Abstract
Noroviruses (NoVs) are now considered the most common cause of outbreaks of nonbacterial gastroenteritis, but the factors which control the incidence of NoVs are poorly understood. In 2006, the pattern of NoV outbreak epidemics in Victoria, Australia, changed compared to the pattern for 2002 to 2005 and 2007. This study examined molecular correlates of the changed NoV periodicity. For the period of 2002 to 2007, 8,507 fecal specimens from 1,495 gastroenteritis outbreaks were tested for NoV by reverse transcription-PCR, and 1,018 NoV outbreaks were identified. Nucleotide sequence analysis was used to define genotypes and GII.4 variants. For 2002 to 2007, GII.4 was the predominant genotype. For the period of 2002 to 2005 and 2007, a single NoV outbreak epidemic occurred in warmer months of each year, but in 2006 two epidemics occurred in 1 year, one in colder months and one in warmer months of the year. For 2002 to 2007, four major GII.4 variants, “2002 Oxford/Farmington Hills,” “2004 Hunter,” “2006a,” and “2006b,” were identified. Each NoV outbreak epidemic was linked principally to one of these four variants, and there was a time link, a delay of 2 to 6 months, between the first detection of a GII.4 variant and the first outbreak epidemic in which it was the principal variant. The unusual 2006 pattern of outbreak epidemics can then be correlated with the appearance of two GII.4 variants within a short space of time, resulting in two outbreak epidemics in a short space of time, i.e., in the 1 year. This study provides a potentially greater ability to predict the characteristics of NoV epidemics.
The noroviruses (NoVs) are single-stranded, positive-sense RNA viruses classified as belonging to the genus Norovirus within the family Caliciviridae. The NoV genome is approximately 7.6 kb in length and comprises three open reading frames (ORFs). ORF 1 encodes the nonstructural polyprotein which includes the RNA helicase, protease, and polymerase proteins; ORF 2 encodes the major structural capsid protein; and ORF 3 encodes a small virion-associated protein (9).
NoVs are currently classified into five genogroups. Three of these, GI, GII, and GIV, occur in human infections, though most NoVs affecting humans belong to genogroup GI or GII (15). Within each genogroup one or more genotypes have been identified (15), although the GII.4 genotype is the most common NoV genotype detected (1, 3, 8, 12).
NoVs are now considered the most common cause of outbreaks of nonbacterial gastroenteritis (4). NoV outbreaks can occur as epidemics (2), i.e., peaks in the incidence of outbreaks above the general level (2, 7). Furthermore, there is now some evidence to suggest that two sets of factors control the dynamics of NoV outbreak epidemics, and these are probably a combination of molecular and environmental variables (2).
It is now well established that GII.4 strains periodically undergo genetic changes, and these altered forms are termed “variants” (8, 20). Furthermore, there is some evidence that the emergence of new GII.4 variants can correlate with the appearance of NoV epidemics. For example, the appearance of the GII.4 “Hunter” variant in Australia in 2004 coincided with an increase in GII activity (3). Kroneman et al. (13) reported that an increase in gastroenteritis outbreaks in the Netherlands in 2004 was linked to the appearance of the GII.4 “GII.4-2004” variant. Ho et al. (10) reported that a NoV epidemic in 2006 in Hong Kong was linked to the emergence of the GII.4 “95/96”-like variant.
Studies in a number of countries indicate epidemics of NoV outbreaks tend to occur each year with a peak in colder months in northern hemisphere countries (19) and a peak in warmer months in southern hemisphere countries, such as Australia (2, 16). However, NoV epidemics that occur out of the normal synchrony have been reported. For example, Lopman et al. (14) noted an aberrant peak in NoV reports in the summer of 2002 in England and Wales. Studies in this laboratory for the period of 2001 to 2005 indicated a regular periodicity in NoV outbreaks each year, with a single epidemic in warmer months of the year (1, 2, 16). In 2006, however, as indicated in the current study, two epidemics of NoV outbreaks took place in 1 year, the first in winter (the middle of the year) and the second in the more usual time period, later, in the warmer part of the year. In 2007, the usual pattern of NoV outbreak epidemics returned, with a single epidemic later, in the warmer part of the year.
Thus, in summary, NoV outbreak epidemics generally follow a predictable pattern in which there is a single marked peak at a particular time of the year (warmer weather in the southern hemisphere and colder weather in the northern hemisphere). On rare occasions, however, this pattern can break down and two epidemics rather than one can occur in a given year. Furthermore, the timing of one of these two epidemics now becomes aberrant in that it occurs in the colder part of the year in the southern hemisphere and the warmer part of the year in the northern hemisphere. It is not currently understood exactly what factors influence the appearance of the yearly NoV outbreak epidemic, although there is a suggestion that a mixture of genetic and environmental factors interact to facilitate the epidemic.
The hypothesis tested in the current study is that each yearly epidemic is principally linked to a particular GII.4 variant, but if two major GII.4 variants arise more or less at the same time, then it can be expected that two epidemics will occur in a given calendar year, with one epidemic linked principally to one GII.4 variant and the other epidemic linked to the other GII.4 variant.
MATERIALS AND METHODS
Outbreak definition and investigation.
For the purposes of this study, an outbreak of gastroenteritis was defined as an incident, apparently associated with a common event or location, in which four or more individuals had symptoms of gastroenteritis.
All outbreaks included in this study were those for which fecal specimens were sent to the Victorian Infectious Diseases Reference Laboratory in Victoria, Australia, for testing in the period of 2002 to 2007. The Victorian Infectious Diseases Reference Laboratory is the main public health laboratory for viral identification in Victoria. As such, it receives fecal material from gastroenteritis outbreaks reported to the Victorian Department of Human Services where a viral etiology is suspected. Outbreak specimens are also occasionally sent by other institutions, such as hospitals. Outbreak documentation and management were carried out by staff linked to the Victorian Department of Human Services and/or staff at the affected setting. Only outbreaks which occurred in the state of Victoria, Australia, were included in the study.
The date of an outbreak was taken as the onset date. If this was not available, the date the outbreak was first notified or the earliest date of collection of a specimen from the outbreak was taken as the date of the outbreak. Only one fecal specimen per person per outbreak was tested.
Fecal processing and RNA extraction.
For the period of 2002 to 2007, a total of 8,507 fecal specimens from 1,495 gastroenteritis outbreaks were received for testing. Fecal specimens were prepared as a 20% (vol/vol) suspension in Hanks' complete balanced salt solution, and the suspension was vigorously shaken and then clarified by centrifugation (22). The clarified fluid was then collected for RNA extraction.
RNA extraction, prior to NoV ORF 1 reverse transcription-PCR (RT-PCR), was then carried out using either the manual QIAamp viral RNA kit (23, 24) or the Corbett automated nucleic acid extraction procedure (23).
NoV ORF 1 RT-PCR.
ORF 1 RT-PCR testing for NoV was carried out on all fecal specimens from the gastroenteritis outbreaks received for testing in the period of 2002 to 2007. A two-round heminested procedure, essentially as given by Yuen et al. (24), was utilized.
NoV nucleotide sequencing and phylogenetic analysis.
The NoV ORF 1 genotype definition for all NoV outbreaks involved sequencing analysis of one NoV specimen, chosen without bias, from each outbreak where a positive NoV specimen was identified. If both GI and GII specimens were identified in a particular outbreak, then an attempt was made to sequence both a GI and a GII specimen.
The second-round PCR product of the NoV ORF 1 RT-PCR test procedure was purified and sequenced essentially as described by Marshall et al. (17). A region of 440 nucleotides within the RNA polymerase region, which corresponded to nucleotides 4484 to 4923 of Camberwell virus (AF145896), was then utilized for phylogenetic analysis and genotype definition.
MacVector software versions 7.2 and 8.0 (Oxford Molecular Limited, Madison, WI) were used for analysis of nucleotide sequences. Phylogenetic analysis was carried out using the software PHYLIP versions 3.572, 3.65, and 3.68 (6). The distance-based (Kimura matrix) neighbor-joining method was the principal algorithm used for analysis (functions DNADIST and NEIGHBOR), and the trees were constructed with the software TreeView version 1.5 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) or FigTree version 1.2.2 (http://tree.bio.ed.ac.uk/software/figtree/). The robustness of the neighbor-joining method tree generated was assessed by bootstrap resampling and by comparison with trees constructed using alternate algorithms as described previously (17).
Initially, to define all ORF 1 genotype clusters, separate ORF 1 phylogenetic trees were generated for each year of the study (results not shown). Cluster definition for all trees was carried out essentially as described by Marshall et al. (16).
NoV GII.4 variants were identified after examination of the GII.4 portion of the phylogenetic tree generated for each year of the study. The classification of these subclusters as a particular variant type was dependent on the reference strains that fell into a particular subcluster. The reference strains used for GII.4 variant classification are listed in Table 1.
TABLE 1.
NoV ORF 1 GII.4 reference strains utilized for definition of GII.4 variants
| Variant | Predominant variant yr | Strain | GenBank accession no. | Reference(s) |
|---|---|---|---|---|
| Pre-1996 | Pre-1996 | Camberwell/1994/AUS | AF145896 | 21 |
| Pre-1996 | Pre-1996 | Lordsdale/1993/UK | X86557 | 18, 21 |
| 2002 Oxford/Farmington Hills | 2002-2003 | Farmington Hills/2002/USA | AY502023 | 21 |
| 2002 Oxford/Farmington Hills | 2002-2003 | Oxford/2002/UK | AY588005 | 5 |
| 2004 Hunter | 2004-2005 | Hunter532D/2004/AU | DQ078801 | 21 |
| 2006a | 2006 | Terneuzen70/2006/NL | EF126964 | 11, 18 |
| 2006a | 2006 | Aomori1/2006/JP | AB447432 | 18 |
| 2006b | 2006-2007 | Nijmegen115/2006/NL | EF126966 | 11, 18 |
| 2006b | 2006-2007 | Hokkaido1/2006/JP | AB447427 | 18 |
Quantitative analysis.
The study required quantitative analysis of NoV genotypes and GII.4 variants in an epidemic and the time delay between the appearance of a variant and the onset of an epidemic. For these purposes, the time period of an epidemic was taken as the three consecutive months of highest NoV outbreak incidence in the epidemic.
RESULTS
NoV outbreak epidemics from 2002 to 2007.
During the period of January 2002 to December 2007, 8,507 fecal specimens from 1,495 gastroenteritis outbreaks were received for testing for NoV. NoV was detected by ORF 1 RT-PCR in one or more specimens in 1,018 outbreaks (“NoV outbreaks”). NoV GI was detected in 42 NoV outbreaks, NoV GII in 972 NoV outbreaks, and both NoV GI and GII in 4 NoV outbreaks.
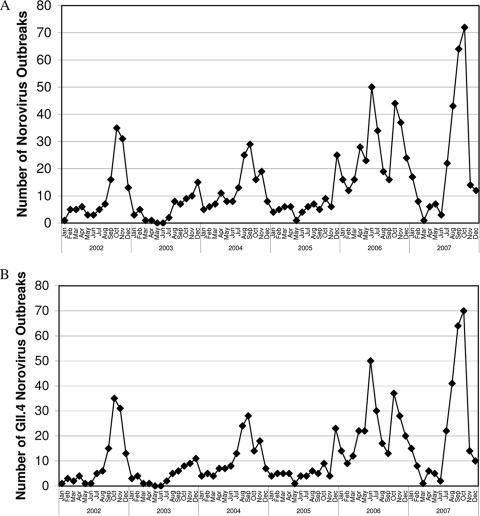
The periodicity of NoV outbreaks for the period of 2002 to 2007 is shown in Fig. 1A. It can be seen that for each calendar year of the study there was a single epidemic in warmer months (i.e., toward the end) of the year. However, it can also be seen that in 2006 there was an additional epidemic that occurred in colder months (i.e., in the middle) of the year.
FIG. 1.
Periodicity of NoV outbreaks in Victoria, Australia, in 2002 to 2007. (A) All NoV outbreaks. (B) GII.4 NoV outbreaks.
Nucleotide sequence analysis results for all NoV outbreak epidemics (2002 to 2007) are listed in Table 2. It can be seen that GII.4 was by far the most common NoV genotype detected in each epidemic for the period of 2002 to 2007 (Table 3). The periodicity of GII.4 NoV outbreaks (Fig. 1B) was essentially the same as that of all NoV outbreaks (Fig. 1A).
TABLE 2.
NoV outbreak genotypes (2002 to 2007)
| Genotype | No. of NoV outbreaks per yr with indicated genotype |
% of all NoV outbreaks | Total no. of outbreaks with genogroup | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | Total | |||
| GI.2 | 4 | 6 | 0 | 0 | 2 | 0 | 12 | 1.2 | |
| GI.3b | 1 | 1 | 0 | 0 | 1 | 4 | 7 | 0.7 | |
| GI.4 | 1 | 0 | 0 | 0 | 15 | 2 | 18 | 1.8 | |
| GI.?aa | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0.1 | |
| GI.?ba | 0 | 0 | 0 | 2 | 1 | 0 | 3 | 0.3 | |
| Unsequenced GI | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0.1 | 42 |
| GII.1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.1 | |
| GII.3a | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0.2 | |
| GII.3c | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.1 | |
| GII.4 | 117 | 50 | 139 | 75 | 274 | 258 | 913 | 89.7 | |
| GIIb | 3 | 3 | 14 | 1 | 17 | 5 | 43 | 4.2 | |
| Unsequenced GII | 1 | 0 | 1 | 4 | 6 | 0 | 12 | 1.2 | 972 |
| GI.2 + GII.4 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0.2 | 2 |
| GI.4 + GII.4 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0.2 | 2 |
| Total | 130 | 61 | 155 | 84 | 319 | 269 | 1,018 | 100.0 | 1,018 |
A BLAST search revealed no matches to known genotypes. A pairwise nucleotide alignment showed that the 2004 cluster (GI.?a) is different from the 2005 and 2006 cluster (GI.?b).
TABLE 3.
NoV genotypes detected in each peak 3-month period
| Yr, epidemic no. | Peak 3 moa | No. of NoV outbreaks with indicated genotype |
% of peak outbreaks with genotype GII.4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI.2 | GI.3b | GI.4 | Unclassified GI | GII.3a | GII.4 | GIIb | GI.2 + GII.4 | GI.4 + GII.4 | Not sequenceable | Total | |||
| 2002 | Sep to Nov | 0 | 0 | 0 | 0 | 0 | 81 | 0 | 1 | 0 | 0 | 82 | 98.8 |
| 2003 | Oct to Dec | 3 | 0 | 0 | 0 | 0 | 28 | 3 | 0 | 0 | 0 | 34 | 82.4 |
| 2004 | Aug to Oct | 0 | 0 | 0 | 1 | 0 | 66 | 3 | 0 | 0 | 0 | 70 | 94.3 |
| 2005 | Oct to Dec | 0 | 0 | 0 | 1 | 2 | 36 | 0 | 0 | 0 | 1 | 40 | 90.0 |
| 2006, 1 | May to Jul | 1 | 0 | 1 | 0 | 0 | 102 | 3 | 0 | 0 | 0 | 107 | 95.3 |
| 2006, 2 | Oct to Dec | 0 | 0 | 13 | 0 | 0 | 85 | 1 | 0 | 1 | 5 | 105 | 81.0 |
| 2007 | Aug to Oct | 0 | 2 | 0 | 0 | 0 | 175 | 2 | 0 | 0 | 0 | 179 | 97.8 |
| Total | 4 | 2 | 14 | 2 | 2 | 573 | 12 | 1 | 1 | 6 | 617 | 92.9 | |
Jul, July; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December.
NoV GII.4 variant analysis.
The predominant GII.4 variants for NoV epidemics for the period of 2002 to 2007 are listed in Table 4. It can be seen that epidemic 1 (May to July) in 2006 was anomalous in that “variant 2006a” contributed in a significant way to only the one epidemic, in contrast to all the other GII.4 variant forms, which contributed in a significant way to two successive epidemics (“2002 Oxford/Farmington Hills” in 2002 and 2003, “2004 Hunter” in 2004 and 2005, and “2006b” in 2006 [epidemic 2, in October to December] and 2007).
TABLE 4.
NoV GII.4 variants found during each peak 3-month period
| Yr, epidemic no. | Peak 3 moa | No. (%) of indicated variantsb |
||||||
|---|---|---|---|---|---|---|---|---|
| Pre-1996 | 2002 Oxford/ Farmington Hills | 2004 Hunter | 2006a | 2006b | Unclassified | Total | ||
| 2002 | Sep to Nov | 0 (0.0) | 75 (92.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (7.4) | 81 |
| 2003 | Oct to Dec | 4 (14.3) | 24 (85.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 28 |
| 2004 | Aug to Oct | 1 (1.5) | 0 (0.0) | 65 (98.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 66 |
| 2005 | Oct to Dec | 0 (0.0) | 4 (11.1) | 21 (58.3) | 11 (30.6) | 0 (0.0) | 0 (0.0) | 36 |
| 2006, 1 | May to Jul | 0 (0.0) | 0 (0.0) | 3 (2.9) | 96 (94.1) | 2 (2.0) | 1 (1.0) | 102 |
| 2006, 2 | Oct to Dec | 0 (0.0) | 0 (0.0) | 0 (0.0) | 35 (41.2) | 50 (58.8) | 0 (0.0) | 85 |
| 2007 | Aug to Oct | 0 (0.0) | 0 (0.0) | 1 (0.6) | 19 (10.9) | 154 (88.0) | 1 (0.6) | 175 |
| Total | 5 | 103 | 90 | 161 | 206 | 8 | 573 | |
Jul, July; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December.
Bold indicates the predominant variant for a given epidemic.
The time delay between the first detection of a new GII.4 variant and its first manifestation as a NoV outbreak epidemic was also determined. For the four major GII.4 variants found, the time of their first detection in a NoV outbreak was as follows: 2002 Oxford/Farmington Hills in July 2002, 2004 Hunter in February 2004, 2006a in December 2005, and 2006b in June 2006. From Table 4 it can be determined that the time delay between the appearance of each variant and its first manifestation as an epidemic was 2, 6, 5, and 4 months, respectively, i.e., a range of 2 to 6 months.
DISCUSSION
Studies of the periodicity of NoV outbreaks in Victoria, Australia, indicate that NoV outbreak epidemics tend to follow a regular periodicity with a single epidemic each year, typically toward the end (i.e., in warmer months) of the year (2). In 2006, this pattern of NoV outbreak periodicity changed dramatically in that there were two epidemics in 1 year and the first epidemic occurred in the middle (i.e., in colder months) of the year. In 2007, the usual pattern resumed, with one epidemic toward the end (i.e., in warmer months) of the year. The unusual change in pattern in 2006 provides an opportunity to examine the role of genetic factors in these events.
Analysis of GII.4 variants for the period of 2002 to 2007 showed two things. First, each NoV outbreak epidemic was linked principally to a single GII.4 variant. Thus, the variant 2002 Oxford/Farmington Hills was the predominant variant for the NoV outbreak epidemics for the years 2002 and 2003 and variant 2004 Hunter was the predominant variant for the NoV outbreak epidemics for the years 2004 and 2005. For the first 2006 NoV outbreak epidemic, variant 2006a was the predominant variant, whereas for the second NoV outbreak epidemic in 2006, variant 2006b was the predominant variant. In 2007, variant 2006b was the predominant variant for the NoV outbreak epidemic.
Second, there is a relationship between the first detection of a major new GII.4 variant and its first manifestation as the principal variant in an outbreak epidemic. This was found to be a time delay of 2 to 6 months between the first detection and the beginning of the epidemic. Thus, if two new variants emerge in close proximity, as occurred in late 2005 and in 2006, it would be predicted that there would be two epidemics, each linked to one of the variants, and the epidemics would occur in the order in which the variants were first detected. This was in fact the case. The 2006a variant (first detected in December 2005) was the principal variant associated with the epidemic beginning in May 2006, and the 2006b variant (first detected in June 2006) was the principal variant associated with the epidemic beginning in October 2006.
The key findings of the current study are first that each NoV outbreak epidemic is linked principally to a particular GII.4 variant and second that there is a time link (a delay of 2 to 6 months) between the first detection of a major GII.4 variant and the occurrence of the first epidemic in which it is the principal variant. In 2006 in Victoria, Australia, the normal pattern of a single outbreak epidemic per year was broken. The findings of the current study explain this. In 2005 and 2006, the appearance of two major GII.4 variants within a short space of time resulted in two outbreak epidemics occurring in 2006 in a short space of time. The findings of the current study offer a potentially greater ability to predict the characteristics of NoV epidemics in a given year.
Footnotes
Published ahead of print on 20 January 2010.
REFERENCES
- 1.Bruggink, L., and J. Marshall. 2008. Molecular changes in the norovirus polymerase gene and their association with incidence of GII. 4 norovirus-associated gastroenteritis outbreaks in Victoria, Australia, 2001-2005. Arch. Virol. 153:729-732. [DOI] [PubMed] [Google Scholar]
- 2.Bruggink, L. D., and J. A. Marshall. 2009. Norovirus epidemics are linked to two distinct sets of controlling factors. Int. J. Infect. Dis. 13:e125-e126. [DOI] [PubMed] [Google Scholar]
- 3.Bull, R. A., E. T. V. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, B., and M. McKendrick. 2004. A review of viral gastroenteritis. Curr. Opin. Infect. Dis. 17:461-469. [DOI] [PubMed] [Google Scholar]
- 5.Dingle, K. E., and Norovirus Infection Control in Oxfordshire Communities Hospitals. 2004. Mutation in a Lordsdale norovirus epidemic strain as a potential indicator of transmission routes. J. Clin. Microbiol. 42:3950-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 7.Friedman, G. D. 2004. Primer of epidemiology, 5th ed., p. 79. McGraw-Hill, New York, NY.
- 8.Gallimore, C. I., M. Iturriza-Gomara, J. Xerry, J. Adigwe, and J. J. Gray. 2007. Inter-seasonal diversity of norovirus genotypes: emergence and selection of virus variants. Arch. Virol. 152:1295-1303. [DOI] [PubMed] [Google Scholar]
- 9.Green, K. Y. 2007. Caliciviridae: the noroviruses, p. 949-979. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 1, Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Ho, E. C. M., P. K. C. Cheng, A. W. L. Lau, A. H. Wong, and W. W. L. Lim. 2007. Atypical norovirus epidemic in Hong Kong during summer of 2006 caused by a new genogroup II/4 variant. J. Clin. Microbiol. 45:2205-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanerva, M., L. Maunula, M. Lappalainen, L. Mannonen, C.-H. von Bonsdorff, and V.-J. Anttila. 2009. Prolonged norovirus outbreak in a Finnish tertiary care hospital caused by GII.4-2006b subvariants. J. Hosp. Infect. 71:206-213. [DOI] [PubMed] [Google Scholar]
- 12.Kirkwood, C. 2004. Viral gastroenteritis in Europe: a new norovirus variant? Lancet 363:671-672. [DOI] [PubMed] [Google Scholar]
- 13.Kroneman, A., H. Vennema, Y. van Duijnhoven, E. Duizer, and M. Koopmans. 2004. High number of norovirus outbreaks associated with a GGII.4 variant in the Netherlands and elsewhere: does this herald a worldwide increase? Eurosurveillance 8(52).
- 14.Lopman, B. A., M. Reacher, C. Gallimore, G. K. Adak, J. J. Gray, and D. W. G. Brown. 2003. A summertime peak of “winter vomiting disease”: surveillance of noroviruses in England and Wales, 1995 to 2002. BMC Public Health 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, J. A., and L. D. Bruggink. 2006. Laboratory diagnosis of norovirus. Clin. Lab. 52:571-581. [PubMed] [Google Scholar]
- 16.Marshall, J. A., A. Dimitriadis, and P. J. Wright. 2005. Molecular and epidemiological features of norovirus-associated gastroenteritis outbreaks in Victoria, Australia in 2001. J. Med. Virol. 75:321-331. [DOI] [PubMed] [Google Scholar]
- 17.Marshall, J. A., M. E. Hellard, M. I. Sinclair, C. K. Fairley, B. J. Cox, M. G. Catton, H. Kelly, and P. J. Wright. 2003. Incidence and characteristics of endemic Norwalk-like virus-associated gastroenteritis. J. Med. Virol. 69:568-578. [DOI] [PubMed] [Google Scholar]
- 18.Motomura, K., T. Oka, M. Yokoyama, H. Nakamura, H. Mori, H. Ode, G. S. Hansman, K. Katayama, T. Kanda, T. Tanaka, N. Takeda, H. Sato, and the Norovirus Surveillance Group of Japan. 2008. Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history. J. Virol. 82:11247-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S284-S287. [DOI] [PubMed] [Google Scholar]
- 20.Siebenga, J., A. Kroneman, H. Vennema, E. Duizer, and M. Koopmans. 2008. Food-borne viruses in Europe network report: the norovirus GII. 4 2006B (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Eurosurveillance 13:1-4. [PubMed] [Google Scholar]
- 21.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witlox, K. J., T. Karapanagiotidis, L. D. Bruggink, and J. A. Marshall. 2010. The effect of fecal turbidity on norovirus detection by reverse transcriptase polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 66:230-232. [DOI] [PubMed] [Google Scholar]
- 23.Witlox, K. J., T. N. Nguyen, L. D. Bruggink, M. G. Catton, and J. A. Marshall. 2008. A comparative evaluation of the sensitivity of two automated and two manual nucleic acid extraction methods for the detection of norovirus by RT-PCR. J. Virol. Methods 150:70-72. [DOI] [PubMed] [Google Scholar]
- 24.Yuen, L. K. W., M. G. Catton, B. J. Cox, P. J. Wright, and J. A. Marshall. 2001. Heminested multiplex reverse transcription-PCR for detection and differentiation of Norwalk-like virus genogroups 1 and 2 in fecal samples. J. Clin. Microbiol. 39:2690-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]