Abstract
The aim of the present study was to analyze the importance of nontypeable Haemophilus influenzae (NTHi) isolated from patients with sepsis (invasive isolates) compared to nasopharyngeal isolates from patients with upper respiratory tract infection for resistance to complement-mediated attack in human serum and to correlate this result with disease severity. We studied and characterized cases of invasive NTHi disease in detail. All patients with invasive NTHi isolates were adults, and 35% had a clinical presentation of severe sepsis according to the ACCP/SCCM classification of sepsis grading. Moreover, 41% of the patients had evidence of immune deficiency. The different isolates were analyzed for survival in human serum and for binding of 125I-labeled, purified human complement inhibitors C4b-binding protein (C4BP), factor H, and vitronectin, in addition to binding of regulators directly from serum. No significant differences were found when blood-derived and nasopharyngeal isolates were compared, suggesting that interactions with the complement system are equally important for NTHi strains, irrespective of isolation site. Interestingly, a correlation between serum resistance and invasive disease severity was found. The ability to resist the attack of the complement system seems to be important for NTHi strains infecting the respiratory tract as well as the bloodstream.
Haemophilus influenzae is an important human-specific pathogen that can be classified according to the presence of a polysaccharide capsule (20). The encapsulated strains cause invasive diseases, whereas the unencapsulated and hence nontypeable H. influenzae (NTHi) strains are mainly found in local upper and lower respiratory tract infections (2, 35). However, NTHi is, after Streptococcus pneumoniae, the most common microbe found in children with acute otitis media and is the main cause of exacerbations in patients suffering from chronic obstructive pulmonary disease and bronchiectasis (3, 4, 13, 28, 32, 34). NTHi can also cause sinusitis, conjunctivitis, and pneumonia in children (25). Thus, NTHi is a heterogenous species, capable of great variation in virulence, and is found in the airway either as a commensal or as a pathogen with the capacity to invade the airway epithelium.
Invasive disease caused by H. influenzae type b (Hib) mainly affects infants and children, causing potentially life-threatening conditions, such as meningitis, epiglottitis, and severe sepsis. After introduction of the conjugate vaccine against Hib in the early 1990s, the incidence of invasive disease caused by Hib has decreased substantially in the Western hemisphere (5). In contrast, it has been suggested that the incidence of NTHi septicemia is increasing (36). Most clinical studies of invasive Haemophilus infections have been about Hib, and less is known about the clinical characteristics of invasive disease caused by NTHi. In relation to its extensive presence in nasopharyngeal and sputum cultures, NTHi is infrequently found in the bloodstream and it seems likely that host factors are equally important as specific bacterial virulence in patients with NTHi sepsis.
The complement system is the first line of defense against pathogenic microorganisms (7). Activation of the complement system leads to a cascade of protein activation and deposition on the surface of the pathogen, resulting in formation of the membrane attack complex (MAC) and opsonization of the pathogen, followed by phagocytosis (38). The classical pathway of the complement system is activated by target-bound antibodies and C-reactive protein (37), whereas the alternative pathway is spontaneously activated through direct contact with foreign particles or cells (38). Both pathways lead to the formation of the C3 convertases, with subsequent cleavage of C3 to C3a and C3b. Thereafter, the C5 convertases are formed and the terminal pathway is activated, which results in the formation of the MAC and lysis of the cell. To prevent nonspecific damage from excess complement activation, the complement cascade is tightly regulated. Important regulators of the complement system are C4b-binding protein (C4BP) (governing the classical/lectin pathway) (6), factor H and factor H-like protein 1 (alternative pathway) (41), and vitronectin and complement factor H-related protein 1 (terminal pathway) (18, 33).
The complement system is classified as a part of serum, but there are several studies demonstrating the presence of complement in various sites of the body. Reports of the presence of complement components in the respiratory tracts of healthy individuals are scarce. There are several studies, however, indicating the importance of complement in the respiratory tract during infections. The permeability of the mucosa increases during inflammation, and plasma, including complement proteins and immunoglobulins, enters the airway lumen (11, 12, 29). This process, designated plasma exudation, has been suggested to be the first line of the mucosal defense.
The pathogenesis of many microorganisms relies on their capacity to avoid, resist, or neutralize the host defense, including the complement system. Therefore, many pathogens have evolved different mechanisms to avoid complement-mediated killing. A frequent strategy used by some pathogens is binding of complement inhibitors such as C4BP, factor H, and vitronectin, which all protect from complement-mediated attacks (7, 22, 31). These inhibitors are captured on the bacterial surface in such a way that they are still functionally active. In previous studies, we have shown that NTHi binds C4BP and factor H and that these interactions significantly contribute to bacterial serum resistance (15, 17). In addition, Haemophilus surface fibrils that can be found solely in Hib, and protein E, which exists in both encapsulated and nontypeable strains (30), interact with vitronectin and thereby prevent complement-induced lysis, resulting in increased bacterial survival in normal human serum (NHS) (14, 16).
In the present study, the characteristics of invasive NTHi infections, including evidence of immune deficiency in the individual patient and the clinical presentation of the septic event, were studied. We correlated these findings with the capacity to bind specific complement regulators and the in vitro serum resistance of the individual isolates. The invasive NTHi isolates obtained from patients with sepsis were compared to nasopharyngeal strains from patients with upper respiratory tract infection. There was no clear difference in serum resistance or binding to complement inhibitors between the two groups of NTHi. Our findings also demonstrate that binding of complement regulators and resistance to human serum are important for NTHi isolates from the upper respiratory tract as well as those from blood samples. Furthermore, a significant correlation between disease severity and in vitro serum resistance was identified in cases of NTHi invasive disease.
MATERIALS AND METHODS
Patient data.
Complete medical records, including clinical presentation, patient history, and laboratory results, from 17 out of 21 identified patients with invasive NTHi disease (bacteremia or meningitis) were studied and registered. The severity of the sepsis was graded according to the ACCP/SCCM classification of sepsis grading (8). Randomly selected nasopharyngeal cultures positive for NTHi (n = 21) were used as controls. All of these cultures had a referral history of airway infection.
Bacterial strains and culture conditions.
Blood-derived NTHi isolates (n = 21) were obtained from patients with invasive NTHi disease (bacteremia or meningitis) in the county of Southwest Skane, Sweden, from 2001 to 2007. In addition, NTHi strains isolated from the nasopharynxes (n = 21) of patients (Southwest Skane in 2007) suffering from upper respiratory tract infection were included for comparison. Bacteria were routinely cultured in brain heart infusion (BHI) liquid broth supplemented with NAD and hemin (both at 10 μg/ml) or on chocolate agar plates at 37°C in a humid atmosphere containing 5% CO2. All strains were characterized by standard bacteriological techniques, including oxidase, fermentation, satellite, and XV tests. Thereafter, the H. influenzae strains were typed by PCR. To exclude that H. haemolyticus was among the isolates, PCR and 16S rRNA sequencing was done (24).
Proteins and antibodies.
C4BP and factor H from human plasma were purchased from Complement technology (Tyler, TX), and vitronectin from human plasma was from Sigma (Sigma-Aldrich, St. Louis, MO). The polyclonal rabbit anti-C4BP and goat anti-human factor H antisera were purchased from Complement technology or Calbiochem (La Jolla, CA). The horseradish peroxidase (HRP)-conjugated donkey anti-goat polyclonal antibody (pAb) was obtained from Serotech (Oxford, United Kingdom). The goat anti-human vitronectin and fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat pAbs were from Serotec (Oxford, United Kingdom). An antiserum obtained from a rabbit immunized with the clinical isolate NTHi 772 for 3 times at 2-week intervals was also included, as a control (1).
Serum bactericidal assay.
Pooled NHS was obtained from healthy blood donors (n = 5) with informed consent. The NTHi strains were diluted in DGVB++ (2.5 mM Veronal buffer, pH 7.3, containing 70 mM NaCl, 140 mM glucose, 0.1% [wt/vol] gelatin, 1 mM MgCl2, and 0.15 mM CaCl2). Bacteria (104 CFU) were incubated in NHS or heat-inactivated NHS (hiNHS) in a final volume of 100 μl at 37°C. After 20 min, 10-μl aliquots were removed and spread onto chocolate agar plates. After 18 h of incubation at 37°C, numbers of CFU were determined.
Flow cytometry.
To analyze whether the NHS contained IgG directed against NTHi and if there were any differences in IgG deposition on NTHi isolates from blood and nasopharynx samples, NHS was tested for reactivity by flow cytometry using a FITC-conjugated mouse anti-human IgG pAb (Dakopatts). A rabbit anti-NTHi pAb was included as a positive control, and in these experiments, a FITC-conjugated goat anti-rabbit pAb (Dakopatts) was used as a secondary layer. Briefly, bacteria (107) were incubated with 50% hiNHS for 1 h on ice. Thereafter, bacteria were washed, followed by addition of the FITC-conjugated detection antibodies, additional washes, and finally flow cytometry analysis. All incubations were done in phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA), and washings were done with the same buffer. Secondary antibodies were added separately as negative controls for each strain analyzed.
Protein labeling and direct binding assay.
Purified C4BP, factor H, and vitronectin were labeled with 0.05 mol iodine (GE Healthcare, Buckinghamshire, United Kingdom) per mol protein, using the chloramine T method (10). The different H. influenzae strains were grown in BHI liquid broth overnight and washed in PBS containing 1% BSA (Saveen Werner, Malmo, Sweden) (PBS-BSA). Bacteria (2 × 107) were incubated with 125I-labeled complement regulators at 37°C for 1 h. After incubation, bacteria were centrifuged (10,000 × g) through a 20% sucrose column. The tubes were frozen and cut, and levels of radioactivity in the pellet and supernatant were measured using a gamma counter. Binding was calculated as amount of bound radioactivity (pellet) versus total radioactivity (pellet plus supernatant).
Serum binding assay.
To analyze whether NTHi from different isolation sites bound C4BP or factor H directly from NHS, bacteria were grown overnight in BHI broth. NTHi bacteria (109) were incubated with hiNHS and buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.4) for 1 h at 37°C. To remove unbound proteins, bacteria were washed 5 times with the same buffer. Thereafter, the bacterial pellet was resuspended in 150 μl of 0.1 M glycine-HCl, pH 2.0, in order to elute bound proteins (15, 17, 21). Bacteria without NHS were used as a negative control. After 15 min of incubation at 37°C with shaking, bacteria were centrifuged and the supernatants were subjected to SDS-PAGE (10%).
To analyze whether NTHi bound vitronectin from NHS, bacteria were grown overnight and incubated with hiNHS and PBS. To remove unbound proteins, NTHi bacteria were washed 5 times with the same buffer. Thereafter, the bacterial pellet was resuspended in 50 μl of 0.1% Triton X-100 (Darmstadt, Germany) and protease inhibitors (Complete; Roche, Mannheim, Germany). After 30 min of incubation at 4°C, bacteria were centrifuged and the supernatants were subjected to SDS-PAGE (10%). Electrophoretic transfer of protein bands from the gel to an Immobilon-P membrane (Millipore, Bedford, MA) was done at 35 V for 2 h. After transfer, the Immobilon-P membrane was blocked in PBS with 0.1% Tween 20 (PBS-Tween) containing 5% milk powder. After several washings, the membrane was incubated with rabbit anti-human C4BP, goat anti-human factor H, or goat anti-human vitronectin pAb, followed by incubation with HRP-conjugated swine anti-rabbit or donkey-anti-goat pAb. After incubation and additional washings in PBS-Tween, development was performed with enhanced chemiluminescence (ECL) Western blotting detection reagents (Pierce, Rockford, IL). No cross-reactivity of the anti-C4BP, anti-factor H, or anti-vitronectin pAbs was seen with bacteria incubated in the absence of NHS.
Statistical analysis.
To test for differences in survival in serum and levels of complement inhibitor binding between the groups, a two-sided Mann-Whitney U test was used. This was done in consideration of small sample sizes and nonparametric data.
RESULTS
Clinical characteristics of invasive NTHi disease.
Twenty-one cases of invasive NTHi were identified during the period from 2001 to 2007, and complete medical records were obtained from 17 patients, as shown in Table 1. In contrast to invasive Hib disease, which in unvaccinated populations is mainly associated with younger patients, all patients with invasive NTHi disease were adults. The cases were divided into separate groups by severity of clinical disease according to the ACCP/SCCM classification (Table 1). Eight of the patients (47%) (patients 1 to 8) had a clinical presentation of sepsis, while nine (53%) (patients 9 to 17) had a presentation of sepsis with meningitis or severe sepsis. Interestingly, seven of the patients (41%) had evidence of immune deficiency. Out of these 7 patients, 6 presented secondary immune deficiencies such as leukemia and treatment-induced neutropenia. However, severe clinical presentations were not linked to immune deficiency. One patient died from the septic episode (28-day mortality; 6%), while 5 patients died within a year (1-year mortality; 29%). All but one of the patients that died within a year from their septic episode presented a mild clinical picture, suggesting the importance of underlying disease.
TABLE 1.
Clinical and epidemiological data from 17 cases of invasive NTHi infection and survival of isolates in serum in vitro
| Patient | Age (yr) | Gender | Immune status at time of sepsis | Sepsis gradinga | % Survival of NTHi strain in serum in vitrob |
|---|---|---|---|---|---|
| 1 | 52 | Male | Agammaglobulinemia | Sepsis | 5 ± 3 |
| 2 | 69 | Male | No sign of immune deficiency | Sepsis | 2 ± 3 |
| 3 | 60 | Female | Metastatic breast cancer | Sepsis | 18 ± 14 |
| 4 | 75 | Male | Hypogammaglobulinemia | Sepsis | 66 ± 31 |
| 5 | 75 | Male | No sign of immune deficiency | Sepsis | 8 ± 7 |
| 6 | 37 | Female | Chemotherapy | Sepsis | 23 ± 7 |
| 7 | 58 | Male | Leukemia, neutropenia | Sepsis | 63 ± 15 |
| 8 | 74 | Female | No sign of immune deficiency | Sepsis | 62 ± 0 |
| 9 | 36 | Male | No sign of immune deficiency | Sepsis/meningitis | 17 ± 8 |
| 10 | 73 | Male | NAc | Sepsis/meningitis | 44 ± 10 |
| 11 | 60 | Male | No sign of immune deficiency | Sepsis/meningitis | 79 ± 24 |
| 12 | 42 | Female | No sign of immune deficiency | Severe sepsis | 89 ± 2 |
| 13 | 39 | Male | No sign of immune deficiency | Severe sepsis | 81 ± 25 |
| 14 | 64 | Female | No sign of immune deficiency | Severe sepsis | 52 ± 4 |
| 15 | 89 | Female | Chronic leukemia | Severe sepsis | 86 ± 19 |
| 16 | 83 | Male | No sign of immune deficiency | Severe sepsis | 64 ± 40 |
| 17 | 74 | Female | No sign of immune deficiency | Severe sepsis | 58 ± 12 |
Graded according to the ACCP/SCCM classification of sepsis grading.
Values shown represent levels observed after 20 min of incubation with 5% NHS and are means ± SD of results from three experiments.
NA, not available.
Serum resistance correlates with sepsis severity.
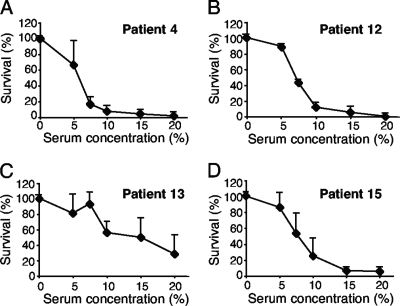
Eight strains isolated from blood samples were studied with regard to survival at increasing concentrations of pooled NHS from 5 donors to determine the appropriate serum concentration for further serum resistance experiments (Fig. 1 and data not shown). When a titration of the serum concentration was performed, all strains except the isolate from patient 13 (Fig. 1C) were killed by 15% NHS after 20 min of incubation. All strains survived in 5% NHS, and therefore, this concentration was used for further studies.
FIG. 1.
Bactericidal activity against a series of randomly selected blood-derived isolates. NTHi bacteria were incubated in the presence of increasing concentrations of NHS (5 to 20%). Four typical strains are shown in panels A to D. After incubation for 20 min, bacteria were spread on chocolate agar plates to allow determination of the number of surviving bacteria. The number of bacteria (CFU) at the initiation of the experiment was defined as 100%. The mean values from three independent experiments are shown, with error bars indicating standard deviations (SD).
When the cases of invasive disease were sorted according to disease severity, we considered the cases of sepsis with meningitis clinically severe and pooled these with the cases of severe sepsis according to the ACCP/SCCM grading. All NTHi strains were tested for serum resistance in 5% NHS for 20 min. Interestingly, NTHi isolates from patients with severe sepsis and meningitis (n = 9) had a significantly (P = 0.04) higher degree of serum resistance (mean, 63%) than NTHi isolates from patients with sepsis (n = 8) (mean, 31%) (Table 1). When we excluded the cases of sepsis with meningitis from the analysis, the level of serum resistance among the patients with severe sepsis (n = 6) (mean, 72%) was still significantly higher than that among the patients with sepsis (P = 0.04).
No difference in serum resistance was found between invasive and nasopharyngeal NTHi isolates.
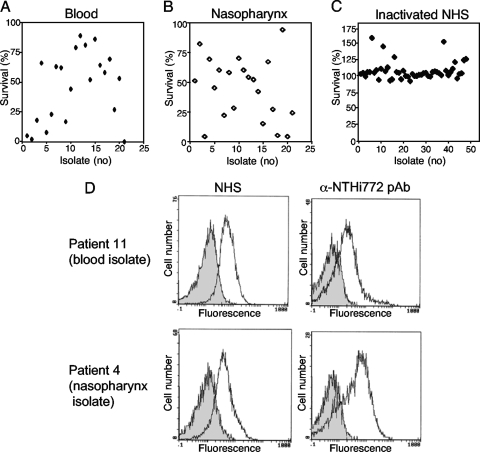
To test whether there was a difference regarding serum resistance between NTHi isolates from separate patients suffering from sepsis (blood-derived isolates) (Table 1) and upper respiratory tract infection (nasopharyngeal isolates) (Table 2), bacteria from the two sites of isolation were compared in a serum bactericidal assay. The nasopharyngeal strains were isolated from immunocompetent patients with suspected bacterial upper respiratory tract infection. Primary viral infection could not, however, be excluded. The age distribution of the patients from whom the nasopharyngeal control isolates were obtained slightly differed from that of the patients with invasive NTHi infection, reflecting the local clinical tradition of taking nasopharyngeal swabs primarily from children. After incubation in NHS (5%), the survival rates of the different NTHi isolates were determined (Fig. 2A and B). Survival rates varied between 2 and 89% for the blood-derived isolates and 4 and 94% for the nasopharyngeal isolates. No significant difference in survival rate was found between the two different sites of isolation, however. All strains included in the study were resistant to hiNHS (Fig. 2C).
TABLE 2.
Demographics of 21 cases of upper respiratory tract infection and serum resistance of NTHi nasopharyngeal isolates in vitro
| Patient | Age (yr) | Gender | % Survival of NTHi strain in serum in vitroa |
|---|---|---|---|
| 1 | 1 | Male | 51 ± 31 |
| 2 | 4 | Female | 82 ± 32 |
| 3 | 3 | Female | 4 ± 5 |
| 4 | 35 | Male | 69 ± 8 |
| 5 | 5 | Male | 45 ± 5 |
| 6 | 36 | Male | 60 ± 41 |
| 7 | 4 | Male | 22 ± 12 |
| 8 | 5 | Female | 58 ± 51 |
| 9 | 8 | Male | 28 ± 4 |
| 10 | 3 | Female | 70 ± 15 |
| 11 | 23 | Female | 60 ± 7 |
| 12 | 0 | Female | 54 ± 40 |
| 13 | 17 | Male | 52 ± 22 |
| 14 | 35 | Male | 42 ± 25 |
| 15 | 4 | Male | 15 ± 1 |
| 16 | 2 | Male | 67 ± 15 |
| 17 | 17 | Male | 27 ± 13 |
| 18 | 2 | Female | 5 ± 3 |
| 19 | 3 | Female | 94 ± 12 |
| 20 | 2 | Male | 4 ± 4 |
| 21 | 1 | Male | 24 ± 19 |
Values shown represent levels observed after 20 min of incubation with 5% NHS and are means ± SD of results from three experiments.
FIG. 2.
NTHi isolates from blood samples (invasive disease) or nasopharynxes of patients with upper respiratory tract infection show variation in serum resistance independent of the presence of IgG in NHS. (A and B) No significant difference in survival rate in NHS was observed between NTHi isolates from blood samples and those from nasopharynxes. (C) All strains were resistant to hiNHS. NTHi isolates from blood samples (n = 21) (A) and nasopharynx (n = 21) (B) were analyzed for survival in human serum. The strains were incubated in the presence of 5% NHS or 5% hiNHS for 20 min. Thereafter, bacteria were spread on chocolate agar plates to allow determination of the number of surviving bacteria. The numbers of bacteria (CFU) at the initiation of the experiment was defined as 100%. (D) The NHS contained IgG directed against NTHi strains independently of whether they were isolated from blood samples or nasopharynxes. Flow cytometry profiles show the deposition of IgG and the binding of a rabbit anti-NTHi antiserum to the surfaces of the NTHi strains from patients 11 (blood) and 4 (nasopharynx). Bacteria were incubated with mouse anti-human IgG or rabbit anti-NTHi antiserum, followed by FITC-conjugated anti-mouse pAb or anti-rabbit pAb. Results for one representative experiment out of three independent ones performed are shown.
To confirm that the NHS used in this study contained antibodies directed against NTHi isolates from both blood samples and nasopharynxes and whether there was a difference in binding of IgG between strains from blood samples and those from nasopharynxes, we analyzed IgG deposition on randomly selected blood-derived (n = 3) and nasopharyngeal (n = 3) isolates. Two typical strains are shown in Fig. 2D. Human IgG was deposited on the blood-derived isolates and the nasopharyngeal isolates to similar extents, suggesting that there were no significant difference in antibody binding to strains from the two anatomical sites of isolation. The anti-NTHi 772 rabbit antiserum was included as a positive control and bound all strains tested. Thus, invasive NTHi strains are not more serum resistant than strains isolated from the upper respiratory tract.
NTHi isolates from blood samples or nasopharynxes bind complement regulators.
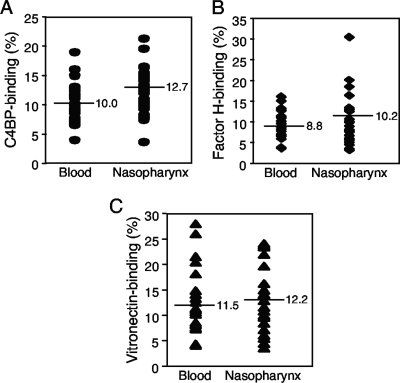
Several bacterial species have been shown to bind complement regulators and thereby counteract the different pathways of the complement system (7, 22, 31). To determine whether there was a difference in binding of various complement inhibitors between NTHi isolates from patients with invasive disease (blood-derived isolates) and those from patients with upper respiratory tract infection (nasopharyngeal isolates), a collection of NTHi strains was incubated with 125I-labeled C4BP, factor H, or vitronectin, followed by separation of unbound ligand. Binding was calculated as the ratio of bound radioactivity versus total radioactivity added. Interestingly, there was no significant difference in binding between blood-derived and nasopharyngeal isolates (Fig. 3). Binding of the complement regulators varied between different strains, independently of isolation sites. The blood-derived and nasopharyngeal strains with the highest levels of C4BP binding bound 19.0% and 21.3%, respectively (Fig. 3A). When factor H was added, the blood-derived and nasopharyngeal strains with the highest levels of factor H binding bound 16% and 30.4% factor H, respectively (Fig. 3B). Finally, the blood-derived strain with the highest level of vitronectin binding bound 28.0% vitronectin, and the nasopharyngeal strain with the highest level of binding bound 24.1% vitronectin.
FIG. 3.
Both blood-derived and nasopharyngeal NTHi isolates bind complement regulators. A series of clinical strains isolated from blood samples (n = 21; cases with invasive disease and an available patient history are listed in Table 1) or nasopharynxes (n = 21; patients with upper respiratory tract infection) was tested for C4BP (A), factor H (B), and vitronectin (C) binding. The different NTHi strains were grown overnight and incubated with 125I-labeled C4BP, factor H, or vitronectin. The binding level was calculated as the percentage of bound radioactivity relative to the level of added radioactivity, measured after separation of free and bound 125I-labeled protein over a sucrose column. The mean values of results from three independent experiments are shown. The mean value for all isolates in the same group is indicated by a line.
In order to analyze whether the strains isolated from blood samples or nasopharynxes also bound the three different complement inhibitors directly from NHS, bacteria were incubated with hiNHS for 1 h at 37°C. Thereafter, bound proteins were eluted, separated by SDS-PAGE, and analyzed by Western blot using specific antibodies directed against human C4BP, factor H, or vitronectin. In these experiments, binding of the three different regulators corresponded with the results from the direct binding assay. Moreover, no significant difference in binding of complement inhibitors could be detected when blood-derived and nasopharyngeal NTHi isolates were compared (data not shown). Taken together, there was no significant difference in binding of the three different complement regulators between blood-derived isolates and nasopharyngeal strains from patients with upper respiratory tract infection, suggesting the importance of the complement regulator binding capacity of NTHi in the upper respiratory tract as well.
DISCUSSION
The results in this study imply that invasive disease caused by NTHi is heterogenous in character. Although it has been reported that NTHi sepsis can have a severe clinical presentation mimicking that of encapsulated strains (40), the proportion of cases in the present study with a severe clinical picture was surprisingly high. The patients affected were all adults, and most cases could be sorted into one of two typical presentations, comprising patients with evidence of immune deficiency and mild clinical presentation and adults with no prior evidence of immune deficiency and severe clinical presentation. In the first category, host factors seem decisive for infection, but in the second category, the virulence of specific bacterial strains could be important, as supported by the higher degree of serum resistance in vitro in this group. Thus, invasive NTHi disease differs from invasive disease caused by Hib in epidemiology as well as general clinical presentation. NTHi can, however, still readily cause severe sepsis in previously healthy individuals. When the individual cases of sepsis were graded by severity according to the ACCP/SCCM grading system, a correlation between severe clinical sepsis and bacterial in vitro serum resistance was found. This indicates that serum resistance is of great importance for the severity of the invasive disease. This finding is consistent with the theory that resistance to human serum facilitates spread of bacteria in the body but has not been demonstrated for NTHi sepsis prior to this study.
An analysis of the clinical data from the sepsis patients showed that many of the cases of NTHi sepsis occurred in immunocompromised hosts. The finding that the severity of the sepsis cases was significantly related to the survival rate of the NTHi strains in serum in vitro implies that while NTHi seems to act as an opportunistic agent in many cases of invasive disease, once it has caused invasive disease, higher complement resistance and survival rates in serum are related to a more clinically severe sepsis. Our data suggest that NTHi septic disease is heterogeneous, possibly reflecting the NTHi cluster in general. The number of cases of invasive disease in this study was limited, however, and this has to be taken into account in the interpretation of the results.
Studies using pulsed-field gel electrophoresis have shown that the dynamics of NTHi carriage in the airways is rapid and that one individual patient can carry more than a dozen different subtypes of NTHi at the same time (39). This raises the issue of selection bias in nasopharyngeal cultures. However, antibodies raised against one NTHi strain give considerable cross-protection against other strains (23). This could partly explain why the incidence of bloodstream infections caused by NTHi is low. In parallel, a decreased humoral immune competence caused by a disease such as leukemia or by immune-modulating pharmaceutical agents could explain some of the cases of NTHi sepsis seen and presented in this study.
In our initial experiments, we incubated invasive isolates with increasing concentrations of NHS and analyzed survival. All strains except one were killed by 15% NHS (Fig. 1 and data not shown). However, even if the majority of a population is sensitive to NHS, it is likely that just a few bacteria may survive and that this particular subpopulation is able to multiply and initiate an infection. Importantly, no significant difference in survival between the various clinical strains from blood samples and those from nasopharynxes was found when serum resistance was determined, suggesting the need for NTHi, irrespective of isolation site, for resistance of bactericidal activity of human serum.
Previous studies showed that the NTHi strain R2866, which was isolated from a child with meningitis, had a high degree of serum resistance depending on the expression of the lipooligosaccharide biosynthesis gene lgtC (9, 19, 40). The phase-variable lgtC expression was demonstrated to inhibit C4b deposition and render the bacteria more resistant to human serum (19). Phase variation of outer membrane proteins and lipooligosaccharides also contributes to virulence of H. influenzae and is involved in evasion of the immune system (39). Intriguingly, phase variation can affect binding of regulatory proteins and has been shown to affect serum resistance (19). Since phase variation is a common phenomenon and depends on the current bacterial environment, it cannot be excluded that phase variation is a factor affecting the in vitro results of our study.
Another aim was to analyze the difference in binding of complement inhibitors and serum resistance between invasive and nasopharyngeal isolates. We have recently demonstrated that NTHi binds C4BP (15) and that both NTHi and Hib bind factor H (17) and vitronectin (14, 16). However, no major differences in binding of complement regulators were detected between strains from the two isolation sites. This indicates that binding of complement regulators is important and possibly facilitates survival and colonization of NTHi both in the airway and in the bloodstream. However, the variations of complement regulator binding capacity and ability to survive in human serum between the different invasive isolates suggest that additional factors are involved in the ability of NTHi to survive in the respiratory tract and in human serum.
The ability of nasopharyngeal isolates to bind factor H, C4BP, and vitronectin suggests that complement regulators may be present in the nasopharyngeal tract and thus would be utilized by NTHi. In fact, during inflammation, complement proteins as well as immunoglobulins and components of the coagulation system enter the airway lumen (11, 12, 29). In patients with chronic otitis media with effusion, local complement activation in the middle ear mucosa, including an intense deposition of C3, has been observed (27). In addition, factor H, factor H-like protein 1, and factor H-related proteins are complement components found in middle ear effusions of patients with otitis media (26).
In conclusion, NTHi isolates from patients with severe sepsis have a higher capacity to resist the bactericidal effect of human serum than NTHi isolates from patients with mild clinical sepsis. Our results also show that it is of importance for NTHi to bind specific complement regulators, irrespective of whether the bacteria are located in the bloodstream or in the nasopharynx, suggesting that this is an adaption to the innate immunity in the upper respiratory tract. In addition to a variation in host factors, virulence factors other than bacterial components binding complement inhibitors are required for determining the invasive capacity of a particular NTHi strain.
Acknowledgments
This work was supported by grants from the Alfred Österlund, the Anna and Edwin Berger, the Marianne and Marcus Wallenberg, and the Greta and Johan Kock Foundations; the Swedish Medical Research Council; the Swedish Society of Medicine; the Cancer Foundation at the University Hospital in Malmo; and Skane County Council's research and development foundation.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Ahren, I. L., D. L. Williams, P. J. Rice, A. Forsgren, and K. Riesbeck. 2001. The importance of a beta-glucan receptor in the nonopsonic entry of nontypeable Haemophilus influenzae into human monocytic and epithelial cells. J. Infect. Dis. 184:150-158. [DOI] [PubMed] [Google Scholar]
- 2.Aubrey, R., and C. Tang. 2003. The pathogenesis of disease due to type b Haemophilus influenzae. Methods Mol. Med. 71:29-50. [DOI] [PubMed] [Google Scholar]
- 3.Bandi, V., M. A. Apicella, E. Mason, T. F. Murphy, A. Siddiqi, R. L. Atmar, and S. B. Greenberg. 2001. Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am. J. Respir. Crit. Care. Med. 164:2114-2119. [DOI] [PubMed] [Google Scholar]
- 4.Bandi, V., M. Jakubowycz, C. Kinyon, E. O. Mason, R. L. Atmar, S. B. Greenberg, and T. F. Murphy. 2003. Infectious exacerbations of chronic obstructive pulmonary disease associated with respiratory viruses and non-typeable Haemophilus influenzae. FEMS Immunol. Med. Microbiol. 37:69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bath, S. 2002. Progress towards elimination of Haemophilus influenzae type b invasive disease among infants and children—United States 1998-2000. MMWR Morb. Mortal. Wkly. Rep. 51:234-237. [PubMed] [Google Scholar]
- 6.Blom, A. M. 2002. Structural and functional studies of complement inhibitor C4b-binding protein. Biochem. Soc. Trans. 30:978-982. [DOI] [PubMed] [Google Scholar]
- 7.Blom, A. M., T. Hallstrom, and K. Riesbeck. 2009. Complement evasion strategies of pathogens-acquisition of inhibitors and beyond. Mol. Immunol. 46:2808-2817. [DOI] [PubMed] [Google Scholar]
- 8.Bone, R. C., W. J. Sibbald, and C. L. Sprung. 1992. The ACCP-SCCM consensus conference on sepsis and organ failure. Chest 101:1481-1483. [DOI] [PubMed] [Google Scholar]
- 9.Erwin, A. L., S. Allen, D. K. Ho, P. J. Bonthuis, J. Jarisch, K. L. Nelson, D. L. Tsao, W. C. Unrath, M. E. Watson, Jr., B. W. Gibson, M. A. Apicella, and A. L. Smith. 2006. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect. Immun. 74:6226-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenwood, F. C., W. M. Hunter, and J. S. Glover. 1963. The preparation of I-131-labelled human growth hormone of high specific radioactivity. Biochem. J. 89:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greiff, L., M. Andersson, J. S. Erjefalt, C. G. Persson, and P. Wollmer. 2003. Airway microvascular extravasation and luminal entry of plasma. Clin. Physiol. Funct. Imaging 23:301-306. [DOI] [PubMed] [Google Scholar]
- 12.Greiff, L., I. Erjefalt, C. Svensson, P. Wollmer, U. Alkner, M. Andersson, and C. G. Persson. 1993. Plasma exudation and solute absorption across the airway mucosa. Clin. Physiol. 13:219-233. [DOI] [PubMed] [Google Scholar]
- 13.Groenewegen, K. H., and E. F. Wouters. 2003. Bacterial infections in patients requiring admission for an acute exacerbation of COPD; a 1-year prospective study. Respir. Med. 97:770-777. [DOI] [PubMed] [Google Scholar]
- 14.Hallstrom, T., A. M. Blom, P. F. Zipfel, and K. Riesbeck. 2009. Nontypeable Haemophilus influenzae protein E binds vitronectin and is important for serum resistance. J. Immunol. 183:2593-2601. [DOI] [PubMed] [Google Scholar]
- 15.Hallstrom, T., H. Jarva, K. Riesbeck, and A. M. Blom. 2007. Interaction with C4b-binding protein contributes to nontypeable Haemophilus influenzae serum resistance. J. Immunol. 178:6359-6366. [DOI] [PubMed] [Google Scholar]
- 16.Hallstrom, T., E. Trajkovska, A. Forsgren, and K. Riesbeck. 2006. Haemophilus influenzae surface fibrils contribute to serum resistance by interacting with vitronectin. J. Immunol. 177:430-436. [DOI] [PubMed] [Google Scholar]
- 17.Hallstrom, T., P. F. Zipfel, A. M. Blom, N. Lauer, A. Forsgren, and K. Riesbeck. 2008. Haemophilus influenzae interacts with the human complement inhibitor factor H. J. Immunol. 181:537-545. [DOI] [PubMed] [Google Scholar]
- 18.Heinen, S., A. Hartmann, N. Lauer, U. Wiehl, H. M. Dahse, S. Schirmer, K. Gropp, T. Enghardt, R. Wallich, S. Halbich, M. Mihlan, U. Schlotzer-Schrehardt, P. F. Zipfel, and C. Skerka. 2009. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114:2439-2447. [DOI] [PubMed] [Google Scholar]
- 19.Ho, D. K., S. Ram, K. L. Nelson, P. J. Bonthuis, and A. L. Smith. 2007. lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J. Immunol. 178:1002-1012. [DOI] [PubMed] [Google Scholar]
- 20.Kilian, M. 2003. Haemophilus, p. 623-635. In P. R. Murray (ed.), Manual of clinical microbology, vol. 8. ASM Press, Washington, DC. [Google Scholar]
- 21.Kirjavainen, V., H. Jarva, M. Biedzka-Sarek, A. M. Blom, M. Skurnik, and S. Meri. 2008. Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein. PLoS Pathog. 4:e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambris, J. D., D. Ricklin, and B. V. Geisbrecht. 2008. Complement evasion by human pathogens. Nat. Rev. Microbiol. 6:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukundan, D., Z. Ecevit, M. Patel, C. F. Marrs, and J. R. Gilsdorf. 2007. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J. Clin. Microbiol. 45:3207-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, T. F., A. L. Brauer, S. Sethi, M. Kilian, X. Cai, and A. J. Lesse. 2007. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J. Infect. Dis. 195:81-89. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, T. F., H. Faden, L. O. Bakaletz, J. M. Kyd, A. Forsgren, J. Campos, M. Virji, and S. I. Pelton. 2009. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr. Infect. Dis. J. 28:43-48. [DOI] [PubMed] [Google Scholar]
- 26.Narkio-Makela, M., J. Hellwage, O. Tahkokallio, and S. Meri. 2001. Complement-regulator factor H and related proteins in otitis media with effusion. Clin. Immunol. 100:118-126. [DOI] [PubMed] [Google Scholar]
- 27.Narkio-Makela, M., J. Jero, and S. Meri. 1999. Complement activation and expression of membrane regulators in the middle ear mucosa in otitis media with effusion. Clin. Exp. Immunol. 116:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, I. S., T. A. Seemungal, M. Wilks, S. J. Lloyd-Owen, G. C. Donaldson, and J. A. Wedzicha. 2002. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 57:759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persson, C. G., I. Erjefalt, U. Alkner, C. Baumgarten, L. Greiff, B. Gustafsson, A. Luts, U. Pipkorn, F. Sundler, C. Svensson, et al. 1991. Plasma exudation as a first line respiratory mucosal defence. Clin. Exp. Allergy 21:17-24. [DOI] [PubMed] [Google Scholar]
- 30.Ronander, E., M. Brant, E. Eriksson, M. Morgelin, O. Hallgren, G. Westergren-Thorsson, A. Forsgren, and K. Riesbeck. 2009. Nontypeable Haemophilus influenzae adhesin protein E: characterization and biological activity. J. Infect. Dis. 199:522-531. [DOI] [PubMed] [Google Scholar]
- 31.Rooijakkers, S. H., and J. A. van Strijp. 2007. Bacterial complement evasion. Mol. Immunol. 44:23-32. [DOI] [PubMed] [Google Scholar]
- 32.Rosell, A., E. Monso, N. Soler, F. Torres, J. Angrill, G. Riise, R. Zalacain, J. Morera, and A. Torres. 2005. Microbiologic determinants of exacerbation in chronic obstructive pulmonary disease. Arch. Intern. Med. 165:891-897. [DOI] [PubMed] [Google Scholar]
- 33.Schvartz, I., D. Seger, and S. Shaltiel. 1999. Vitronectin. Int. J. Biochem. Cell Biol. 31:539-544. [DOI] [PubMed] [Google Scholar]
- 34.Sethi, S., and T. F. Murphy. 2008. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 359:2355-2365. [DOI] [PubMed] [Google Scholar]
- 35.St. Geme, J. W., III. 1993. Nontypeable Haemophilus influenzae disease: epidemiology, pathogenesis, and prospects for prevention. Infect. Agents Dis. 2:1-16. [PubMed] [Google Scholar]
- 36.Tsang, R. S., M. L. Sill, S. J. Skinner, D. K. Law, J. Zhou, and J. Wylie. 2007. Characterization of invasive Haemophilus influenzae disease in Manitoba, Canada, 2000-2006: invasive disease due to non-type b strains. Clin. Infect. Dis. 44:1611-1614. [DOI] [PubMed] [Google Scholar]
- 37.Volanakis, J. E. 2001. Human C-reactive protein: expression, structure, and function. Mol. Immunol. 38:189-197. [DOI] [PubMed] [Google Scholar]
- 38.Walport, M. J. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058-1066. [DOI] [PubMed] [Google Scholar]
- 39.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 40.Williams, B. J., G. Morlin, N. Valentine, and A. L. Smith. 2001. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zipfel, P. F., C. Skerka, J. Hellwage, S. T. Jokiranta, S. Meri, V. Brade, P. Kraiczy, M. Noris, and G. Remuzzi. 2002. Factor H family proteins: on complement, microbes and human diseases. Biochem. Soc. Trans. 30:971-978. [DOI] [PubMed] [Google Scholar]