hashimoto, I. Tokimatsu, and M. Nasu. 1996. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J. Clin. Microbiol. 34:2464-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]