Abstract
BBL CHROMagar VanRE (CVRE) was compared with bile esculin azide agar plus vancomycin to screen for vancomycin-resistant enterococcus (VRE) colonization. CVRE distinguishes Enterococcus faecalis (green colonies) from Enterococcus faecium (mauve colonies) on the basis of chromogenic substrate use. CVRE sensitivity and specificity were 98.6% and 99.1%. Positive and negative predictive values were 95.9% and 99.7%.
Vancomycin-resistant enterococci (VRE) commonly colonize the gastrointestinal tract of humans and can cause outbreaks of increased colonization rates and occasional extraintestinal infections, including bacteremia and peritonitis in pediatric, elderly, or immunocompromised patients (2). Liver transplant patients and patients with hematologic malignancies are at particular risk for severe disease (3, 6, 7). The prevalence of VRE colonization increases with age and exposure to antibiotics (2). Treatment regimens that include exposure of patients to cephalosporins or glycopeptides that eradicate normal bowel flora often allow VRE to proliferate. Although the practice is controversial (4), many hospitals as a part of their infection prevention measures currently screen at-risk patients for VRE colonization. Contact precautions are typically implemented for VRE-colonized patients to control nosocomial spread of VRE among noncolonized or other high-risk patients.
Screening methods for VRE detection rely on either a culture-based method, generally consisting of an agar medium containing 4 to 8 μg of vancomycin and bile esculin (BEAV) to detect bile esculin-positive vancomycin-resistant organisms, or nucleic acid amplification testing (NAAT). Culture-based screening methods using BEAV require 24 to 48 h to identify positive colonies and another 24 to 48 h to confirm identification and vancomycin resistance (5). Several formulations of media have been developed in recent years that reduce the turnaround time for VRE screening. Many of these media contain chromogenic substrates that allow visual confirmation of positive colonies, thereby reducing or eliminating the need for extensive secondary testing (1). NAAT is more rapid, but increased cost and complexity make this type of testing more burdensome than agar plate screening.
For this study, 400 remnant deidentified consecutive nonduplicated stool specimens were simultaneously cultured on CHROMagar VanRE (CVRE) and BEAV and evaluated for the presence of VRE after 24 and 48 h of incubation at 35 to 37°C in a 5% CO2-enriched atmosphere and ambient air, respectively. Colonies recovered on BEAV were presumptively identified as VRE by the results of testing that included Gram stain morphology analysis, a negative catalase reaction, and PYR (l-pyrrolidonyl-β-naphthylamide) hydrolysis. Isolates were subcultured for phenotypic identification (VITEK2, bioMérieux, Durham, NC) and vancomycin susceptibility testing by broth microdilution. Similarly, colonies producing a characteristic green or mauve color on CVRE (Fig. 1) were subcultured for identification and microbroth susceptibility testing. Sensitivity, specificity, positive predictive values (PPV), and negative predictive values (NPV) were calculated and are presented in Table 1. Quality control testing was performed daily and included both vancomycin-sensitive and vancomycin-resistant Enterococcus spp.
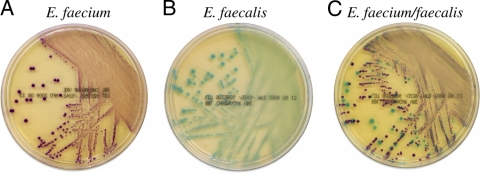
FIG. 1.
Growth of E. faecium (A), E. faecalis (B), and mixed E. faecalis and E. faecium (C) on BBL CHROMagar VanRE.
TABLE 1.
VRE testing resultsa
| Incubation duration (h) and test category | Species | No. of isolates with indicated result |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | n | % Sensitivity | % Specificity | PPV (%) | NPV (%) | ||
| 24 | ||||||||||
| CVRE alone | VREfm | 107 | 273 | 3 | 17 | 400 | 86.3 | 98.9 | 97.3 | 94.1 |
| VREfs | 16 | 379 | 3 | 2 | 400 | 88.9 | 99.2 | 84.2 | 99.5 | |
| VREfm and VREfss | 123 | 652 | 6 | 19 | 800 | 86.6 | 99.1 | 95.3 | 97.2 | |
| CVRE + catalase, | VREfm | 107 | 274 | 2 | 17 | 400 | 86.3 | 99.3 | 98.2 | 94.2 |
| Gram stain | VREfs | 16 | 379 | 3 | 2 | 400 | 88.9 | 99.2 | 84.2 | 99.5 |
| VREfm and VREfs | 123 | 653 | 5 | 19 | 800 | 86.6 | 99.2 | 96.1 | 97.2 | |
| 48 | ||||||||||
| CVRE alone | VREfm | 123 | 272 | 4 | 1 | 400 | 99.2 | 98.6 | 96.9 | 99.6 |
| VREfs | 17 | 373 | 10 | 0 | 400 | 100.0 | 97.4 | 63.0 | 100.0 | |
| VREfm and VREfs | 140 | 651 | 7 | 2 | 800 | 98.6 | 98.9 | 95.2 | 99.7 | |
| CVRE + catalase, | VREfm | 123 | 274 | 2 | 1 | 400 | 99.2 | 99.3 | 98.4 | 99.6 |
| Gram stain | VREfs | 17 | 378 | 5 | 0 | 400 | 100.0 | 98.7 | 77.3 | 100.0 |
| VREfm and VREfs | 140 | 652 | 6 | 2 | 800 | 98.6 | 99.1 | 95.9 | 99.7 | |
TP (true positive), mauve or green colonies on CVRE, black on BEAV, confirmed by Vitek and MIC testing to represent VRE; TN (true negative), no mauve or green colonies on CVRE, no VRE from BEAV; FP (false positive), mauve or green colonies on CVRE, no VRE isolated on BEAV; FN (false negative), no mauve or green colonies on CVRE, VRE isolated on BEAV. “CVRE alone” data do not include catalase and Gram stain results. “CVRE + catalase + Gram stain” data include CVRE results along with catalase and Gram stain results.
One hundred and twenty-four VRE E. faecium (VREfm) and 18 VRE E. faecalis (VREfs)-positive specimens were recovered from the traditional media. With no additional testing, CVRE provided a sensitivity and specificity of 86.3% and 98.9% for the detection of VREfm and 88.9% and 99.2% for the detection of VREfs when read after 24 h of incubation. If the incubation was extended to 48 h, the sensitivity and specificity of the medium were 99.2% and 98.6% for VREfm and 100% and 97.4% for VREfs. Performing catalase and Gram stain testing of presumptively positive isolates increased the specificity slightly for both E. faecium and E. faecalis. The prevalence of VREfm colonization at our institution was 31%, while VREfs was present in 4.5% of the specimens tested. PPV and NPV values for each are shown in Table 1. The cumulative PPV values for all VRE were 96.1% (24 h) and 95.9% (48 h). Cumulative NPV for all VRE were 97.2% (24 h) and 99.7% (48 h).
False-positive results for VREfs (green colonies) were predominantly caused by yeast colonies (5 of 10 false-positive colonies), which came up slowly and turned the medium green by 48 h of incubation and led to a fairly low positive predictive value for VREfs at 48 h. However, when Gram stain and catalase testing were included in the analysis, the PPV improved from 63.0% to 77.3%. Three additional false-positive results for VREfs were found for single isolates of E. casseliflavus, Streptococcus mutans, and E. raffinosis. Simple additional testing could have eliminated all of these false-positive results (a motility test for E. casseliflavus, PYR testing for S. mutans, and raffinose utilization for E. raffinosis). There were one E. faecium isolate and one E. faecalis isolate that produced green colonies on CVRE and not found on BEAV. The false-positive Enterococcus spp. were confirmed to be VRE but were not isolated from BEAV, so they were counted as false positives in the analysis. There were four “false-positive” VREfm (mauve colony) isolates, namely, one Staphylococcus sciuri isolate, one Gram-negative rod isolate, and two VREfm isolates not identified on BEAV. Of these, Gram staining would have eliminated the Gram-negative rod isolate and a positive catalase test result would have eliminated the S. sciuri isolate. Performing Gram stain and catalase testing alone increased the PPV for VREfm from 96.9% to 98.4%.
We conclude that CVRE is a rapid, sensitive, and specific medium for the detection of VRE from stool specimens at 24 h. A further gain in sensitivity can be attained by holding the plates for a 48-h evaluation. Performing rapid catalase and Gram stain testing increases the specificity of the medium. Screening for VRE by the use of CVRE medium is more rapid than traditional screening. This medium performs as well as the gold standard BEAV medium and caneliminate the need for extensive biochemical identification and antibiotic susceptibility testing.
Acknowledgments
This study was funded by Becton Dickinson, Baltimore, MD.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Ledeboer, N. A., R. J. Tibbetts, and W. M. Dunne. 2007. A new chromogenic agar medium, chromID VRE, to screen for vancomycin-resistant Enterococcus faecium and Enterococcus faecalis. Diagn. Microbiol. Infect. Dis. 59:477-479. [DOI] [PubMed] [Google Scholar]
- 2.Bonten, M. J., S. Slaughter, A. W. Ambergen, M. K. Hayden, J. van Voorhis, C. Nathan, and R. A. Weinstein. 1998. The role of “colonization pressure” in the spread of vancomycin-resistant enterococci: an important infection control variable. Arch. Intern. Med. 158:1127-1132. [DOI] [PubMed] [Google Scholar]
- 3.Russell, D. L., A. Flood, T. E. Zaroda, C. Acosta, M. M. Riley, R. W. Busuttil, and D. A. Pegues. 2008. Outcomes of colonization with MRSA and VRE among liver transplant candidates and recipients. Am. J. Transplant. 8:1737-1743. [DOI] [PubMed] [Google Scholar]
- 4.Weber, S. G., S. S. Huang, S. Oriola, W. C. Huskins, G. A. Noskin, K. Harriman, R. N. Olmsted, M. Bonten, T. Lundstrom, M. W. Climo, M. C. Roghmann, C. L. Murphy, and T. B. Karchmer. 2007. Legislative mandates for use of active surveillance cultures to screen for methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: position statement from the Joint SHEA and APIC Task Force. Am. J. Infect. Control 35:73-85. [DOI] [PubMed] [Google Scholar]
- 5.Winn, W. C., and E. W. Koneman. 2006. Streptococci, enterococci and the streptococcus-like bacteria, p. 672-764. Koneman's color atlas and textbook of diagnostic microbiology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 6.Worth, L. J., K. A. Thursky, J. F. Seymour, and M. A. Slavin. 2007. Vancomycin-resistant Enterococcus faecium infection in patients with hematologic malignancy: patients with acute myeloid leukemia are at high-risk. Eur. J. Haematol. 79:226-233. [DOI] [PubMed] [Google Scholar]
- 7.Zaas, A. K., X. Song, P. Tucker, and T. M. Perl. 2002. Risk factors for development of vancomycin-resistant enterococcal bloodstream infection in patients with cancer who are colonized with vancomycin-resistant enterococci. Clin. Infect. Dis. 35:1139-1146. [DOI] [PubMed] [Google Scholar]