Abstract
Stabilization of mRNA by the ubiquitous RNA binding protein human antigen R (HuR), a member of the embryonic lethal abnormal vision (ELAV) protein family, requires canonical binding to AU-rich element (ARE)-bearing target mRNA and export of nuclear HuR-mRNA complexes to the cytoplasm. In human mesangial cells (HMC) both processes are induced by angiotensin II (AngII) via protein kinase Cδ (PKCδ)-triggered serine phosphorylation of HuR. By testing different point-mutated Flag-tagged HuR proteins, we found that Ser 318 within RNA recognition motif 3 (RRM3) is essential for AngII-induced binding to ARE-bearing mRNA but irrelevant for nucleocytoplasmic HuR shuttling. Conversely, mutation at Ser 221 within the HuR hinge region prevents AngII-triggered HuR export without affecting mRNA binding of HuR. Using phosphorylation state-specific antibodies, we found a transient increase in HuR phosphorylation at both serines by AngII. Functionally, PKCδ mediates the AngII-induced stabilization of prominent HuR target mRNAs, including those of cyclin A, cyclin D1, and cyclooxygenase-2 (COX-2), and is indispensable for AngII-triggered migration and wound healing of HMC. Our data suggest a regulatory paradigm wherein a simultaneous phosphorylation at different domains by PKCδ coordinates mRNA binding and nucleocytoplasmic shuttling of HuR, both of which events are essentially involved in the stabilization of HuR target mRNAs and relevant cell functions.
The tight control of mRNA turnover is increasingly recognized as an important paradigm of eukaryotic gene expression, allowing cells to respond appropriately to environmental changes (22, 44). In this context, destabilizing AU-rich elements (AREs) present in the 3′ untranslated regions (UTR) of a subset of many labile mRNAs have been recognized as important cis-regulatory determinants of exosomal mRNA decay and were denoted posttranscriptional operons (31, 46). Through binding to AREs, different RNA binding proteins (ARE-BPs) not only convey control of mRNA decay but, in addition, can modulate the translation and intracellular trafficking of their cargo mRNAs (44). Prominent examples of ARE-BPs with a predominant destabilizing effect on ARE-mRNAs are poly(A)/poly(U) binding degradation factor (AUF), tristetraprolin (TTP), and K-homology-type splicing regulatory protein (KSRP) (9, 24, 61). In contrast, the ubiquitously expressed embryonic lethal abnormal vision (ELAV) protein human antigen R (HuR) is one of the best-characterized ARE-BPs, with a predominant stabilizing effect on many ARE-bearing mRNAs coding, e.g., for cell cycle regulators, growth factors, proto-oncogenes, apoptosis-regulatory proteins, cytokines, and various proinflammatory enzymes (2, 15, 35). Functionally, overexpression of HuR results in a marked increase in the half-lives of ARE-mRNAs, some of which are critically involved in human pathologies, including chronic inflammation, cardiovascular diseases, and cancer (13, 46).
Three main levels of HuR regulation have been identified so far. A decrease in the HuR expression level in response to cyclic GMP (cGMP)-elevating agents was found (5, 36), and, complementary to these observations, a reduction in HuR mRNA stability through the action of microRNA (miR)-519 has previously been identified as a novel mode of regulating HuR expression (3). In addition, caspase-dependent degradation of HuR protein, which is critically involved in the regulation of apoptosis, was found as a further posttranslational mechanism of HuR control (4, 42). Importantly, the latter studies nicely illustrate that HuR itself is a target of posttranscriptional and posttranslational regulation. Since HuR is most abundantly localized within the cell nucleus, the export of nuclear HuR to the cytoplasm (nucleocytoplasmic HuR shuttling) represents the major mode of regulating HuR activity (19). Concomitantly, various reports have demonstrated that the content of cytoplasmic HuR is an eligible indicator for a poor survival prognosis of patients suffering from specific cancers (10, 11, 13), although the molecular details of pathologically increased cytoplasmic HuR levels are largely unknown (13, 28). Physiologically, HuR shuttling from the nucleus to the cytoplasm can be induced by numerous external stimuli which themselves activate a diverse set of signaling pathways, including different mitogen-activated protein kinases (MAPKs), AMP-activated kinase (AMPK), cyclin dependent kinase 1 (Cdk1), cell cycle checkpoint kinase 2 (Chk2), coactivator-associated arginine methyltransferase 1 (CARM1), and different members of the protein kinase C (PKC) family (15, 35). Despite the variety of kinases involved in HuR shuttling, a direct posttranslational modification of HuR was reported only for CARM1 (37), Chk2 (1), Cdk1 (33), PKCα (14), and PKCδ (16). The opposite outcome of specific HuR phosphorylation by different kinases indicates that HuR phosphorylation resembles a master switch for regulating HuR activity and HuR-triggered cell functions.
Structurally, a basic hinge region (HR) within the HuR protein bears a sequence denoted HuR nucleocytoplasmic shuttling sequence (HNS) which is critical for nuclear export of HuR to the cytoplasm (19, 32). In addition, via specific association with transportin 2, the HR is involved in HuR import to the nucleus (25). Functionally, PKCδ-triggered HuR phosphorylation in the nucleus is indispensable for AngII-evoked stabilization of inducible cyclooxygenase (COX-2) mRNA in human mesangial cells (HMC) and in the rat kidney, resulting in elevated prostaglandin E2 formation (16, 17). PKCδ is a ubiquitously expressed member of the Ca2+-independent family of new PKC isoforms implicated in diverse cell functions, including cell proliferation, cell migration, and cell survival (48). The translocation of PKCδ to the nucleus is critical for cell responses induced by AngII (16, 45) and various genotoxins and apoptotic stimuli (48) and relies on a bipartite nuclear localization sequence (NLS) in the PKCδ protein (60). Although the stabilizing effects of HuR on ARE-bearing mRNA are important for protection of mRNA from cytoplasmic decay by the exosomal complex, HuR binding to target mRNA already occurs in the nucleus (19, 32). In this study we asked how the completely distinct events of HuR mRNA binding and HuR trafficking to the cytoplasm are temporally coordinated by PKCδ. We present evidence that PKCδ, by triggering an elaborate series of phosphorylation events at different HuR domains, activates and coordinates the two events that are indispensable for stabilization of HuR target mRNAs. Furthermore, we demonstrate that a dual phosphorylation of HuR by PKCδ is critically involved in AngII-induced stabilization of cyclin A and D1 mRNAs and functionally relevant for HuR-triggered cell migration.
MATERIALS AND METHODS
Reagents.
Angiotensin II and anti-FlagM2 antibody-coupled agarose were purchased from Sigma-Aldrich (Deisenhofen, Germany). Fibronectin and rottlerin were obtained from Calbiochem (Schwalbach, Germany). Ribonucleotides and modifying enzymes were purchased from Life Technologies (Karlsruhe, Germany). Antibodies raised against β-actin, HuR, histone deacetylase 1 (HDAC-1), PKCδ, and 14-3-3θ; a pan-specific 14-3-3 antibody; and anti-rabbit and anti-mouse horseradish peroxidase-linked IgGs were purchased from Santa Cruz Biotechnology (Heidelberg, Germany). Antibodies against cyclin D1 and green fluorescent protein (GFP) were obtained from New England Biolabs (Frankfurt am Main, Germany). Fluorescently labeled antibodies were from Molecular Probes (Karlsruhe, Germany). Human recombinant PKCδ was obtained from Biomol GmbH (Hamburg, Germany). All cell culture media and supplements were purchased from Life Technologies (Karlsruhe, Germany).
Cell culture.
Human mesangial cells (HMC) (product code CC-2559) were derived from Cambrex Bio Science (Walkersville, MD). For experiments, serum-free preincubations were performed in Dulbecco's modified Eagle's medium supplemented with bovine serum albumin (BSA). HMC at between passages 4 and 10 were used.
RNA interference.
Gene silencing was performed using small interfering RNAs (siRNAs) for human PKCδ (sc-36253) and HuR (sc-35619) and a control siRNA (sc-37007) from Santa Cruz Biotechnology. Transfection of subconfluent HMC was performed by use of the Oligofectamine reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer′s instructions.
Western blot analysis.
Cytoplasmic and nuclear fractions from HMC were prepared according to a protocol from Schreiber et al. (54). To ensure equal sample loading of nuclear proteins, the blots were reprobed with an anti-histone deacetylase 1 (HDAC-1)-specific antibody. Total protein extracts were obtained by using standard procedures as described previously (18). After 1 h of blocking in 2% fat-free milk powder in Tris-buffered saline containing 0.5% Tween, Western blots were probed with the primary antibody overnight at 4°C. Following incubation with a horseradish peroxidase-conjugated secondary antibody, signals were detected with an ECL system (Amersham Bioscience, Freiburg, Germany).
Expression and purification of recombinant protein.
The plasmid pQE-30-His-HuRwt and related single-point-mutated pQE-His-HuRΔSer158/-ΔSer221/-ΔSer318/-ΔSer324 or, double mutated pQE-30-HuRΔSer158/ΔSer221 and pQE-30-HuRΔSer221/ΔSer318 constructs, which each contain either a single or a double serine-to-alanine substitution, were generated by use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as described previously (16).
RNA-electrophoretic mobility shift assay (EMSA) of in vitro-phosphorylated HuR.
Phosphorylation of different His-tagged HuR proteins (pQE-30-His-HuRwt, pQE-His-HuRΔSer158/-ΔSer221/-ΔSer318/-ΔSer324, pQE-30-HuRΔSer158/ΔSer221, and pQE-30-HuRΔSer221/ΔSer318) by human recombinant PKCδ was tested by an in vitro kinase assay using HuR as a substrate and was performed by using a modified protocol described by Geiges et al. (23). Briefly, 5 μg of recombinant PKCδ was incubated in PKC assay buffer containing 20 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 10 μM Na2ATP, 25 μg/ml phosphatidylserine, 2.5 μg/ml diolein, 1 μl of [γ-32P]dATP (3,000 Ci/mmol), 2 μg of recombinant His-tagged HuR constructs, and 100 μM CaCl2. Reaction mixtures were incubated at 32°C for 10 min, and half of the PKC reaction mixture was directly subjected to SDS-PAGE for monitoring in vitro phosphorylation. After fixing, the gels were vacuum dried and radioactive signals visualized with a PhosphorImager. The other half of the reaction was directly used for RNA gel shift assay as described previously (14). Briefly, each of phosphorylated His-tagged HuR protein was incubated with a single-stranded T4 polynucleotide kinase-labeled (30 kcpm/reaction) RNA oligonucleotide at room temperature for 15 min in a buffer containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 40 mM KCl, 2 mM dithiothreitol (DTT), 5% glycerol, and 0.5% Nonidet P-40. To reduce nonspecific binding, total yeast RNA (200-ng/ml final concentration) was added. Subsequently protein complexes were separated in 6% nondenaturing polyacrylamide gels and run in Tris-borate-EDTA. The sequence of the RNA oligonucleotide was according to the “site III” within the 3′ UTR of the human COX-2 gene (51) and represents a typical class III ARE referred to as COX-2-ARE-2: 5′-GCAUGCUGUUCCUUUUCUUUUCU-3′.
CD spectroscopy.
Circular dichroism (CD) analysis was performed on a Jasco J-810 spectropolarimeter (Jasco) under an N2 atmosphere. CD steady-state spectra in the far-UV region were recorded using a 2-mm quartz cuvette, a bandwidth of 1 nm, and a response time of 1s. The scanning speed was 50 nm per minute. Each spectrum was the result of five accumulations, and protein concentrations were 5 μM. Spectra were taken from 50 mM Na2HPO4 (pH 8.7) at 25°C. In all CD experiments, the background spectrum of buffer without protein was subtracted from the protein spectra, and CD spectra were initially analyzed by the software accompanying the spectrophotometer.
Construction of Flag-tagged HuR.
Flag-tagged recombinant wild-type HuR (pCMV-Flag-HuRwt) was generated by subcloning the pQE-His-wild-type HuR plasmid DNA into the pCMV-Flag-N3 expression vector as described previously (16). Accordingly, the plasmids pCMV-Flag-HuRΔSer221 and pCMV-Flag-HuRΔSer318, each bearing a single serine-to-alanine substitution at the depicted positions, were generated by changing a single base pair (TCC to GCC) using the following (sense) primers: for pCMV-Flag-HuRΔSer221, 5′-GAGATTCAGGTTCGCCCCCATGGGCGTCG-3′ (corresponding to nucleotides 648 to 676), and for pCMV-Flag-HuRΔSer318, 5′-AAATCTTAC AGGTTGCCTTCAAAACCAAC-3′ (corresponding to nucleotides 938 to 966). For generation of double-mutated pCMV-Flag-HuRΔSer221/ΔSer318 bearing two serine-to-alanine substitutions, the plasmid pCMV-Flag-HuRΔSer221 was used as a template and amplified by PCR with the same primers which had been used for generation of pCMV-Flag-HuRΔSer318. All mutants were generated by use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and HMC were transiently overexpressed by using the Lipofectamine reagent from Invitrogen.
Generation of phospho-HuR-specific antibodies.
Antibodies recognizing phosphorylated HuR at either Ser 221 or Ser 318 were custom made by Eurogentec (Brussels, Belgium) using the keyhole limpet hemocyanin (KLH)-coupled peptides AQRFRF(p)SPMGVDH (anti-HuR-pSer221) and DKILQV(p)SFKTNK (anti-HuR-pSer318).
The ability of each antibody to recognize only its specific antigen was confirmed by Western blot analysis using either pQE-His-HuRwt (HuRwt) or corresponding recombinant HuR proteins bearing a single point mutation either in Ser 221 (HuRΔSer221) or in Ser 318 (HuRΔSer318) for in vitro kinase assay with recombinant PKCδ as described above.
The specificity of each phospho-antibody was confirmed by testing the blocking capacity of the corresponding peptide. Therefore, 100 ng of each peptide used for immunization was preincubated for 30 min with approximately 100 ng of in vitro-phosphorylated wild-type or point-mutated pQE-His-HuR proteins before the protein mixture was subjected to SDS-PAGE.
IP.
For immunoprecipitation (IP) of endogenous or Flag-tagged HuR, nuclear or cytoplasmic extracts (400 μg) were incubated overnight at 4°C either with 2 μg of a monoclonal anti-HuR antibody or, alternatively, with 50 μl of 50% anti-FlagM2-antibody coupled agarose or with the same amount of mouse IgG (both diluted in lysis buffer containing 5% fetal calf serum [FCS]). Subsequently, protein G-Sepharose CL-4B beads (Amersham-Biosciences, Freiburg, Germany) were added and incubated for another 2 h. After centrifugation for 5 min at 3,000 × g, the precipitated complexes were successively washed three times with low-salt buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 2 mM EGTA, 0.1% SDS) and once with high-salt buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.2% Triton X-100, 2 mM EDTA, 2 mM EGTA, 0.1% SDS). After these washing steps, beads were subjected to SDS-PAGE and coimmunoprecipitated proteins detected by Western blot analysis.
Immunoprecipitation and qRT-PCR analysis (pulldown RT assay).
HuR-bound mRNA was isolated by using a pulldown RT assay as described previously (14). In brief, cells were treated with lysis buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.1% NP-40, 50 mM NaF, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM disodium glycerol phosphate, and 100 U/ml RNasin) and cell lysates immunoprecipitated with 2 μg of either an HuR-specific antibody or anti-Flag-coupled agarose overnight at 4°C. Alternatively, the same amount of mouse IgG was used in control IP reactions. Subsequently, protein G-Sepharose CL-4B beads were added and incubated for a further 2 h. After a short centrifugation (3,000 × g), the beads were successively washed with low- and high-salt buffers before total RNA was extracted by using the Tri reagent (Sigma-Aldrich). The HuR-bound RNA was reverse transcribed using SuperScript reverse transcriptase (Invitrogen, Karlsruhe, Germany) and subjected to quantitative reverse transcription-PCR (qRT-PCR). Normalization of input RNA was confirmed by RT reaction of total cellular RNA isolated from an equal amount of cell extract as was used for IP and subsequent assessment of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels.
Real-time RT-PCR.
Two-step real-time PCR was performed using a TaqMan (ABI 7500) instrument from Perkin-Elmer. The mRNA levels for cyclin A, cyclin D1, COX-2, and GAPDH were determined by using a protocol according to the “hot-start” real-time PCR procedure with “Quanti-Tec” SYBR green (Qiagen, Hilden, Germany). Total RNA was extracted using the Tri reagent, and reverse transcription was performed using 0.5 μg of total RNA and reverse transcriptase from Invitrogen (Invitrogen, Karlsruhe, Germany) with random hexamer primers. The following oligonucleotides were used for PCR: cyclin A forward, 5′-ATTAGTTTACCTGGACCCAG-3′; cyclin A reverse, 5′-CACAAACTCTGCTACTTCTG-3′; 3′. cyclin D1 forward, 5′-GCTGCTCCTGGTGAACAAGC-3′; cyclin D1 reverse, 5′-TTCAATGAAATCGTGCGGG-3′; COX-2 forward, 5′-TTCAAATGAGATTGTGGGAAAATTGCT-3′; COX-2 reverse, 5′-AGATCATCTCTGCCTGAGTATCTT-3′; GAPDH forward, 5′-CACCATCTTCCAGGAGCGAG-3′; and GAPDH reverse, 5′-GCAGGAGGCATTGCTGAT-3′. Calculation of relative COX-2, cyclin A, and cyclin D1 mRNA levels was done by using the 2−ΔΔCT method (40). According to this method, the threshold cycle (CT) values of cyclin A, cyclin D1, and COX-2 mRNA levels were normalized to the CT values of GAPDH mRNA within the same sample.
Detection of mRNA by standard PCR.
For standard RT-PCR, the same conditions were used as described for qRT-PCR. The following primer sets were used: HuR forward: 5′-CACAGCTTGGGCTACGGCTTTGTG-3′ and HuR reverse: 5′-AGGACCCGCGAGTTGATGATCCG-3′; β-actin forward: 5′-TTGCCGACAGGATGCAGAAGGA-3′ and β-actin reverse: 5′-AGGTGGACAGCGAGGCCAGGAT-3′; PKCδ forward: 5′-AGCCGGGACACTATATTCCAGA-3′ and PKCδ reverse 5′-CTTGCCGTAGGTCCCACTGTTG-3′. The different PCR products (HuR, 238 bp; β-actin, 128 bp; PKCδ, 365 bp) were separated on a 1% agarose gel containing 0.5 μg/ml ethidium bromide. The identity of amplicons was confirmed by DNA sequencing using the AbiPrism 310 Genetic Analyzer from Applied Biosystems.
Transwell cell migration.
For quantitative analysis of cell migration, the chemotaxis of HMC across 5-μm-pore-size polycarbonate filters (6.5 mm in diameter) was measured in 24-well transwell chambers (Costar Corning Co., Cambridge, MA). The lower sides of the transwell filters were coated with fibronectin (10 μg/ml) and washed with phosphate-buffered saline (PBS) before siRNA-transfected HMC were seeded at 1.0 × 105 cells/well in serum-free medium containing 0.1% BSA on the upper chamber of the transwell. Thereafter, cells were treated with vehicle or with the different stimuli for 24 h at 37°C before nonmigrated cells were removed with a cotton swab. For quantification of cell migration, the remaining cells were fixed with 4% paraformaldehyde in PBS, stained with 0.1% crystal violet in 0.1 M borate (pH 9.0)-2% ethanol, and extracted in 10% acetic acid before the absorbance of the eluted dye was measured at 595 nm in an enzyme-linked immunosorbent assay (ELISA) reader. Each assay was done in triplicate, and the data given are mean values ± standard errors of the means (SEM) from three independent experiments.
In vitro wound scratch assay.
At 24 h after transfection of siRNAs, HMC were seeded on 10-cm culture dishes, and once cells had reached 70 to 90% confluence, cell monolayers were mechanically wounded by creating a linear scratch of ∼0.2 to 0.3 mm in width using a sterile P-200 pipette tip. Immediately thereafter, cells were washed with serum-free medium to remove cell debris, and fresh starvation medium was added to each plate before the cells were stimulated with the appropriate reagents. The sizes of the scratches were imaged immediately (0 h) and 24 h after stimulation with a BZ-7000 microscope (Biozero, Keyence, Neu-Isenburg, Germany) equipped with a Zeiss Apo 20×/0.75 numerical aperture (NA), objective and images were analyzed by using the BZ-H1TL8 software from Biozero. Each assay was performed in duplicate, and representative results from three independent experiments are shown.
Proliferation assay.
Cell growth was monitored after 24, 48, and 72 h using a CellTiter 96 nonradioactive cell proliferation assay (Promega, Mannheim, Germany). The formation of formazan through a cleavage of the tetrazolium salt 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) in metabolically active cells was measured as absorbance at 570 nm by use of a spectrophotometer. All assays were performed as triplicates, and data given are mean values ± SEM of three independent results.
Indirect immunofluorescence microscopy.
For indirect immunofluorescence analysis, cells were grown on coverslips and subsequently fixed for 20 min at room temperature with 4% paraformaldehyde in PBS, and monitoring of shuttling of Flag-tagged HuR protein was performed as described previously (14). Stained cells were finally monitored using a BZ-7000 inverse immunofluorescence microscope (Biozero, Keyence, Neu-Isenburg, Germany) equipped with a Zeiss Apo 20×/0.75 NA objective, and images were analyzed by using the BZ-H1TL software from Biozero.
Statistical analysis.
Results are expressed as means ± standard deviations (SD). Data from migration assays are shown as box plots, with 5 to 95% percentiles indicated by open boxes and SEM depicted by error bars. The data are presented as fold induction compared to untreated control values. Statistical analysis was performed using Student's t test and analysis of variance (ANOVA) with the Bonferroni multiple-comparison test. P values of <0.05, <0.01 and <0.005 were considered significant.
RESULTS
Phosphorylation at Ser 221 by PKCδ confers nucleocytoplasmic HuR shuttling.
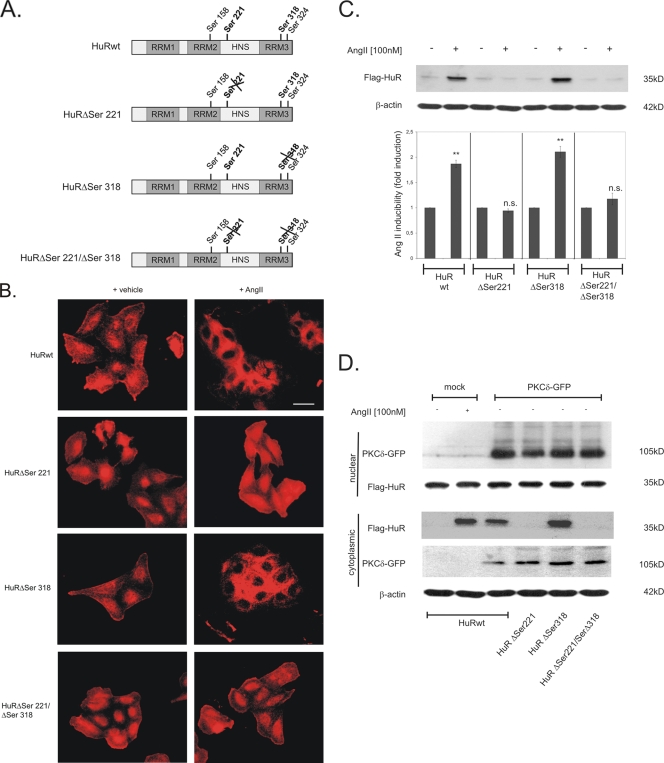
Previously, by use of kinase assays, we have identified serine 221 (Ser 221) and serine 318 (Ser 318) as being critical target sites for PKCδ-mediated in vitro phosphorylation of HuR (16). Furthermore, we found that mutations in both serine residues totally impaired the AngII-induced stabilization of COX-2 mRNA in renal MC (16). Here, we aimed to further delineate whether these different phosphorylation sites might have different mechanistic roles in the activation of HuR. To this end, we generated a series of N-terminally Flag-tagged wild-type (HuRwt) and point-mutated HuR proteins bearing either a serine-to-alanine mutation at position 221 (HuRΔSer221) that lacks the phosphorylation site or, alternatively, a comparable mutation in Ser 318 (HuRΔSer318) or a mutation of both serines (HuRΔSer221/Δ318), as illustrated in Fig. 1A. Determination of the subcellular localization of Flag-tagged HuR proteins in transiently transfected HMC by indirect immunofluorescence revealed that in accordance to its predominant nuclear localization, all HuR proteins under basal conditions were preferentially detected within cell nuclei (Fig. 1B, +vehicle). AngII stimulation of HMC transiently transfected with Flag-tagged wild-type HuR (HuRwt) caused an almost complete redistribution of HuR from the nucleus to the cytoplasm (Fig. 1B). Importantly, a similar change in intracellular HuR localization caused by AngII was observed with HuRΔSer318 (Fig. 1B). In contrast, AngII-induced nucleocytoplasmic shuttling was totally impaired in cells which had been transfected with the HuRΔSer221 construct, and a similar loss in HuR export was observed with the construct bearing mutations in both serines (HuRΔSer221/ΔSer318) (Fig. 1B). These results indicate that Ser 221 is the critical phosphorylation site in HuR which is functionally involved in AngII-induced export of HuR. In line with these findings from indirect immunofluorescence, Western blot analysis of cytoplasmic cell fractions with an anti-Flag-specific antibody revealed that an increase in cytoplasmic HuR levels caused by AngII is not found in HMC which have been transfected either with HuRΔSer221 or with HuRΔSer221/ΔSer318 (Fig. 1C). Again, a point mutation in Ser 318 had no significant effect on the AngII-induced increase of cytoplasmic HuR (Fig. 1C, HuRΔSer318). Furthermore, results from cycloheximide experiments indicate that differences in cytoplasmic Flag-HuR abundance were caused neither by differences in expression levels nor by variations in the protein stabilities of the different Flag-HuR constructs (data not shown).
FIG. 1.
Analysis of AngII-induced nucleocytoplasmic shuttling of wild-type HuR and HuR proteins carrying point mutations at PKCδ phosphorylation sites. (A) Schematic diagram of putative PKC phosphorylation sites within HuR; the ones critical for phosphorylation by the PKCδ are depicted in bold letters. Flag-tagged HuR constructs used to express wild-type HuR (HuRwt) and corresponding Ser-mutated HuR proteins with either a PKCδ-nonphosphorylatable Ser 221 (HuRΔSer221) or Ser 318 (HuRΔSer318) or mutations in serines 221 and 318 (HuRΔSer221/ΔSer318) are shown and marked by a cross. RRM, RNA recognition motif; HNS, HuR nucleocytoplasmic shuttling sequence. (B) Indirect immunofluorescence was applied to visualize AngII-induced changes in the subcellular localization of different Flag-tagged HuR proteins. Flag-HuR was detected by immunofluorescence at 24 h after transfection of HMC with pCMV-Flag plasmids. Images show Flag-HuR in quiescent HMC either left untreated (+ vehicle) or stimulated for 2 h with 100 nM AngII (+ AngII) before cells were fixed and stained with the anti-Flag antibody and the secondary anti-mouse Cyc 3 antibody. Bar, 50 μm. Data are representative of three independent experiments giving similar results. (C) HMC cells were transfected with plasmids to express the Flag-tagged proteins depicted in panel A, and the levels of ectopic HuR in cytoplasmic fractions were monitored by Western blot analysis. To confirm equal protein contents within cytoplasmic extracts, the blots were stripped and reprobed with an anti-β-actin antibody. Data represent means ± SD (n = 3) from three independent experiments. **, P ≤ 0.01 compared with unstimulated vehicle controls. n.s., not significant. (D) HMC cells were cotransfected with plasmids encoding the different Flag-tagged proteins depicted in panel A together with a GFP-tagged PKCδ expression plasmid (PKCδ-GFP). At 24 h after transfection, cytosolic and nuclear extracts were prepared and the contents of GFP-tagged PKCδ and Flag-tagged HuR (Flag-HuR) were monitored with their corresponding anti-tag antisera. To correct for variations in cytoplasmic protein loading, the blots were stripped and additionally probed with an anti-β-actin antibody. Data are representative of two independent experiments giving similar results.
Next, we tested whether ectopic expression of PKCδ could mimic AngII effects on HuR shuttling in ΗMC. Interestingly, GFP-tagged PKCδ displayed a strong constitutive nuclear localization (Fig. 1D), indicating that the NLS of ectopically expressed PKCδ is already exposed and allows translocation to the nucleus without any external stimulus.
Similar to the effects of AngII, an increase in cytoplasmic Flag-HuR content was found in PKCδ-GFP-transfected cells (Fig. 1D, lower panel). Consistent with the results from experiments with AngII, the nuclear export of HuR by ectopic PKCδ expression was impaired in cells which were cotransfected with Flag-HuR constructs bearing a mutation in Ser221 (HuRΔSer221 and HuRΔSer221/ΔSer318) (Fig. 1D, lower panel). These results demonstrate that HuR activation by AngII is fully mimicked by ectopic PKCδ expression.
Phosphorylation of Ser 318 promotes HuR binding to ARE.
In contrast to Ser 221, which lies within the HNS of HuR, Ser 318 is located in the distal RNA recognition motif 3 (RRM3), suggesting that the two phosphorylation sites may have different roles in HuR activation. To address this issue, we compared the RNA binding affinity of bacterially expressed wild-type histidine (His)-tagged HuR to an ARE-mRNA with the binding affinity of different point-mutated His-tagged HuR constructs bearing either a single or two serine-to-alanine substitutions (Fig. 1A). Prior to EMSA, the different HuR proteins were phosphorylated by an in vitro kinase assay with human recombinant PKCδ (Fig. 2, upper panel). For the gel shift assay we used an RNA oligonucleotide with an ARE from the human 3′ UTR of COX-2 (COX-2 ARE-II) which encompasses a typical nonpentameric destabilizing class III ARE (8) and which displays a high binding affinity for HuR (14, 51). Importantly, when assessing the RNA binding profiles, we observed that in vitro phosphorylation caused a robust increase in the RNA binding affinity of wild-type HuR (HuRwt) compared to unphosphorylated HuR (vehicle) (Fig. 2A). Furthermore, we found that His-HuRΔSer221 displayed an mRNA binding affinity to COX-2-ARE-II that was similar to that of HuRwt, although phosphorylation of this construct was less intense (Fig. 2A). Strikingly, a replacement of serine 318 by an alanine (HuRΔSer318) not only caused a marked reduction in the in vitro phosphorylation by PKCδ but also resulted in a complete loss of RNA binding (Fig. 2A). Consistently, a total loss of HuR phosphorylation and RNA binding was observed with the HuRΔSer221/ΔSer318 mutant. In contrast, a doubly mutated HuRΔSer158/ΔSer221 construct retained full RNA binding affinity despite the strikingly reduced phosphorylation of this construct (Fig. 2A). These results indicate that a phosphorylation at Ser 318 is indispensable for maximal HuR binding. These results also demonstrate that phosphorylation of HuR does potentiate its binding affinity to ARE-bearing mRNA. Furthermore, our study shows that Ser 318 within RRM3 of HuR selectively confers AngII-induced binding to ARE-mRNA.
FIG. 2.
(A) Phosphorylation of HuR at Ser 318 is required for AngII-induced HuR binding to ARE-mRNA. For in vitro phosphorylation assay, 2 μg of bacterially expressed and histidine-tagged pQE-30-His-HuR proteins was incubated with 5 μg of recombinant PKCδ in a cell-free assay with [γ-32P]ATP. Loading of equal protein amounts was ascertained by Coomassie blue staining (data not shown). RNA binding affinity of unphosphorylated (vehicle) and in vitro-phosphorylated wild-type HuR (HuRwt) and the indicated point-mutated pQE-30-His-HuR proteins was analyzed by EMSA using a gene-specific ARE oligonucleotide from the human COX-2 3′ UTR as described in Materials and Methods. RNA-protein complexes were resolved from unbound RNA (free probe) by nondenaturing gel electrophoresis. The conditions for RNA binding were as described in Materials and Methods. The EMSA shown is representative of three independent experiments giving similar results. (B) Analysis of wild-type HuR and HuR ΔSer318 by far-UV CD. The mean molar ellipticity, Φ, was measured from 190 to 250 nm by using approximately 5 μM each protein diluted in phosphate buffer.
Since Ser 318 is located within the boundaries of RRM3 and in close proximity to the C terminus of the protein, a point mutation at this position, independent of its phosphorylation status, may affect domain stability of RRM3. In order to test whether the serine-to-alanine mutation at Ser318 would cause any changes in protein folding in full-length HuR, we used CD spectroscopy. As shown in Fig. 2B, no significant structural changes could be determined when comparing the CD spectra of wild-type HuR and HuRΔSer318 proteins, indicating that the amino acid exchange in RRM3 did not interfere with the folding of the HuR protein.
Critical roles of Ser 221 and Ser 318 in HuR binding to target mRNA.
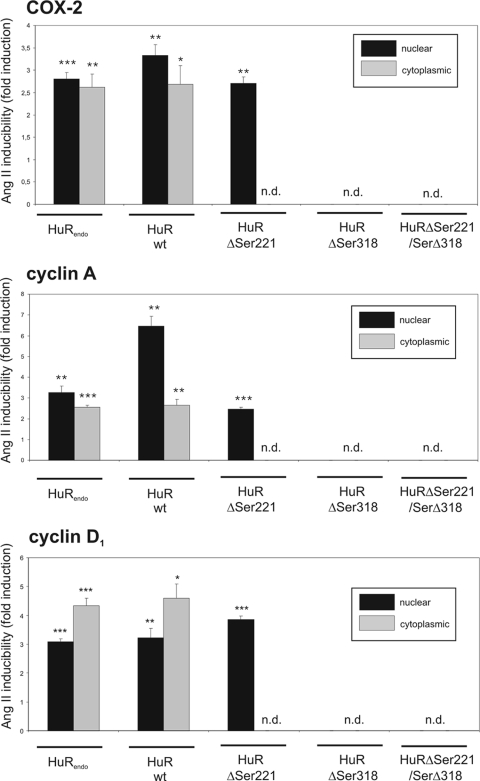
To prove the functional relevance of Ser 318 in the AngII-induced HuR binding to ARE-containing mRNA, we performed a ribonucleoprotein immunoprecipitation (RNP-IP) assay in which either endogenous or different Flag-HuR proteins were immunoprecipitated from different cell fractions of either untransfected controls or HMC transiently transfected with the different Flag-HuR proteins illustrated in Fig. 1A. We focused on the AngII-induced HuR binding to COX-2, cyclin A, and cyclin D1 mRNA species by specifically isolating HuR-associated mRNA by means of HuR immunoprecipitation. We have chosen these target mRNAs since they represent some of the best-characterized examples of HuR target mRNAs (39, 59) and, more importantly, their expression is modulated by AngII (6, 16, 49). Stimulation of HMC with AngII for 16 h significantly induced weak and constitutive binding (set as 1.0-fold) of endogenous HuR to all the tested transcripts in both the nuclear (black bars) and the cytoplasmic (open bars) fractions (HuRendo in Fig. 3). It is important to note that cell fractions used for the IP contained comparable amounts of GAPDH mRNA (data not shown). Similar to the increase in pulldown mRNA associated with endogenous HuR, the content of all mRNA species bound to wild-type HuR protein is clearly induced by AngII (HuRwt in Fig. 3). Consistently, HuR and Flag precipitates from nuclear fractions (black bars) yielded an overall larger amount of COX-2- and cyclin A-encoding mRNAs than precipitates from cytoplasmic extracts (gray bars). Similar to the IPs from nuclear wild-type HuR, AngII-mediated HuR binding of all mRNAs was also detectable in the Flag-IPs from HuRΔSer221-transfected HMC, although the absolute amount of bound mRNA varied between the different HuR mutants (HuRΔSer221 in Fig. 3). In contrast, corresponding Flag-IPs from cytoplasmic extracts of HuRΔSer221 transfectants contained no mRNA (Fig. 3, gray bars). These results again underline the functional importance of Ser 221 in AngII-induced export of HuR together with its bound cargo mRNA to the cytoplasm. Consistent with our hypothesis that Ser 318 is indispensable for HuR binding to mRNA, none of the tested transcripts was detectable in HuRΔSer318-transfected HMC independent of which fraction was used for the IP (n.d. in Fig. 3). Also, none of the tested mRNAs was detectable in the RNP-IPs from cells expressing an HuR protein with mutations in both serines (HuRΔSer221/ΔSer318 in Fig. 3). Collectively, these results confirm that the AngII-evoked mRNA binding of HuR to ARE-mRNA critically depends on Ser 318. In contrast, mutation of Ser 221, while having no effect on AngII-induced ARE-mRNA binding, is critical for AngII-induced nucleocytoplasmic HuR shuttling.
FIG. 3.
The intracellular allocation of HuR-bound mRNA is serine phosphorylation dependent. HMC were transfected with different Flag-HuR plasmids to express the Flag-tagged proteins depicted in Fig. 1A. Following treatment with AngII (100 nM) for 16 h, nuclear and cytoplasmic extracts were prepared and assayed by IP using control mouse IgG (data not shown), an anti-Flag antibody for ectopic HuR proteins, or an anti-HuR antibody for detection of endogenous HuR (HuRendo). HuR-bound cyclin A, cyclin D1, and COX-2 mRNA in the nuclear (black bars) and cytoplasmic (gray bars) fractions were determined by pulldown RT assays as described in Materials and Methods. Results shown are means ± SD from three independent experiments and are depicted as fold induction versus unstimulated controls. P values: <0.05 (*), <0.01 (**), and <0.005 (***). n.d., not detectable.
HuR is phosphorylated at Ser 221 and Ser 318 in vivo.
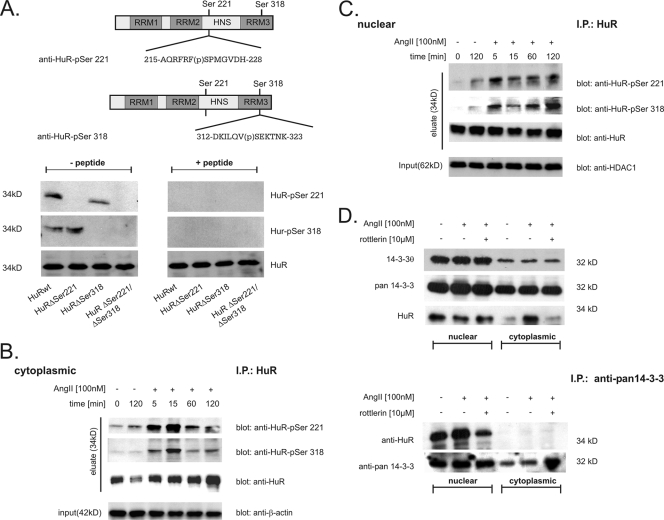
To extend our findings to endogenous HuR, we generated phospho-specific HuR antibodies either directed against a phosphopeptide encompassing a region from amino acid 215 to 226 (anti-HuR-pSer221) or raised against a phosphopeptide spanning amino acids 312 to 323 (anti-HuR-pSer318), as depicted in Fig. 4A (upper panel). Western blot analysis of different in vitro-phosphorylated HuR proteins with either of the serine-specific phospho-HuR antibodies confirmed that the two affinity-purified antibodies displayed no cross-reactivity (Fig. 4A, lower panel). Furthermore, positive signals in the Western blot disappeared after addition of corresponding phosphopeptides, whereas the detection of unphosphorylated HuR was not impaired by the addition of competitor phosphopeptides (Fig. 4A, lower panel, + peptide).
FIG. 4.
HuR is phosphorylated in vivo at serines 221 and 318 in a time-dependent manner. (A) Upper panel, schematic overview of HuR and serines critically involved in PKCδ-dependent HuR phosphorylation. Sequences of peptides used for generation of the indicated HuR phosphorylation-specific antibodies and the positions of phosphorylated (p) serine residues are indicated. Lower panel, for cell-free in vitro phosphorylation assays, the indicated HuR proteins were incubated with PKCδ for 10 min at 32°C and processed for Western blot analysis. Phosphorylation of HuR at Ser 221 (HuR-pSer221) and Ser 318 (HuR-pSer318) in comparison with unphosphorylated HuR (HuR) was monitored by successive incubation with either of the depicted phospho-specific antibodies and a commercial antibody against total HuR as indicated. Both phospho-specific HuR antibodies (anti-HuR-pSer221 and anti-HuR-pSer318) were preincubated on ice for 30 min without (− peptide) or with (+ peptide) 5 μg of the peptide used for immunization before being subjected to PAGE. (B and C). HMC were serum starved overnight and subsequently treated with vehicle (−) or with AngII (100 nM, +) for the indicated times before cells were lysed for cytoplasmic (B) and nuclear (C) fractions. For IP of HuR, a total protein amount of 500 μg from each fraction was incubated overnight with 2 μg of anti-HuR antibody (IP: HuR), and HuR precipitates were assessed by Western blot analysis by successive incubation with the indicated antibodies (eluate). To ascertain equal protein contents within the fractions, the blots were stripped and reprobed with either an anti-β-actin (input in panel B) or anti-HDAC-1 (input in panel C) antibody. AngII increases association of nuclear HuR with 14-3-3 proteins. (D) Upper panel, AngII-induced increase in cytoplasmic HuR level is not accompanied by a change in the intracellular redistribution of 14-3-3θ and pan-14-3-3. Serum-starved HMC were treated with vehicle (−), AngII (+), or AngII plus the PKCδ-specific inhibitor rottlerin (10 μM) for 2 h before cells were lysed for nuclear extracts and cytoplasmic fractions. Subsequently, different cell fractions from HMC were successively probed with the indicated antibodies. The data shown are representative of two independent experiments giving similar results. Lower panel, for IP, a total protein amount of 200 μg of either nuclear or cytoplasmic extract was incubated overnight with 2 μg of a pan-specific 14-3-3 antibody (IP: anti-pan 14-3-3), and precipitates were subsequently analyzed by Western blot analysis using an anti-HuR-specific antibody. To confirm equal amounts of input protein, the IPs were reprobed with the anti-pan 14-3-3 antibody as indicated.
To extend this finding to endogenous HuR, we monitored the phosphorylation of both serine residues in HuR in the nuclear and cytoplasmic fractions of AngII-treated HMC. Since detection of native phospho-Ser HuR in both cell fractions gave unsatisfactory results, we aimed to improve the sensitivity by enriching HuR via an immunoprecipitation prior to Western blot analysis (Fig. 4B and C). Time course experiments performed with both phospho-antibodies showed that stimulation of HMC with AngII (100 nM) induced a rapid but transient increase in cytoplasmic phospho-HuR levels, with a maximal increase in cytoplasmic phospho-HuR levels found after 15 min of AngII stimulation, independent of which phospho-HuR-specific antibody was used for Western blot analysis (Fig. 4B, eluate). Importantly, this is exactly the time point after AngII treatment when cytoplasmic HuR is firstly detectable by Western blot analysis (A. Doller and W. Eberhardt, unpublished data). A similar kinetics of serine phosphorylation of HuR at both sites was found in the precipitates from corresponding nuclear extracts (Fig. 4C, eluate). Collectively, these data clearly demonstrate a dual phosphorylation at serines 221 and 318 of both nuclear and cytoplasmic HuR by AngII in vivo.
AngII enhances the constitutive HuR binding to 14-3-3 proteins.
In a previous study Kim et al. (33) demonstrated that phosphorylation of HuR by Cdk1 causes HuR to reside within the nucleus and that this blockade in HuR export is due to HuR binding to the chaperon 14-3-3θ. A similar regulatory paradigm where a phosphorylated ARE-BP specifically binds to members of the 14-3-3 scaffold protein family, thereby preventing its intracellular trafficking, has been proposed for p37AUF-1 (26), TTP (30, 56), BRF1 (52), and KSRP (12, 24). Since in HMC, AngII treatment induces a redistribution of HuR from the nucleus to the cytoplasm via a PKCδ-dependent phosphorylation, we hypothesized that the disrupted interaction of nuclear HuR with one or more members of the 14-3-3 family could be a main reason for AngII-induced export of HuR to the cytoplasm. However, Western blot analysis with either a pan-specific 14-3-3 antibody or an antibody specifically raised against the θ isoform of 14-3-3 chaperons revealed that the increase in cytoplasmic HuR caused by AngII is not accompanied by any changes in the contents of nuclear or cytoplasmic 14-3-3 proteins (Fig. 4D, upper panel).
In a next step, we tested for an association of HuR with 14-3-3 in HMC by IP. Using a pan-specific 14-3-3 antibody, we found that the immunoprecipitated 14-3-3 proteins displayed a strong constitutive association exclusively to nuclear HuR. Interestingly, the binding of HuR to 14-3-3 was further increased when cells were treated with AngII, whereas HuR binding to 14-3-3 was reduced when cells were additionally treated with the PKCδ inhibitor rottlerin (Fig. 4D, lower panel). A similar increase in HuR affinity to 14-3-3 was already observed at 20 min (data not shown). Collectively, these data indicate that AngII causes an increase in nuclear HuR association with 14-3-3 chaperons in a PKCδ-dependent manner.
AngII-induced expression of cyclin A and D1 depends on HuR and PKCδ.
Το address the question whether the AngII-induced association of HuR with cyclin A and D1 mRNAs shown in Fig. 3 would have any impact on their expression levels, we measured steady-state mRNA levels of the corresponding genes by qRT-PCR. In order to prove the functional role of HuR and PKCδ in AngII-induced expression of both cyclins, we silenced either HuR or PKCδ by transfection of specific siRNAs. Assessment of steady-state HuR and PKCδ levels by RT-PCR demonstrated that the expression of both mRNA species was strongly reduced by their specific siRNAs but unaffected by a control siRNA (Fig. 5A, lower panel). Furthermore, GAPDH mRNA levels were not affected by the siRNA transfections, thus excluding an unspecific interference by the siRNA (Fig. 5A, lower panel). Treatment of HMC with AngII (100 nM) for 16 h caused a strong and significant increase in cyclin A and D1 mRNA levels in vehicle-treated HMC, and a similar increase in steady-state mRNA levels was measured in cells transfected with a control siRNA (Fig. 5A, upper panel). Interestingly, depletion of either HuR or PKCδ by specific siRNA constructs completely abrogated the AngII-induced increase in both cyclins without diminishing the constitutive levels of both cyclins (Fig. 5A, upper panel). Time course experiments showed that concomitant with alterations in mRNA levels, AngII caused a clear increase in cyclin A and D1 protein levels, with a clear peak observed at 16 h of AngII stimulation (Fig. 5B). Collectively, these data indicate that in HMC, HuR and PKCδ are both indispensable for AngII-induced expression of cyclin A and D1.
FIG. 5.
The AngII-triggered increase in cyclin A and D1 mRNA levels depends on HuR and PKCδ. (A) Serum-starved HMC were either left untransfected (vehicle) or were transfected with control duplex siRNA (siRNA-control), with duplex siRNA of human HuR (siRNA-HuR), or with duplex siRNA of human PKCδ (siRNA-PKCδ) as described in Materials and Methods. After transfection, cells were serum starved for 16 h before being stimulated for further 16 h with vehicle (−) or AngII (+). Upper panel, changes in the mRNA levels of different cyclins were determined by qRT-PCR by normalizing cyclin mRNAs to GAPDH mRNA. The results are means ± SD (n = 3) and are presented as fold induction versus nonstimulated controls of cyclin A (**, P ≤ 0.01) or cyclin D1 (§, P ≤ 0.05). n.s., not significant. Lower panel, the efficiency of silencing of HuR and PKCδ was monitored by assessment of the steady-state mRNA levels of the genes by RT-PCR analysis using either HuR- or PKCδ-specific primers, respectively. To correct for variations, β-actin-specific primers were used for PCR. (B) Time course induction of cyclin A and D1 in response to AngII. HMC were serum starved for 16 h and subsequently treated with either vehicle (−) or AngII (+) for the indicated times. Protein lysates (50 μg) of whole-cell extracts were subjected to SDS-PAGE and immunoblotted with anti-cyclin A or anti-cyclin D1 antibodies. To confirm an equal protein content within the fractions, the blots were stripped and reprobed with an anti-β-actin antibody. The Western blots shown are representative of two independent experiments giving similar results.
Critical role of PKCδ in AngII-induced cell migration.
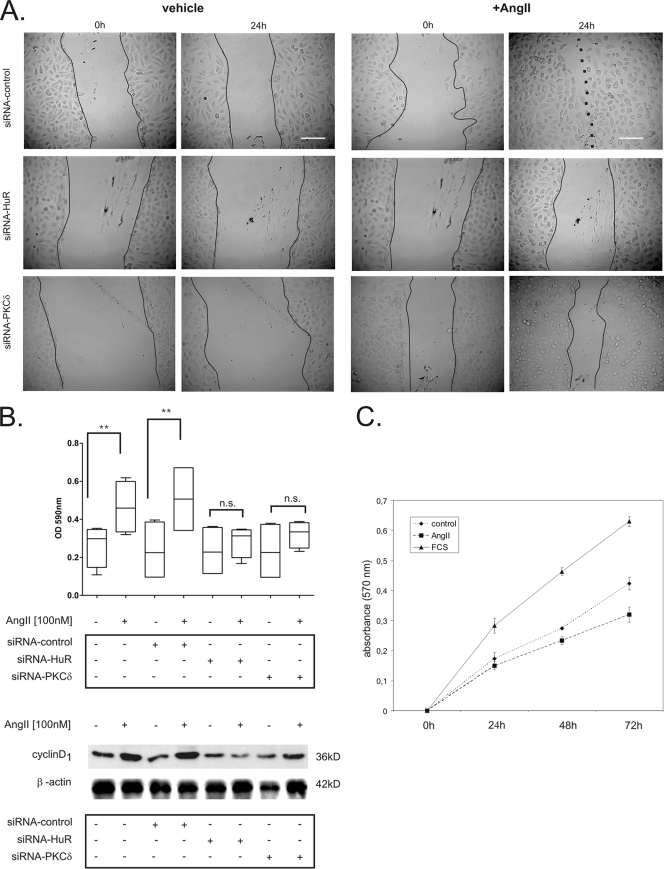
Given the critical role of cyclins in cell cycle progression and cell migration, we next aimed to validate a potential functional impact of HuR and PKCδ on these cell responses, and we therefore performed a wound scratch assay. This assay is commonly used for simultaneously monitoring changes in cell proliferation and cell migration. To this end, HMC were transfected either with a control siRNA (siRNA-control), with an siRNA specifically raised against HuR (siRNA-HuR), or with an siRNA construct directed against PKCδ (siRNA-PKCδ) before their ability to migrate across a denuded area of the monolayer was monitored throughout a 24-h stimulation. As shown in Fig. 6A, linearly wounded HMC transfected with a control siRNA (siRNA-control) showed an accelerated wound closure after AngII treatment compared to vehicle-treated control cells (Fig. 6A, upper panel). Interestingly, depletion of HuR by specific siRNA strongly attenuated AngII-induced wound closure (Fig. 6A, middle panel). Similarly, attenuation of PKCδ expression by specific siRNA strongly impaired the closure of the scratched area by AngII (Fig. 6A, lower panel).
FIG. 6.
AngII-induced migration of HMC depends on HuR and PKCδ. (A) Downregulation of HuR or PKCδ attenuates AngII-induced motility of HMC in a wound scratch assay. HMC were transfected either with control duplex siRNA (siRNA-control), with duplex siRNA for human HuR (siRNA-HuR) or with duplex siRNA for human PKCδ (siRNA-PKCδ) as described in Materials and Methods. After transfection, cells were cultured to a confluent monolayer and subsequently serum starved for 16 h before being treated for a further 24 h with vehicle (control) or AngII (100 nM) before the remaining cell-free space was analyzed by microscopy. Photographs of representative microscopic fields immediately (0 h) and 24 h after wounding are shown. Full lines indicate scratch borders, and the dotted line demarcates the attached cell fronts. The photographs shown are representative of two independent experiments giving similar results. Bars, 100 μm. (B) Upper panel, changes in migration of siRNA-transfected cells were determined by a transwell assay as described in Materials and Methods. Box plots show data summarized from three independent experiments performed in replicates. **, P ≤ 0.01 compared with untreated controls. n.s., not significant. Lower panel, the effects of silencing HuR or PKCδ on cyclin D1 expression were monitored by assessment of the total cyclin D1 levels by Western blot analysis using an anti-cyclin D1-specific antibody. To correct for variations in protein loading, the blot was stripped and probed with an anti-β-actin antibody. (C). Proliferation analysis of HMC stimulated for the indicated times points with vehicle (control), AngII, or 10% FCS by MTT assay. Data represent means ± SEM (n = 3).
In a next step, we tried to dissect the AngII-induced effects on cell migration from alterations of cell proliferation. To this end, cell migration was separately monitored by a transwell migration assay. Similar to the effects observed in the wound scratch assay, AngII caused a significant increase in cell migration after 24 h, and importantly, this stimulatory effect was completely prevented in cells which had been transfected with a HuR-siRNA (Fig. 6B, upper panel). Similar to the case for the HuR-siRNA-transfected cells, AngII-induced cell migration was significantly impaired in PKCδ-depleted HMC (Fig. 6B, upper panel). In contrast, AngII-triggered cell migration was not affected by transfection of a negative-control siRNA (siRNA-control), thus indicating that the inhibition of AngII-mediated cell migration is specifically caused by the knockdown of HuR or PKCδ. Furthermore, to test whether the expression of cyclin D1, which is known to promote cell migration mainly by inhibiting the Rho/ROCK-dependent signaling cascades (for a review, see reference 38), is affected by the HuR or PKCδ siRNA transfections, we monitored total cyclin D1 levels by Western blot analysis (Fig. 6B, lower panel). In accordance with the effects observed for cyclin D1 mRNA levels (Fig. 5A), AngII clearly increased the content of cyclin D1. Again, this increase was abrogated by the transfection of either HuR- or PKCδ-specific siRNA. In contrast, control siRNA had no effects on cyclin D1 protein levels (Fig. 6B, lower panel).
Since cyclin A and D1 in most cells exert a predominant proliferative action, we finally tested whether the observed effects in the in vitro wound closure assay could additionally result from an AngII-induced increase in cell proliferation. To this end, we performed a nonradioactive cell proliferation assay. Synchronized HMC were plated at a low density and treated with either vehicle (control), AngII (100 nM), or 20% FCS, and cell proliferation was assessed by the MTT assay (Fig. 6C). Stimulation of HMC with FCS caused a marked and linear increase in cell proliferation (Fig. 6C). In contrast, AngII exerted a moderate inhibitory effect on HMC proliferation (Fig. 6C). This is in agreement with the nonmitogenic and hypertrophic effects of AngII triggered by the AT2 receptor (29, 53). In summary, our data suggest that the PKCδ-induced and HuR-dependent stabilization of ARE-mRNA critically contributes to the promigratory effects of AngII in HMC.
DISCUSSION
HuR-mediated effects on mRNA stability and translation are tightly linked to its cytoplasmic levels, and, importantly, an increase in cytoplasmic HuR abundance directly correlates with the incidence of various human cancers (13, 27, 41). Meanwhile, there is compelling evidence that posttranslational modification of HuR, namely, phosphorylation at either serine or threonine residues located within different domains of the protein, plays a critical role in HuR regulation (1, 14, 16, 33, 34). Given the fact that HuR in most quiescent cells is localized predominantly within the nucleus and only a minor portion of this nuclear HuR pool is mobilized to the cytoplasm, the direct posttranslational modification of nuclear HuR may represent an important mode of activating HuR-triggered functions on ARE-bearing mRNA species. Although some previous studies have demonstrated that HuR is a target of different posttranslational modifications, the question of how these modifications may coordinate mRNA binding and HuR shuttling to the cytoplasm are not well understood. In this study, we provide first evidence that a tandem phosphorylation at two distinct HuR domains ensures a tight coordination of both key steps involved in HuR activation. A single phosphorylation of the distal RRM3 at Ser 318 modulates specific RNA binding to ARE-mRNA, whereas a phosphorylation at Ser 221 within the HNS exclusively promotes AngII-induced HuR export to the cytoplasm. Importantly, both phosphorylation sites act in independent manners, since a point mutation in Ser 221 did not impair RNA binding affinity mediated by Ser 318 and, vice versa, replacement of Ser 318 by an alanine did not affect AngII-induced HuR export to the cytosol. Simultaneous HuR phosphorylation at both serine residues in vivo is furthermore proven by the rapid phosphorylation at both sites upon AngII stimulation as documented by using phospho-specific antibodies (Fig. 4). Furthermore, we found that AngII-mediated phosphorylation of cytoplasmic HuR is more transient and declines much earlier than the phosphorylation of nuclear HuR. These differences in phosphorylation kinetics indicate the involvement of a cytosolic phosphatase in the shutdown of the stimulus-induced HuR shuttling. It furthermore indicates that dephosphorylation of cytoplasmic HuR precedes the reimport of cytosolic HuR back to the nucleus. Further investigations are needed to definitely prove the functional role of phosphatases in the coordination of HuR shuttling.
Structurally, the nucleocytoplasmic shuttling of HuR relies on a HNS spanning residues 205 to 237 and located in the hinge region (HR) between RRM2 and RRM3 (20). Importantly, similar to the RRMs, the HR of HuR is a target of different posttranslational modifications (1, 16, 33, 34, 37). In contrast to the inducible effects on HuR shuttling which we observed for phosphorylation of Ser 221 by either PKCα or PKCδ (14, 16), a phosphorylation of HuR at serine residue Ser 202 or Ser 242 by Cdk1 leads HuR to reside in the nucleus and thereby also prevents a recruitment of its target mRNA to the translation machinery (33, 34). In contrast to both of these serine positions lying adjacent to the HNS, the PKC target Ser 221 is located directly within the HNS. Similar to Ser 221, Arg 217, the target site for CARM1-dependent HuR methylation, lies within the HNS, and it is also critical for increased HuR export upon lipopolysaccharide (LPS) stimulation (37). In view of these observations, Kim and Gorospe (35) have suggested a model in which the relative position of a specific posttranslational modification toward the HNS resembles a master switch for HuR export. According to this model, we suggest that a single modification in or adjacent to the HNS causes a conformational perturbation which aggravates further posttranslational modifications in this region. Experiments to test the validity of this model are ongoing.
Another important question arising from our data is whether an increase in cytoplasmic HuR abundance caused by AngII is attributable mainly to an increased nuclear export or, alternatively, results from an inhibited HuR (re)import back to the nucleus. Mechanistically, the nuclear export of HuR and its cargo mRNA is predicted to occur via a RanGTP-dependent export receptor and CRM1-dependent transport, which depends on a direct interaction of HuR with the nuclear ligands APRIL and pp32 (7, 47). Similarly to HuR export, the import of HuR to the nucleus is mediated by an interaction of cytoplasmic HuR with transportin 2 via the HNS, which exposes a typical nuclear import signal which is also present in various other RBPs (25). The observation that Ser 221 HuR-Flag mutants, in contrast to the wild-type HuR-Flag or endogenous HuR, even after AngII stimulation fully reside within the nucleus (Fig. 1B) indicates that the increase in cytoplasmic HuR levels caused by AngII results from an increased export of HuR from the nucleus to the cytoplasm rather than from a blockade of HuR import back to the nucleus. Furthermore, our finding from RNP-IP demonstrating that the AngII-induced increase in HuR binding to nuclear target mRNAs is not impaired in Ser 221-mutated HuR constructs implies that the HuR-mRNA complex induced by AngII is initially formed already within the nucleus. Further experimental evidence is needed to test whether some of the known nuclear HuR receptors could be additional targets of PKCδ.
Our study provides clear evidence that PKCδ-dependent phosphorylation within the highly conserved RRM3 promotes HuR binding to ARE-mRNA in a direct manner, thus indicating that modification at this RRM is an important mechanism of controlling stimulus-dependent HuR binding to ARE-mRNA. It is worth mentioning that in contrast to the PKCδ-triggered increase in RNA binding, Chk2-mediated phosphorylation of HuR reduces its association with SIRT1 mRNA and other target mRNAs (1). Besides Ser 221, we have previously identified Ser 158 as a further critical phosphorylation site, which is indispensable for HuR phosphorylation by the conventional PKCα isoenzyme (14). Notably, Ser 158 resides within RRM2, and phosphorylation of this residue by PKCα likewise results in an increased HuR binding and stabilization of COX-2 mRNA, which bears both class II and class III AREs (14). This raises the important question whether phosphorylation-specific modification of different RRMs may confer target specificity in HuR binding toward a certain class of ARE.
By elucidating specific functions of different HuR domains, Fialcowitz-White et al. (21) could demonstrate that removal of RRM3, in contrast to the effects caused by RRM1 deletion, did not affect the binding affinity of HuR to synthetic AREs but instead had a strong negative influence on the formation of multisubunit protein complexes on ARE substrates. However, since the study did not investigate the possible impact of posttranslational HuR modifications on RNA binding affinity, their findings are not necessarily discrepant with our observations. Furthermore, by employing CD spectroscopy, we could demonstrate that a point mutation of Ser 318 had no significant effect on the protein folding of full-length HuR (Fig. 2B), thus substantiating our hypothesis that the loss in RNA binding observed with HuRΔSer318 is mainly due to an inefficient phosphorylation by PKCδ. Similar to the case for HuR, a posttranslational modification of the hnRNP K-homology (KH) domain 3 of the RNA binding protein hnRNP K by the c-Src kinase negatively affects its interaction with the differentiation control element (DICE) (43).
An emerging model of regulation of RBP functions is that the transient association of the phosphorylated forms of regulatory proteins, including TTP (50, 57), KSRP (12, 24), and HuR (33) with 14-3-3 scaffold proteins hinders their subcellular distribution and posttranscriptional influence on target ARE-mRNA. Similar to the phosphorylation-induced interaction of HuR with 14-3-3θ by Cdk1 (33), the association of KSRP, TTP, and BRF1 (52) to 14-3-3 chaperons is triggered through phosphorylation of the relevant RBP. Furthermore, in all cases, an association with the relevant chaperon was found predominantly with the phosphorylated RBP but not with the unphosphorylated protein. We found that AngII induces a moderate increase in the constitutive binding of nuclear HuR to 14-3-3 chaperons, although we could not further identify which specific member of the 14-3-3 protein family is modulated by AngII. 14-3-3, by virtue of its acidic nature, is known to promote PKC activity like acidic lipids do (58). In addition, it is tempting to hypothesize that 14-3-3 chaperons may selectively help to couple HuR to other nuclear target proteins involved in AngII-induced export of HuR. Further studies are needed to unravel the functional role of 14-3-3 in PKCδ-mediated HuR shuttling.
According to the antagonistic actions on HuR shuttling, PKCδ and Cdk1 have opposing effects on the mRNA stability of cyclins and thus may exert an antagonistic influence on cyclin-controlled cell functions (35, 59). Although AngII in many cell types exerts a predominant mitogenic activity, in some cell types, including vascular smooth muscle cells and MC, AngII inhibits cell proliferation but acts to trigger a hypertrophic response, mainly through activation of the AT2 receptor (6, 29, 53, 55). Accordingly, we found that AngII had no positive influence on mesangial cell proliferation but exerted a strong migratory influence on HMC, which was markedly impaired by depletion of either HuR or PKCδ (Fig. 6), thus indicating a functional link between the PKCδ-mediated and HuR-dependent stabilization of cyclins and cell migration by AngII. This is in agreement with the well-established functions of some cyclins, such as cyclin D1, which promotes cell migration mainly by inhibiting the Rho/ROCK-dependent signaling cascades (reviewed in reference 38).
In summary, our data establish the regulatory role of posttranslational HuR modification by PKCδ, which acts as a master switch in HuR and HuR-controlled cell responses such as cell migration and others triggered by AngII.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft grants EB 257/2-2, PF 361/6-1, GRK757, GRK1172 FOG 784, and EXC 147/1.
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Abdelmohsen, K., R. Pullmann, Jr., A. Lal, H. H. Kim, S. Galban, X. Yang, J. D. Blethrow, M. Walker, J. Shubert, D. A. Gillespie, H. Furneaux, and M. Gorospe. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25:543-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelmohsen, K., Y. Kuwano, H. H. Kim, and M. Gorospe. 2008. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 389:243-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelmohsen, K., S. Srikantan, Y. Kuwano, and M. Gorospe. 2008. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U. S. A. 105:20297-20302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelmohsen, K., S. Srikantan, X. Yang, A. Lal, H. H. Kim, Y. Kuwano, S. Galban, K. G. Becker, D. Kamara, R. de Cabo, and M. Gorospe. 2009. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 28:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akool, el-S., H. Kleinert, F. M. Hamada, M. H. Abdelwahab, U. Förstermann, J. Pfeilschifter, and W. Eberhardt. 2003. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA-stabilizing factor HuR. Mol. Cell. Biol. 23:4901-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun-Dullaeus, R. C., M. J. Mann, A. Ziegler, H. E. von der Leyen, and V. J. Dzau. 1999. A novel role for the cyclin-dependent kinase inhibitor p27(Kip1) in angiotensin II-stimulated vascular smooth muscle cell hypertrophy. J. Clin. Invest. 104:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y., N. Xu, and A. B. Shyu. 2002. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol. Cell. Biol. 22:7268-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denkert, C., W. Weichert, S. Pest, I. Koch, D. Licht, M. Kobel, A. Reles, J. Sehouli, M. Dietel, and S. Hauptmann. 2004. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 64:189-195. [DOI] [PubMed] [Google Scholar]
- 11.Denkert, C., W. Weichert, K. J. Winzer, B. M. Muller, A. Noske, S. Niesporek, G. Kristiansen, H. Guski, M. Dietel, and S. Hauptmann. 2004. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin. Cancer Res. 10:5580-5586. [DOI] [PubMed] [Google Scholar]
- 12.Díaz-Moreno, I., D. Hollingworth, T. A. Frenkiel, G. Kelly, S. Martin, S. Howell, M. García-Mayoral, R. Gherzi, P. Briata, and A. Ramos. 2009. Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat. Struct. Mol. Biol. 16:238-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon, D. A., N. D. Tolley, P. H. King, L. B. Nabors, T. M. McIntyre, G. A. Zimmerman, and S. M. Prescott. 2001. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 108:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doller, A., A. Huwiler, R. Müller, H. H. Radeke, J. Pfeilschifter, and W. Eberhardt. 2007. Protein kinase C α-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell 18:2137-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doller, A., J. Pfeilschifter, and W. Eberhardt. 2008. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 20:2165-2173. [DOI] [PubMed] [Google Scholar]
- 16.Doller, A., el-S. Akool, A. Huwiler, R. Müller, H. H. Radeke, J. Pfeilschifter, and W. Eberhardt. 2008. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cδ elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol. Cell. Biol. 28:2608-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doller, A., S. Gauer, E. Sobkowiak, H. Geiger, J. Pfeilschifter, and W. Eberhardt. 2009. Angiotensin II induces renal plasminogen activator inhibitor-1 and cyclooxygenase-2 expression post-transcriptionally via activation of the mRNA-stabilizing factor human-antigen R. Am. J. Pathol. 174:1252-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberhardt, W., M. Schulze, C. Engels, E. Klasmeier, and J. Pfeilschifter. 2002. Glucocorticoid-mediated suppression of cytokine-induced matrix metalloproteinase-9 expression in rat mesangial cells: involvement of nuclear factor-κB and Ets transcription factors. Mol. Endocrinol. 6:1752-1766. [DOI] [PubMed] [Google Scholar]
- 19.Fan, X. C., and J. A. Steitz. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17:3448-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, X. C., and J. A. Steitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U. S. A. 95:15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fialcowitz-White, E. J., B. Y. Brewer, J. D. Ballin, C. D. Willis, E. A. Toth, and G. M. Wilson. 2007. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J. Biol. Chem. 282:20948-20959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Martínez, J., A. Aranda, and J. E. Pérez-Ortín. 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Geiges, D., T. Meyer, B. Marte, M. Vanek, G. Weissgerber, S. Stabel, J. Pfeilschifter, D. Fabbro, and A. Huwiler. 1997. Activation of protein kinase C subtypes alpha, gamma, delta, epsilon, zeta, and eta by tumor-promoting and nontumor-promoting agents. Biochem. Pharmacol. 53:865-875. [DOI] [PubMed] [Google Scholar]
- 24.Gherzi, R., M. Trabucchi, M. Ponassi, T. Ruggiero, G. Corte, C. Moroni, C. Y. Chen, K. S. Khabar, J. S. Andersen, and P. Briata. 2006. The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Güttinger, S., P. Mühlhäusser, R. Koller-Eichhorn, J. Brennecke, and U. Kutay. 2004. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. U. S. A. 101:2918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, C., and R. Schneider. 2006. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 25:3823-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinonen, M., P. Bono, K. Narko, S. H. Chang, J. Lundin, H. Joensuu, H. Furneaux, T. Hla, C. Haglund, and A. Ristimäki. 2005. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 65:2157-2161. [DOI] [PubMed] [Google Scholar]
- 28.Hollams, E. M., K. M. Giles, A. M. Thomson, and P. J. Leedman. 2002. mRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 27:957-980. [DOI] [PubMed] [Google Scholar]
- 29.Huwiler, A., S. Stabel, D. Fabbro, and J. Pfeilschifter. 1995. Platelet-derived growth factor and angiotensin II stimulate the mitogen-activated protein kinase cascade in renal mesangial cells: comparison of hypertrophic and hyperplastic agonists. Biochem. J. 305:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, B. A., J. R. Stehn, M. B. Yaffe, and T. K. Blackwell. 2002. Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 277:18029-18036. [DOI] [PubMed] [Google Scholar]
- 31.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9:1161-1167. [DOI] [PubMed] [Google Scholar]
- 32.Keene, J. D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. U. S. A. 96:5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, H. H., K. Abdelmohsen, A. Lal, R. Jr. Pullmann, X. Yang, S. Galban, S. Srikantan, J. L. Martindale, J. Blethrow, K. M. Shokat, and M. Gorospe. 2008. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 22:1804-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, H. H., X. Yang, Y. Kuwano, and M. Gorospe. 2008. Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle 7:3371-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, H. H., and M. Gorospe. 2008. Phosphorylated HuR shuttles in cycles. Cell Cycle 7:3124-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloss, S., H. Furneaux, and A. Mülsch. 2003. Post-transcriptional regulation of soluble guanylyl cyclase expression in rat aorta. J. Biol. Chem. 278:2377-2383. [DOI] [PubMed] [Google Scholar]
- 37.Li, H., S. Park, B. Kilburn, M. A. Jelinek, A. Henschen-Edman, D. W. Aswad, M. R. Stallcup, and I. A. Laird-Offringa. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 277:44623-44630. [DOI] [PubMed] [Google Scholar]
- 38.Li, Z., C. Wang, G. C. Prendergast, and R. G. Pestell. 2006. Cyclin D1 functions in cell migration. Cell Cycle 5:2440-2442. [DOI] [PubMed] [Google Scholar]
- 39.Lin, S., W. Wang, G. M. Wilson, X. Yang, G. Brewer, N. J. Holbrook, and M. Gorospe. 2000. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 20:7903-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 41.Mazan-Mamczarz, K., P. R. Hagner, S. Corl, S. Srikantan, W. H. Wood, K. G. Becker, M. Gorospe, J. D. Keene, A. S. Levenson, and R. B. Gartenhaus. 2008. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 27:6151-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazroui, R., S. Di Marco, E. Clair, C. von Roretz, S. A. Tenenbaum, J. D. Keene, M. Saleh, and I. E. Gallouzi. 2008. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J. Cell Biol. 180:113-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messias, A. C., C. Harnisch, A. Ostareck-Lederer, M. Sattler, and D. H. Ostareck. 2006. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-src-mediated tyrosine phosphorlyation. J. Mol. Biol. 361:470-481. [DOI] [PubMed] [Google Scholar]
- 44.Moore, M. J. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309:1514-1518. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima, H., G. D. Frank, H. Shirai, A. Hinoki, S. Higuchi, H. Ohtsu, K. Eguchi, A. Sanjay, M. E. Reyland, P. J. Dempsey, T. Inagami, and S. Eguchi. 2008. Novel role of protein kinase C-δ Tyr 311 phosphorylation in vascular smooth muscle cell hypertrophy by angiotensin II. Hypertension 51:232-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newbury, S. F., O. Mühlemann, and G. Stoecklin. 2006. Turnover in the Alps: an mRNA perspective. Workshops on mechanisms and regulation of mRNA turnover. EMBO Rep. 7:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebane, A., A. Aab, and J. A. Steitz. 2004. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10:590-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyland, M. E. 2007. Protein kinase Cδ and apoptosis. Biochem. Soc. Trans. 35:1001-1004. [DOI] [PubMed] [Google Scholar]
- 49.Sadoshima, J., H. Aoki, and S. Izumo. 1997. Angiotensin II and serum differentially regulate expression of cyclins, activity of cyclin-dependent kinases, and phosphorylation of retinoblastoma gene product in neonatal cardiac myocytes. Circ. Res. 80:228-241. [DOI] [PubMed] [Google Scholar]
- 50.Sandler, H., and G. Stoecklin. 2008. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 36:491-496. [DOI] [PubMed] [Google Scholar]
- 51.Sengupta, S., B. C. Jang, M. T. Wu, J. H. Paik, H. Furneaux, and T. Hla. 2003. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 278:25227-25233. [DOI] [PubMed] [Google Scholar]
- 52.Schmidlin, M., M. Lu, S. A. Leuenberger, G. Stoecklin, M. Mallaun, B. Gross, R. Gherzi, D. Hess, B. A. Hemmings, and C. Moroni. 2004. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 23:4760-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schöcklmann, H. O., S. Lang, and R. B. Sterzel. 1999. Regulation of mesangial cell proliferation. Kidney Int. 56:1199-1207. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber, E., P. Matthias, M. M. Müller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulze-Lohoff, E., M. Köhler, H. Fees, N. Reindl, and R. B. Sterzel. 1993. Divergent effects of arginine vasopressin and angiotensin II on proliferation and expression of the immediate early genes c-fos, c-jun and Egr-1 in cultured rat glomerular mesangial cells. J. Hypertens. 11:127-134. [DOI] [PubMed] [Google Scholar]
- 56.Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W. F. Rigby, T. K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-13-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, L., G. Stoecklin, S. Van Way, V. Hinkovska-Galcheva, R. F. Guo, P. Anderson, and T. P. Shanley. 2007. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-α mRNA. J. Biol. Chem. 282:3766-3777. [DOI] [PubMed] [Google Scholar]
- 58.Van Der Hoeven, P. C., J. C. Van der Wal, P. Ruurs, and W. J. Van Blitterswijk. 2000. Protein kinase C activation by acidic proteins including 14-3-3. Biochem. J. 347:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, W., M. C. Caldwell, S. Lin, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida, K., Y. Miki, and D. Kufe. 2002. Activation of SAPK/JNK signaling by protein kinase Cdelta in response to DNA damage. J. Biol. Chem. 277:48372-48378. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]