Abstract
Coagulase-negative staphylococci (CNS) are among the most frequently isolated bacterial species in clinical microbiology, and most CNS-related infections are hospital acquired. Distinguishing between these frequently multiple-antibiotic-resistant isolates is important for both treatment and transmission control. In this study we used isolates of methicillin-resistant coagulase-negative staphylococci (MR-CNS) that were selected from a large surveillance study of the direct spread of MR-CNS. This strain collection was used to evaluate (i) Raman spectroscopy as a typing tool for MR-CNS isolates and (ii) diversity between colonies with identical and different morphologies. Reproducibility was high, with 215 of 216 (99.5%) of the replicate samples for 72 isolates ending up in the same cluster. The concordance with pulsed-field gel electrophoresis (PFGE)-based clusters was 94.4%. We also confirm that the skin of patients can be colonized with multiple MR-CNS types at the same time. Morphological differences between colonies from a single patient sample correlated with differences in Raman and PFGE types. Some morphologically indistinguishable colonies revealed different Raman and PFGE types. This indicates that multiple MR-CNS colonies should be examined to obtain a complete insight into the prevalence of different types and to be able to perform an accurate transmission analysis. Here we show that Raman spectroscopy is a reproducible typing system for MR-CNS isolates. It is a tool for screening variability within a collection of isolates. Because of the high throughput, it enables the analysis of multiple colonies per patient, which will enhance the quality of clinical and epidemiological studies.
Coagulase-negative staphylococci (CNS) are among the most frequently isolated bacterial species in clinical microbiology. They colonize the human skin, throat, nose, and/or gut and represent a major part of the normal bacterial flora of healthy people (15, 20). Although CNS have long been regarded as nonpathogenic, they have now been recognized as relevant opportunistic pathogens (23).
Most CNS-related infections are hospital acquired. Since many nosocomial isolates are able to form biofilms on indwelling devices such as intravascular catheters, the eradication of CNS is difficult, and the removal of the catheter or device can be the only effective intervention. Bloodstream infections (BSI) caused by these organisms are frequently related to the use of medical devices. They account for significant morbidity and mortality, especially in immunocompromised individuals and neonates (16, 17, 20).
A likely source of CNS isolates involved in catheter-related infections appears to be the skin flora. Several studies indicate that subtle morphological differences can be found among CNS colonies cultured from infected catheters, suggesting that multiple CNS types are present (3, 5, 18).
Furthermore, it has been shown that CNS bacteria can be transmitted by hospitalized patients and health care workers (1, 13, 21). Isolates involved in transmission are often characterized by their resistance to many commonly used antibiotics, including methicillin (6, 9, 14). Although several studies suggest that colonized patients and health care workers should be considered reservoirs for these microorganisms, sometimes too little attention is paid to the control of these multiresistant isolates (7, 8, 23).
Distinction between clinically significant and contaminating isolates as well as determining possible transmission routes in infection prevention and surveillance studies can be important. This, however, requires a more detailed characterization of isolates using an appropriate typing method. The most widely used typing method for outbreak analysis and surveillance of CNS is pulsed-field gel electrophoresis (PFGE). However, this technique is laborious and has a long turnaround time and low sample throughput, which makes it unsuitable for routine use and large-scale clinical studies. Currently, the “gold standard” to assess phylogenic relatedness between isolates is multilocus sequence typing (MLST). Recently, Raman spectroscopy has been reported to be a powerful tool for the identification of microorganisms at the species and subspecies levels. The technique is fast and easy to use. Raman spectra of bacteria are a representation of their overall molecular composition and can be used as strain-specific spectroscopic fingerprints (10, 12, 22). By using a collection of Staphylococcus aureus isolates, it was shown that the typing results obtained are comparable to those obtained by PFGE (22). Highly similar groupings compared to amplification fragment length polymorphism (AFLP) were previously documented for a collection of Acinetobacter isolates (10).
In this study we used a collection of methicillin-resistant coagulase-negative staphylococci (MR-CNS) to illustrate that Raman spectroscopy can be used as a rapid and reproducible typing technique for MR-CNS isolates. We confirm that the skin of hospital patients can be colonized with multiple MR-CNS types, and therefore, screening for variability is essential for accurate transmission analysis.
MATERIALS AND METHODS
Culture and storage of isolates.
Isolates were selected from a large surveillance study performed at Erasmus MC (Rotterdam, The Netherlands) to evaluate risk factors for acquiring MR-CNS.
Therefore, patients were sampled daily during their hospital stay by swabbing of the lower part of the arm, just above the inside of the wrist. At this location, it is easy to reproducibly collect samples by different health care workers, and it is not incriminating for the patient. The study was approved by the Erasmus MC ethical review board.
Swabs were incubated at 35°C in phenol red mannitol broth (Becton Dickinson, Franklin Lakes, NJ) containing 5 μg/ml of ceftizoxime and 75 μg/ml of aztreonam. After 48 h, all cultures were plated onto a Columbia blood agar plate (Becton Dickinson) and incubated for 48 h at 35°C. Isolates were stored at −80°C in glycerol-containing brain heart infusion broth (Becton Dickinson) until further use. Colonies with different visual morphologies were stored as different MR-CNS types. For colonies with identical visual morphologies, only one colony was stored.
MR-CNS isolates.
In total, 162 isolates obtained from 19 patients were selected from the surveillance study. These isolates were divided into 3 collections.
Collection I was used as a reference collection to evaluate the reproducibility of Raman spectroscopy and the diversity of MR-CNS types over time. This collection contained 72 MR-CNS isolates obtained from 5 different patients. Isolates were collected over a period of 8 to 27 days, depending on the admission period. To determine the reproducibility of the measurements, so-called full biological replicates were performed. This means that for each replicate, a separate new culture and a separate new sample preparation were performed on separate days.
Collection II was used to evaluate the clonal diversity of CNS isolates showing differences in colony morphology. This collection contained isolates obtained from 4 patients. For each patient, a culture showing two morphotypes was selected. For each morphotype, 5 morphologically identical colonies were selected from the initial growth medium, resulting in 40 isolates.
Collection III was used to evaluate the clonal diversity of CNS isolates with identical colony morphologies. This collection consisted of 50 isolates that were obtained from 10 patients. For each patient, a culture showing one morphotype was selected. For each culture, 5 morphologically identical colonies were selected.
PFGE.
Isolates were cultured on Columbia blood agar plates and incubated overnight at 35°C. PFGE was performed as described previously (4). Briefly, a suspension of bacteria was mixed with 1% InCert agarose (FMC Bioproducts). Agarose plugs were incubated with lysostaphin (Sigma-Aldrich, Zwijndrecht, The Netherlands), and spheroplasts were lysed by using proteinase K (Sigma-Aldrich). DNA was digested by SmaI (Fermentas, St. Leon-Rot, Germany). Macrorestriction fragments were separated by using a Bio-Rad CHEF Mapper apparatus (Bio-Rad, Veenendaal, The Netherlands), with a total run time of 20 h. PFGE type definition was based on the presence of unique banding patterns.
Culture and sample preparation for Raman spectroscopy.
Culturing of bacterial isolates and sample preparation prior to Raman measurements were performed as described previously (22). Briefly, all isolates were grown for 20 h at 35°C on Trypticase soy agar (Becton Dickinson). The biomass was suspended in sterilized distilled water (prepared in-house) and transferred onto a quartz slide (Hellma Benelux, Rijswijk, The Netherlands). A removable silicone isolator (Sigma-Aldrich) containing 24 wells was placed onto this slide. Samples were allowed to dry for 20 min at 35°C.
Raman spectroscopy.
Raman measurements were performed as described previously (2, 22). Raman spectra were collected by using a high-performance Raman Module 2500 apparatus coupled to a custom-built inverted microscope stage with an automated XYZ stage (River Diagnostics BV, Rotterdam, The Netherlands). Automated data collection was performed by using RiverIcon software, version 1.63 (River Diagnostics). To account for sample heterogeneity, for each sample, spectra were measured at multiple positions by using a 1-s signal collection time per measurement. The total measurement time per sample was 40 to 60 s.
Data analysis. (i) Signal preprocessing.
All measured spectra were processed to remove noninformative signal variance, such as the background signal contribution from the fused silica substrate. Small variations in biochemical composition between independent cultures of an isolate were corrected as described previously (11). Software scripts for spectrum pretreatment and data analysis were written with MATLAB, version 7.1 (MathWorks).
(ii) Calculation of similarities between spectra.
The similarity between pairs of spectra was expressed as a percentage and calculated by using the squared Pearson correlation coefficient (R2) multiplied by 100.
(iii) Reproducibility of Raman measurements.
Each isolate of collection I was cultured and measured three times. For each isolate, the R2 values were calculated over the Raman spectra obtained for three replicate measurements. The lowest R2 value was selected as the minimum correlation between replicate samples of an isolate. This value was named the ReproMatch and accounts for any signal variance due to differences in culturing, sample preparation, or actual Raman measurements.
(iv) Hierarchical cluster analysis (HCA).
To analyze spectral relationships between different isolates, a cluster analysis of sets of spectra was performed by using the pairwise similarities as a distance matrix in combination with Ward's cluster algorithm (22). This resulted in dendrograms in which each node defines a cluster and represents the lowest correlation coefficient (or similarity) between isolates in that cluster.
(v) Interpretation of dendrograms.
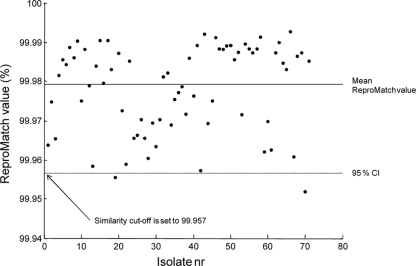
To interpret spectroscopic relationships between isolates as displayed in the dendrograms, a similarity cutoff level must be defined. In this study, the 95% confidence interval (CI) was calculated from all ReproMatch values to exclude clear ReproMatch outliers. The isolate with the lowest ReproMatch value within the CI determined the similarity cutoff level. Clusters of isolates showing a similarity above the cutoff level were considered to be clonally related, and isolates showing a similarity below the cutoff level were considered to be clonally unrelated. All clusters defined with this procedure could then be validated by using epidemiological data, previously known typing results for the isolates, or a collection of reference isolates.
RESULTS
Concordance with PFGE.
All 162 isolates selected for this study were analyzed by using Raman spectroscopy and PFGE. The Raman clustering found was in good concordance with the results obtained by using PFGE, since for 153 out of 162 isolates (94.4%), the classifications found with both techniques were identical. In total, 46 different Raman types were found, compared to 49 different unique PFGE patterns. In four Raman clusters, isolates that showed 2 or 3 different PFGE patterns were combined. On the other hand, it was found for two PFGE patterns that isolates were divided between 2 different Raman clusters.
Reproducibility of Raman spectroscopy results.
The 72 isolates of collection I were cultured and measured in triplicate, leading to 216 samples. For each isolate, the minimal ReproMatch value was calculated as described above. These values are displayed in Fig. 1. The average ReproMatch value was high (99.978%). Based on the 95% CI of the ReproMatch values, the similarity cutoff level was set to 99.957%. After using this cutoff level for the HCA analysis of collection I, 215 out of 216 samples clustered according to the isolate. The reproducibility of the Raman method was therefore calculated to be 99.5%.
FIG. 1.
ReproMatch values and similarity cutoff levels for all MR-CNS isolates of collection I. Isolate nr, isolate number.
Diversity of MR-CNS types over time.
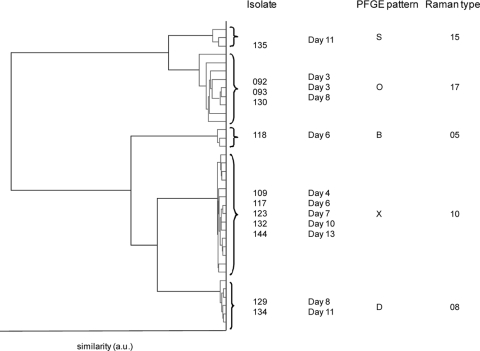
The results obtained for collection I indicate that for the 5 selected patients, multiple Raman and PFGE patterns were found in time. To illustrate this finding, the results of the cluster analysis of the isolates obtained from patient 1 are given in Fig. 2. For this patient, 11 swabs were collected over a period of 13 days. Colonies with identical morphotypes were found in 4 cultures, colonies of two morphotypes were found in 4 cultures, and on 3 days, the culture was negative for MR-CNS. In total, 5 different Raman types and 5 different PFGE patterns were found; the concordance between both methods was 100%. Raman types 10 and 17 were dominantly present and were found more frequently over time (for 5 and 3 isolates, respectively). Raman types 5 and 8 were isolated only once during this period.
FIG. 2.
Raman-based clustering found for triplicate measurements from samples from patient 1. The legend indicates the isolate number, collection date of the skin swab, the PFGE pattern found, and the Raman type found for each isolate. a.u., arbitrary units.
Diversity in colonies with different morphologies.
The results for the 40 isolates of collection II are given in Table 1.
TABLE 1.
Results of Raman spectroscopy and PFGE analysis of MR-CNS isolates of collection IIa
| Patient | Morphotype | Raman type(s) | PFGE pattern(s) |
|---|---|---|---|
| 6 | A | 30 | AG |
| B | 29, 32, 33 | AH, AI, AJ | |
| 7 | C | 23 | Z |
| D | 31 | AA | |
| 8 | E | 24, 26 | AB, AC |
| F | 25 | AD | |
| 9 | G | 27, 28 | AE |
| H | 34 | AF |
For each of the 4 patients, cultures showing 2 morphotypes were selected. For each morphotype, 5 isolates were analyzed.
All four patients showed different Raman types and PFGE patterns for isolates with different morphotypes. These findings point out that in this study, differences in colony morphology correspond to different pheno- or genotyped isolates. When the five selected colonies with identical morphotypes were compared, it was found that identical MR-CNS types were found for 5 out of 8 morphotypes. For morphotypes 6B and 8E, multiple Raman types were found, in concordance with PFGE results. For morphotype 9G, the 5 isolates with PFGE pattern “AE” were divided into 2 Raman clusters. This was further examined with collection III.
Diversity in colonies with identical morphologies.
Table 2 displays the results obtained for the 50 isolates of collection III.
TABLE 2.
Results of Raman spectroscopy and PFGE analysis of MR-CNS isolates of collection IIIa
| Patient | Morphotype | Raman type(s) | PFGE pattern(s) |
|---|---|---|---|
| 10 | I | 35, 46 | AK, AL |
| 11 | J | 38 | AM |
| 12 | K | 36 | AN |
| 13 | L | 41 | AO |
| 14 | M | 37, 42 | AP, AQ |
| 15 | N | 44 | AR |
| 16 | O | 39 | AS |
| 17 | P | 43 | AT |
| 18 | Q | 45 | AU |
| 19 | R | 40 | AV, AW |
For each of the 10 patients, cultures showing 1 morphotype were selected. For each morphotype, 5 isolates were analyzed.
When the Raman clustering and PFGE typing results were compared, only one discrepancy was found, since for patient 19, the 5 isolates with Raman type 40 showed two different PFGE patterns.
For 8 out of 10 patients, the five colonies selected from one culture shared the same Raman type and PFGE pattern, indicating that an identical morphotype corresponded with isolates that were identical at the molecular level. However, for patients 10 and 14, two molecularly distinct isolates were found among the five colonies with an identical morphotype. Results obtained with Raman spectroscopy and PFGE were concordant.
DISCUSSION
In this study, a collection of 162 MR-CNS isolates obtained from skin swabs has been characterized by use of both Raman spectroscopy and PFGE analysis.
The reproducibility of the Raman method was calculated to be 99.5%. The concordance between Raman spectroscopy and PFGE was high since for 94.4% of all isolates, the classification with both techniques was congruent. It was seen that isolates with different PFGE patterns could be found in multiple Raman clusters and also that isolates with identical PFGE patterns were divided among different Raman clusters. This implies that discordances between these methods were probably mutual, as is the case in such comparison between methods of equal quality (19).
Besides the feasibility of Raman spectroscopy as a typing tool for different bacterial species (10, 22), the use of this technique for the fast screening of variability within a collection of MR-CNS was examined. The Raman clustering found for MR-CNS isolates obtained from 5 patients revealed that a patient can be colonized with multiple MR-CNR types during a hospital stay. Although the CNS skin population can be dynamic and resistant isolates can arise due to antibiotic treatment, this pattern of colonization with different MR-CNS isolates seems unlikely. The random occurrence and absence of strains suggest a lack in the reproducibility of the sample collection from patient skin, since only one isolate of each morphotype was stored and analyzed. For the patients in this study, one or two isolate types were found more frequently over time, indicating that the patient was persistently colonized with this isolate. Isolate types that were found only once in the same time period could be transient isolates. Since samples were taken from the arm, the chance of retrieving transient isolates by transmission from a patient, health care worker, or the innate environment is relatively high. On the other hand, these isolate types could be persistent but in a lower number and could be missed.
To confirm the statement that multiple isolate types could be present in one culture, an additional study of 14 patients was performed. Both Raman spectroscopy and PFGE revealed that, indeed, different isolate types were found not only among colonies of different morphotypes but also for some colonies with the same morphotype. These findings indicate that, even for the limited number of colonies tested in this study, the variability in CNS populations on skin is surely higher than might be expected from morphological differences only. In an ideal study design, therefore, several single MR-CNS colonies should be selected and characterized individually to obtain a correct insight into the prevalence of isolate types. We also found that a more reliable determination of colony morphology by visual inspection could be obtained by incubation for at least 72 h (data not shown). However, this will result in a delay of at least 2 days, compared to the typing of isolates after a 20-h incubation time, as we have used here. This is not a problem in a research setting but could be a consideration in a clinical situation, e.g., for the microbiological diagnosis of catheter-related bloodstream infections (CR-BSI) (5, 16). Furthermore, previous studies indicated that polyclonality in CNS cultures from CR-BSI isolates correlates with differences in antimicrobial susceptibility patterns and may have important consequences for the treatment of these infections (5).
Since only a small number of patients were included in this study, it is not possible to draw a significant medical conclusion. However, our findings do provide technical support that characterizing only a limited number of MR-CNS colonies may result in misleading information, as multiple isolates may be present in cultures. A problem for large surveillance studies of the transmission of CNS or for the reliable diagnosis of CR-BSI could result by simply missing significant clonal types. Analysis of multiple colonies will lead to a significant increase in the numbers of samples that need to be analyzed and will demand a typing tool with high sample throughput. Although PFGE analysis is still the gold standard for the typing of CNS isolates, we have shown that Raman spectroscopy could provide typing results that are in good concordance with PFGE results. Due to its high throughput, Raman spectroscopy enables the analysis of multiple colonies per patient, which will increase the quality of clinical and epidemiological studies.
Footnotes
Published ahead of print on 30 December 2009.
REFERENCES
- 1.Ben Saida, N., A. Ferjeni, N. Benhadjtaher, K. Monastiri, and J. Boukadida. 2006. Clonality of clinical methicillin-resistant Staphylococcus epidermidis isolates in a neonatal intensive care unit. Pathol. Biol. (Paris) 54:337-342. [DOI] [PubMed] [Google Scholar]
- 2.Buijtels, P. C., H. F. Willemse-Erix, P. L. Petit, H. P. Endtz, G. J. Puppels, H. A. Verbrugh, A. van Belkum, D. van Soolingen, and K. Maquelin. 2008. Rapid identification of mycobacteria by Raman spectroscopy. J. Clin. Microbiol. 46:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casey, A. L., T. Worthington, P. A. Lambert, and T. S. Elliott. 2007. Evaluation of routine microbiological techniques for establishing the diagnosis of catheter-related bloodstream infection caused by coagulase-negative staphylococci. J. Med. Microbiol. 56:172-176. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeeuwen, A. van Belkum, A. Vindel, J. Garaizer, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus (MRSA) strains: the HARMONY collection. J. Clin. Microbiol. 45:1830-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia de Viedma, D., P. M. Rabadan, M. Diaz, E. Cercenado, and E. Bouza. 2000. Heterogeneous antimicrobial resistance patterns in polyclonal populations of coagulase-negative staphylococci isolated from catheters. J. Clin. Microbiol. 38:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juarez-Verdayes, M. A., M. A. Reyes-Lopez, M. E. Cancino-Diaz, S. Munoz-Salas, S. Rodriguez-Martinez, F. J. de la Serna, C. H. Hernandez-Rodriguez, and J. C. Cancino-Diaz. 2006. Isolation, vancomycin resistance and biofilm production of Staphylococcus epidermidis from patients with conjunctivitis, corneal ulcers, and endophthalmitis. Rev. Latinoam. Microbiol. 48:238-246. [PubMed] [Google Scholar]
- 7.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knauer, A., P. Fladerer, C. Strempfl, R. Krause, and C. Wenisch. 2004. Effect of hospitalization and antimicrobial therapy on antimicrobial resistance of colonizing Staphylococcus epidermidis. Wien. Klin. Wochenschr. 116:489-494. [DOI] [PubMed] [Google Scholar]
- 9.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maquelin, K., L. Dijkshoorn, T. J. van der Reijden, and G. J. Puppels. 2006. Rapid epidemiological analysis of Acinetobacter strains by Raman spectroscopy. J. Microbiol. Methods 64:126-131. [DOI] [PubMed] [Google Scholar]
- 11.Maquelin, K., T. Hoogenboezem, J. Jachtenberg, R. Dumke, E. Jacobs, G. J. Puppels, N. G. Hartwig, and C. Vink. 21 April 2009, posting date. Raman spectroscopic typing reveals the presence of carotenoids in Mycoplasma pneumoniae. Microbiology. doi: 10.1099/mic.0.026724-0. [DOI] [PubMed]
- 12.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann, and G. J. Puppels. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255-271. [DOI] [PubMed] [Google Scholar]
- 13.Milisavljevic, V., F. Wu, J. Cimmotti, J. Haas, P. Della-Latta, E. Larson, and L. Saiman. 2005. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am. J. Infect. Control 33:341-347. [DOI] [PubMed] [Google Scholar]
- 14.Miragaia, M., I. Couto, S. F. Pereira, K. G. Kristinsson, H. Westh, J. O. Jarlov, J. Carrico, J. Almeida, I. Santos-Sanches, and H. de Lencastre. 2002. Molecular characterization of methicillin-resistant Staphylococcus epidermidis clones: evidence of geographic dissemination. J. Clin. Microbiol. 40:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, A. J., M. S. Jackson, and J. Bagg. 2001. The ecology of Staphylococcus species in the oral cavity. J. Med. Microbiol. 50:940-946. [DOI] [PubMed] [Google Scholar]
- 16.Spare, M. K., S. E. Tebbs, S. Lang, P. A. Lambert, T. Worthington, G. W. Lipkin, and T. S. Elliott. 2003. Genotypic and phenotypic properties of coagulase-negative staphylococci causing dialysis catheter-related sepsis. J. Hosp. Infect. 54:272-278. [DOI] [PubMed] [Google Scholar]
- 17.Uckay, I., D. Pittet, P. Vaudaux, H. Sax, D. Lew, and F. Waldvogel. 2009. Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41:109-119. [DOI] [PubMed] [Google Scholar]
- 18.van Belkum, A., J. Kluijtmans, W. van Leeuwen, W. Goessens, E. ter Averst, J. Wielenga, and H. Verbrugh. 1996. Monitoring persistence of coagulase-negative staphylococci in a hematology department using phenotypic and genotypic strategies. Infect. Control Hosp. Epidemiol. 17:660-667. [PubMed] [Google Scholar]
- 19.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, M. Struelens, and the European Society of Clinical Microbiology and Infectious Diseases Study Group on Epidemiological Markers. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. [DOI] [PubMed] [Google Scholar]
- 20.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 21.Widerstrom, M., T. Monsen, C. Karlsson, and J. Wistrom. 2006. Molecular epidemiology of meticillin-resistant coagulase-negative staphylococci in a Swedish county hospital: evidence of intra- and interhospital clonal spread. J. Hosp. Infect. 64:177-183. [DOI] [PubMed] [Google Scholar]
- 22.Willemse-Erix, D. F., M. J. Scholtes-Timmerman, J. W. Jachtenberg, W. B. van Leeuwen, D. Horst-Kreft, T. C. B. Schut, R. H. Deurenberg, G. J. Puppels, A. van Belkum, M. C. Vos, and K. Maquelin. 2009. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J. Clin. Microbiol. 47:652-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebuhr, W., S. Hennig, M. Eckart, H. Kranzler, C. Batzilla, and S. Kozitskaya. 2006. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 28(Suppl. 1):S14-S20. [DOI] [PubMed] [Google Scholar]