Abstract
Novel human adenoviruses (HAdVs) arise from genome recombination. Analysis of HAdV type 55 from an outbreak in China shows a hexon recombination between HAdV-B11 and HAdV-B14, resulting in a genome that is 97.4% HAdV-B14. Sporadic appearances as a re-emergent pathogen and misidentification as “HAdV-B11a” are due to this partial hexon.
Human adenoviruses (HAdVs) were first identified as respiratory pathogens (7, 18) but are now recognized as causing a range of diseases, including those that are ocular, gastrointestinal, and metabolic (4). There are 51 “serotypes” defined by using biological characteristics, including immunochemical methods, e.g., serum neutralization and hemagglutination (4). A novel HAdV was characterized using nonimmunochemical methods, e.g., genomics and bioinformatics, and was named HAdV-G52 (9). Recently, genomics and bioinformatics data have defined two emergent, pathogenic, and recombinant HAdVs (D53 and D54) (8, 21). All three are characterized by genomics and therefore should be termed appropriately as “type” rather than “serotype,” as discussed at the 9th International Adenovirus Meeting (Dobogókő, Hungary, April 2009) (our unpublished data). Described here is a re-emergent respiratory pathogen, HAdV-B55, which contains a partial hexon recombination conferring a change in serotype and, hence, an escape from immune reactivity against HAdV-B14. It was isolated recently from an acute respiratory disease (ARD) outbreak in China and incorrectly termed “HAdV-B11-like” (22, 23).
Genomes of “HAdV-B11 strain QS-DLL,” noted here as HAdV-B55 (accession no. FJ643676), HAdV-11p (accession no. AF532578), and HAdV-B14p (accession no. AY803294), were obtained from GenBank. Computational analysis included sequence comparisons with zPicture (15), restriction enzyme pattern analysis (http://www.acaclone.com/), phylogeny analysis using MAVID (1), and recombination analysis with SimPlot and Bootscan (12). Genome percent identities were determined using zPicture, MAFFT (10), and Chimera (16).
The genome “chassis” of HAdV-B55 is HAdV-B14 (19), an ARD pathogen, with a partial HAdV-B11 hexon providing the misleading serum neutralization result and incorrect, incomplete original identification, as well as an incorrect clinical diagnosis. HAdV-B55 hexon contains 907 nucleotides from HAdV-B11 out of 2,841 nucleotides (31.9%), embedded in the 34,755 base pair genome (2.6%) (Fig. 1A and B). Sequence comparisons reflect the following: HAdV-B55 versus HAdV-B14 at 98.86%; HAdV-B55 versus HAdV-B11 at 97.64%; and HAdV-B11 versus HAdV-B14 at 97.21% (Fig. 1C). HAdV-B55 appears to have evolved from a recombination between HAdV-B14 and -B11 and should be noted as a new type, in the context of both the recombinant HAdVs that are accepted as novel types (8, 21) and its change in serotype (22). In phylogeny analyses, HAdV-B55 groups into a subclade with HAdV-B14 and HAdV-B14a, whereas hexon (loops 1 and 2) groups into a subclade with HAdV-B11. As “HAdV-B11a” did not cross-react with HAdV-B14 antisera, it is not a simple variant of HAdV-B14. Detailed restriction enzyme (RE) digestion pattern analysis, seemingly anachronistic but highly effective visually, confirms this, showing HAdV-B55 as closer to the HAdV-B14 genome than to the HAdV-B11 genome (data not shown), and also matching the original RE analysis for HAdV-B11a (11).
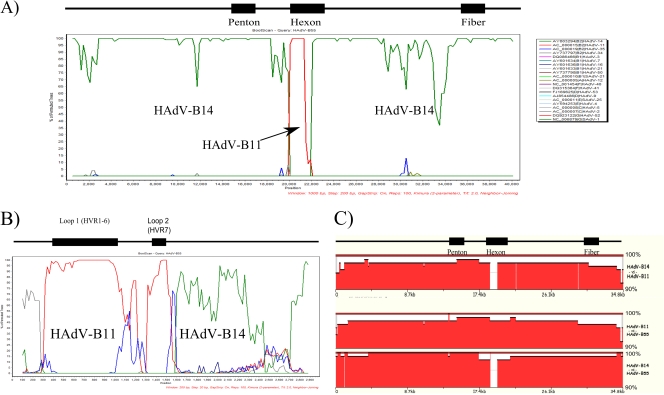
FIG. 1.
(A) Bootscan recombination analysis of the HAdV-B55 genome. The dark green reflects HAdV-B14, and the red indicates HAdV-B11 sequences. Parameters are as follows: 1,000-bp window; 200-bp step; 100 repetitions; neighbor-joining algorithm. The HAdV types analyzed and the colors are the same for both panels A and B, in descending order: HAdV-B14 (top; dark green), B11, B35, B34, B3, B7, B16, B21, B50, simian adenovirus (SAdV)-B21, A12, F40, F41, D53, D9, SAdV-E25, E4, C5, C2, G52, and SAdV-G1 (bottom; light green). (B) Fine-resolution analysis of the partial hexon gene recombination. Bootscan analysis of the hexon gene shows that the proximal region, containing HVR1-6 (“Loop 1”; 407 to 1,034 nucleotide gene location) and HVR7 (“Loop 2”; 1,363 to 1,484 nucleotide gene location), derives from HAdV-B11. The distal portion derives from HAdV-B14, as does the rest of the genome. Parameters are as follows: 200-bp window; 20-bp step; 100 repetitions; neighbor-joining algorithm. (C) Comparative genomics. zPicture compares the nucleotide sequences of pairs of genomes by using a moving overlapping window and scoring percent identities. The y axis is set between 90 and 100%, indicating a very high level of identity.
The divergent hexon comprises two regions of identity indicating a recombination event within the gene (Fig. 1B). Analysis of an earlier described “HAdV-B11a” hexon (GenBank accession no. AY972815) (2) shows it is identical to the one embedded in the “QS” genome (23), suggesting that “HAdV-B11a” was incorrectly characterized earlier as a variant of HAdV-B11 (2, 6, 11, 22) by the use of “then-available” assays. Serum neutralization has the hexon as its target, as does limited molecular typing (3, 13). Since genome rearrangements are missed by serological and molecular assays against a limited repertoire, it is problematic to rely solely on these methods to characterize a virus thoroughly. Thus, due to the limitations of these assays, the original “HAdV-B11a” isolates have been misnamed as well (2, 6, 11).
As “HAdV-B11a,” this virus represented a paradox. HAdV-B11 is a member of subspecies B2 and was not originally associated with respiratory disease (14); only HAdV-B14 in this group is associated with respiratory disease (20). A clue as to the incorrect association of HAdV-B11 as a respiratory pathogen lies in the reports that “HAdV-B11a” was unusual in having cellular tropism to respiratory epithelial cells rather than renal cells, as observed for original and subsequent isolates of HAdV-B11 (11, 23). In addition, the original “HAdV-B11a” did not agglutinate monkey erythrocytes, unlike the true HAdV-B11 field strains and the prototype Slobitski strain (5). If HAdV-B55 is truly a variant of HAdV-B11 and an ARD pathogen, it presents a drastic change in cell and tissue tropism—from renal and urinary tract to lung, with high morbidity and some mortality (11, 23). This paradox is resolved by the analysis of its genome, that it is HAdV-B14-like rather than HAdV-B11-like. By inference, the cell recognition epitope is either the distal 68.1% of the HAdV-B14 hexon or the fiber, rather than the proximal 31.9% encompassing “loops 1 and 2” of the HAdV-B11 hexon.
Although originally identified in sporadic and infrequent historical ARD outbreaks by serum neutralization as “HAdV-B11-like” and recently recharacterized by immunochemistry (serum neutralization and enzyme-linked immunosorbent assay [ELISA]) and limited molecular typing (PCR and hexon sequencing) also as “HAdV-B11-like” (2, 6, 11, 22), genomics and bioinformatics demonstrate that it is a novel type. All of these older techniques, assaying only the hexon gene, simply reconfirmed a HAdV-B11-like hexon epitope. Therefore, it is incorrectly named and noted in GenBank (accession no. FJ643676) as “HAdV-B11a” or “HAdV-B11 strain QS-DLL” (23). Based on the reported and additional computational analyses and within the context of recent reports of HAdV molecular evolution based on recombination (8, 17, 21), this pathogen is a novel adenovirus type that should be named HAdV-B55.
A tsunami of genome sequence information from both newly isolated and presciently archived HAdV strains and their accompanying bioinformatics are leading to an in-depth understanding of the biology of HAdVs. Correct identification and nomenclature of viruses are critical for defining and understanding them as well as their pathogenic variants, particularly in GenBank. The correction of the name of this re-emergent type is urgent, as research groups characterizing other ARD outbreaks by similar or the same HAdV pathogens are on the verge of reporting their findings, adding to the confusion. Genome recombination plays an important role in the molecular evolution of HAdVs, with consequences of newly emerging strains and new understandings of re-emerging pathogens that have tropism change or that become more virulent. An intriguing thought for HAdV-B55 is that perhaps this region of the hexon is a recombination hot spot, driving the evolution, selection, and appearance of a recurrent re-emergent pathogen by altering its serotype and giving it an advantage in a population with seroprevalence against HAdV-B14. As more HAdV genomes are elucidated, they will lead to better understandings of other pathogens that may utilize similar evolution pathways in appearing as emergent and re-emergent pathogens.
Acknowledgments
We thank Qiwei “Kevin” Zhang, The University of Hong Kong, for assistance in obtaining the prepublication genome of HAdV-B55.
This work was supported in part by U.S. Public Health Service NIH grants EY013124 (D.S., M.P.W., M.S.J., and J.C.) and P30EY013104 (J.C.). J.C. is also supported by funding from the Massachusetts Lions Eye Research Fund, Inc., and an unrestricted grant to the Department of Ophthalmology, Harvard Medical School, from Research to Prevent Blindness, Inc. M.S.J. is supported by the United States Air Force Surgeon General, Clinical Investigation no. FDG20040024E.
The views expressed in this material are those of the authors and do not reflect the official policy or position of the U.S. Government, the Department of Defense, or the Department of the Air Force.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Bray, N., and L. Pachter. 2004. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 14:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chmielewicz, B., J. Benzler, G. Pauli, G. Krause, F. Bergmann, and B. Schweiger. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77:232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echavarria, M. 2009. Adenovirus, p. 463-488. In A. J. Zuckerman, J. E. Banatvala, B. D. Schoub, P. D. Griffiths, and P. Mortimer (ed.), Principles and practice of clinical virology, 6th ed. John Wiley and Sons, San Diego, CA.
- 5.Hierholzer, J. C., and A. Pumarola. 1976. Antigenic characterization of intermediate adenovirus 14-11 strains associated with upper respiratory illness in a military camp. Infect. Immun. 13:354-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hierholzer, J. C., A. Pumarola, A. Rodriguez-Torres, and M. Beltran. 1974. Occurrence of respiratory illness due to an atypical strain of adenovirus type 11 during a large outbreak in Spanish military recruits. Am. J. Epidemiol. 99:434-442. [DOI] [PubMed] [Google Scholar]
- 7.Hilleman, M. R., and J. H. Werner. 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 85:183-188. [DOI] [PubMed] [Google Scholar]
- 8.Ishiko, H., and K. Aoki. 2009. Spread of epidemic keratoconjunctivitis due to a novel serotype of human adenovirus in Japan. J. Clin. Microbiol. 47:2678-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones, M. S., II, B. Harrach, R. D. Ganac, M. M. Gozum, W. P. Dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 81:5978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh, K., and H. Toh. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics 9:286-298. [DOI] [PubMed] [Google Scholar]
- 11.Li, Q. G., J. Hambraeus, and G. Wadell. 1991. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology 32:338-350. [DOI] [PubMed] [Google Scholar]
- 12.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madisch, I., G. Harste, H. Pommer, and A. Heim. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 79:15265-15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Numazaki, Y., S. Shigeta, T. Kumasaka, T. Miyazawa, M. Yamanaka, N. Yano, S. Takai, and N. Ishida. 1968. Acute hemorrhagic cystitis in children. Isolation of adenovirus type II. N. Engl. J. Med. 278:700-704. [DOI] [PubMed] [Google Scholar]
- 15.Ovcharenko, I., G. G. Loots, R. C. Hardison, W. Miller, and L. Stubbs. 2004. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res. 14:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 17.Robinson, C. M., J. Rajaiya, M. P. Walsh, D. Seto, D. W. Dyer, M. S. Jones, and J. Chodosh. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J. Virol. 83:8980-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe, W. P., R. J. Huebner, L. K. Gilmore, R. H. Parrott, and T. G. Ward. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570-573. [DOI] [PubMed] [Google Scholar]
- 19.Seto, J., M. P. Walsh, P. Mahadevan, A. Purkayastha, J. M. Clark, C. Tibbetts, and D. Seto. 2009. Genomic and bioinformatics analyses of HAdV-14p, reference strain of a re-emerging respiratory pathogen and analysis of B1/B2. Virus Res. 143:94-105. [DOI] [PubMed] [Google Scholar]
- 20.Van Der Veen, J., and G. Kok. 1957. Isolation and typing of adenoviruses recovered from military recruits with acute respiratory disease in The Netherlands. Am. J. Hyg. 65:119-129. [DOI] [PubMed] [Google Scholar]
- 21.Walsh, M. P., A. Chintakuntlawar, C. M. Robinson, I. Madisch, B. Harrach, N. R. Hudson, D. Schnurr, A. Heim, J. Chodosh, D. Seto, and M. S. Jones. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, Z., Z. Zhu, L. Tang, L. Wang, X. Tan, P. Yu, Y. Zhang, X. Tian, J. Wang, Y. Zhang, D. Li, and W. Xu. 2009. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 47:3082-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu, Z., Y. Zhang, S. Xu, P. Yu, X. Tian, L. Wang, Z. Liu, L. Tang, N. Mao, Y. Ji, C. Li, Z. Yang, S. Wang, J. Wang, D. Li, and W. Xu. 2009. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 47:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]