Abstract
Mitochondrial dysfunction underlying changes in neurodegenerative diseases is often associated with apoptosis and a progressive loss of neurons, and damage to the mitochondrial genome is proposed to be involved in such pathologies. In the present study we designed a mouse model that allows us to specifically induce mitochondrial DNA toxicity in the forebrain neurons of adult mice. This is achieved by CaMKIIα-regulated inducible expression of a mutated version of the mitochondrial UNG DNA repair enzyme (mutUNG1). This enzyme is capable of removing thymine from the mitochondrial genome. We demonstrate that a continual generation of apyrimidinic sites causes apoptosis and neuronal death. These defects are associated with behavioral alterations characterized by increased locomotor activity, impaired cognitive abilities, and lack of anxietylike responses. In summary, whereas mitochondrial base substitution and deletions previously have been shown to correlate with premature and natural aging, respectively, we show that a high level of apyrimidinic sites lead to mitochondrial DNA cytotoxicity, which causes apoptosis, followed by neurodegeneration.
A variety of both exogenous and endogenous reactive compounds present a constant threat to the integrity of DNA in living cells. DNA damage introduced by such compounds can lead to high and deleterious mutation rates as well as DNA cytotoxicity, both to the nuclear and the mitochondrial genome. This has triggered the evolution of several different DNA repair pathways (28). One is the base excision repair (BER) pathway, which repairs small base alterations that do not distort the DNA helix. Repair of such highly abundant lesions by BER is performed by a multistep process that is initiated by a damage-specific DNA glycosylase, which removes the damaged base. One of these glycosylases is uracil-DNA glycosylase (UDG), which acts to preserve the genome by removing mutagenic uracil residues from the DNA. This glycosylase, as well as the OGG1 glycosylase that is specialized for the removal of oxidized bases, exists in a nuclear and mitochondrial splice form (1, 11, 37, 45). Accordingly, BER of a variety of lesions has been observed in mitochondria (26, 31).
Damage to the mitochondrial DNA (mtDNA) can cause respiratory chain deficiency and lead to disorders that have varied phenotypes (35, 41). Many involve neurological features that are often associated with cell loss within specific brain regions. These pathologies, along with the increasing evidence of a decline in mitochondrial function with aging, have raised speculation that key changes in mitochondrial DNA sequences and functions could have a vital role in age-related neurodegenerative diseases (41). This has also been studied in several model organisms. Mouse models with respiratory chain deficient dopamine neurons have demonstrated adult onset Parkinsonism phenotype (16), and cell death induced by mitochondrial toxicity is likely to underlie Alzheimer disease (32). Mitochondrial oxidative stress and accumulation of mtDNA damage are believed to be particularly devastating to postmitotic differentiated tissue, including neurons (30). The mtDNA contains genetic information for 13 polypeptides that are a part of the electron transport chain and for rRNAs and tRNAs that are necessary for mitochondrial protein synthesis. Thus, damage to the mtDNA genome will affect the energetic capacities of the mitochondria and also influence the level of reactive oxygen species (ROS) and ultimately the susceptibility to apoptosis (30, 35).
Some recent influential studies have assessed the effect of mtDNA mutagenesis, including small base-pair substitutions and larger mtDNA deletions, on the life span of mice. It was concluded that a massive increase in the frequency of mtDNA base-pair substitutions are required for inducing premature aging, whereas the number of mtDNA deletions coincides better with natural aging (25, 47-49).
In the present study, we have combined two novel transgenic mouse models, which allow the induction of a high number of apyrimidinic (AP) sites specifically to the mitochondrial genome in adults simply by the addition of doxycycline to the diet. Such AP sites are created by the expression of a mutated version of mitochondrion-targeted human UDG (abbreviated here as mutUNG1), whereby an amino acid substitution results in an enzyme that removes thymine, in addition to uracil, from DNA (23). The CaMKIIα promoter restricts expression of the mutUNG1 to forebrain neurons (34). We demonstrate that a continuous generation of AP sites leads to apoptosis, accelerated neurodegeneration, and impaired behavior.
MATERIALS AND METHODS
Transgenic constructs and mice.
A pGEM7ZF+ plasmid containing human UNG1 (pUNG15) was a gift from Hans Krokan (38). Site-directed mutagenesis was carried out by using a QuikChange site-directed mutagenesis kit (Stratagene). The primers used in the reaction to obtain Tyr147Ala were 5′-GCTTGATTAGGTCCATGAGCTGGATCCTGTCCCAG-3′ and 5′-CTGGGACAGGATCCAGCTCATGGACCTAATCAAGC-3′. The mutUNG1 was cloned into a pBI-L Tet vector (Clontech). Transgenic mice were generated by pronuclear injection of the pBI-l-mutUNG1 construct using the C57BL/6 strain. Positive founder mice were bred with transgenic mice containing the rtTA2S-M2 Tet-on system, under the control of the CaMKIIα promoter (34). The presence of mutUNG1, luciferase, and rtTA2 were verified by PCR with the following primers: mutUNG1, 5′-TTAGAACCTAATTCCCAATTCCCGGACCG-3′ and 5′-GCTTCTTCCAGCTCTCTCCAAAGCC-3′; luciferase, 5′-GTGGATCCCCCCTAGAGTATTACAATAGC-3′ and 5′-TGGAGACATAGCTTACTGGGACGAAGAC-3′; and rtTA2, 5′-TGCCTTTCTCTCCACAGGTGTCC-3′ and 5′-GAGAGCACAGCGGAATGAC-3′. Doxycycline was administered as previously described (34). Transgenic founders were selected by investigating luciferase expression in brains utilizing in vivo and ex vivo imaging. In vivo imaging was performed by administering d-luciferin intraperitoneally (120 to 150 mg/kg) to mice anesthetized with 2.5% isoflurane. Bioluminescence was recorded by the IVIS100 imaging system (Xenogen/Caliper). Images were analyzed by Living Image software (Xenogen/Caliper).
All experimental procedures were approved by the Section for Comparative Medicine at the Oslo University Hospital and by the Norwegian Animal Research Authority and complied with national laws and institutional regulations governing the use of animals in research.
Cell line.
The pBI-l-mutUNG1 construct was stable transfected into a HeLa Tet-On cell line, together with a linear hygromycin marker (Clontech) by using FuGENE 6 transfection reagent (Roche). Cells were grown in the presence of 400 μg of hygromycin B (Clontech)/ml for selection. The presence of the transgene was confirmed by sequencing, and the expression was based on measuring luciferase activity, using a luciferase assay system (Promega) and a TD20/20 luminometer (Turner Designs). The stable transfected cell line was induced by using 1 μg/ml of doxycycline (Clontech).
Mitochondrial extract.
Mitochondria were isolated with the Mitochondria isolation kit (Pierce) and dissolved in a buffer containing 0.5% Igepal CA-630 (Sigma), 2 mM dithiothreitol (DTT), 1% protein inhibitor cocktail (Sigma catalog no. P8340), and 0.5% Triton X-100 (Sigma) in phosphate-buffered saline (PBS). Mitochondria were incubated on ice for 5 min and centrifuged at a maximum speed on a table centrifuge, and the protein concentration was measured by using Bio-Rad protein assay dye reagent concentrate (Bio-Rad).
Western blot analysis.
Western blot analysis was performed using NuPAGE 10% Bis-Tris gels and Invitrolon polyvinylidene difluoride membranes (Invitrogen). A total of 20 μg of protein mitochondrial extract was applied. The presence of UNG1 was confirmed with 2 μg/ml of UNG1 antibody (ProSci, Inc.), and anti-complex II 70-kDa Fp subunit monoclonal antibody diluted 1/1500 (Invitrogen) was used as a mitochondrial marker and loading control. Secondary antibodies used were anti-rabbit IgG (whole molecule)-alkaline phosphatase antibody (Sigma catalog no. A3812), dilution of 1:30,000 or goat anti-mouse IgG-AP (Santa Cruz Biotechnology, Inc., catalog no. sc-2008), dilution of 1:5,000, respectively. Bands were visualized using ECF substrate (GE Healthcare), a Typhoon 9410, and ImageQuant TL v2003.02 software (Amersham/GE Healthcare).
RT-PCR.
Freshly dissected brain tissue was homogenized by using tubes from a FastRNA Pro Green kit (QBiogene) containing lysis buffer from a Dynabeads mRNA Direct kit (Invitrogen) and a FastPrep Instrument (QBiogene). The supernatant was collected, and mRNA was isolated according to the protocol of Dynabeads mRNA Direct kit (Invitrogen). Reverse transcriptase PCR (RT-PCR) was performed by using a Titanium one-step RT-PCR kit (Clontech), with 30 ng of RNA for each reaction. The primers 5′-ATGGGCGTCTTCTGCCTTGGGCCGT-3′ and 5′-AAAATACGGTTTCCCGAACTCCCCGCT-3′ were used. The annealing temperature in the PCR was set to 60°C, and 25 cycles were used. PCR product was run on a 1% agarose gel, and bands were visualized by using an AlphaImager (Alpha Innotech).
Preparation of substrate for oligonucleotide nicking assay.
A 31-mer oligonucleotide containing either a single uracil or a single thymine residue placed at position 16 from the 5′ terminus was used as a substrate; uracil-substrate, 5′-GGGGCCCGGGCCCCGUGGGCCCCGGGGCCCG-3′, thymine-substrate, 5′-GGGGCCCGGGCCCCGTGGGCCCCGGGGCCCG-3′ and complementary strand 5′-CGGGCCCCGGGGCCCACGGGGCCCGGGCCCC-3′ (for both substrates). All oligonucleotides included phosphorothioate linkages at the ultimate and penultimate 5′ and 3′ residues to reduce exonucleolytic attack (MWG Biotech AG).
The oligonucleotides were end labeled at the 5′ terminus. The labeling reaction contained 28 pmol of the oligonucleotide, 1.48 MBq (0.04 mCi) of [γ-32P]ATP (Perkin-Elmer), 20 U of T4 polynucleotide kinase (New England Biolabs), TSE-buffer (20 mM Tris-HCl [pH 9.5], 1 mM spermidine HCl, 0.1 mM EDTA), and appropriate kinase buffer in a total volume of 60 μl (incubation for 1 h at 37°C). End-labeled oligonucleotide substrates were separated and purified on a denaturing 20% PAGE gel. Double molar amounts of the complementary oligonucleotide were added to form duplex DNA substrate.
Nicking assay.
UDG and TDG activity was measured in a 10-μl assay mixture containing (final) 20 mM Tris-HCl (pH 7.5), 10 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% proteinase inhibitor cocktail (Sigma), 28 nM labeled substrate, and 2.5 μg of total protein (brain) extract. The samples were incubated for 30 min at 37°C, followed by the addition of 2.5 μl of NaOH (0.5 M) and incubation for 20 min at 70°C. The samples were neutralized by adding 2.5 μl of HCl (0.5 M). A total of 10 μl of loading buffer was added, and 10 μl was run on a denaturing 20% PAGE gel. Bands were visualized and quantified by using a Typhoon 9410 and ImageQuant TL v2003.02 software (Amersham/GE Healthcare).
The total protein extract was made by freshly dissected mouse brain tissue that was homogenized in a buffer containing 20 mM Tris-HCl (pH 7.5), 10 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% proteinase inhibitor cocktail (Sigma), and 0.5% Triton X-100 (Sigma). The samples were frozen on ethanol-dry ice and thawed at 37°C; this was performed three times. The samples were then incubated on ice for 5 min and centrifuged at maximum speed on a table centrifuge. The protein concentration was measured by using Bio-Rad protein assay dye reagent concentrate.
Mutation studies.
Point mutation studies of total DNA from brain tissue were performed as described previously (47). The cytochrome B gene was PCR amplified by using the primers 5′-TGGGGTGTTTAGTGGATTAGCTGGTATG-3′ and 5′-GGTGAAGGCTTTAATGCTAACCCAAGAC-3′. PCR-amplified mtDNA from three mutUNG1-expressing mice and three wild-type littermates (cerebellum, hippocampus, and caudate putamen for all) were investigated. The TOPO-cloned cytochrome B gene was sequenced completely using vector-specific M13 primers. Sequences were analyzed by using SeqScape software (Applied Biosystems).
Oxygraph.
Mitochondria were isolated from freshly dissected mouse brain tissue and purified by differential centrifugation in isolation buffer, omitting the Percoll gradient purification step as described in the original protocol (2). The mitochondrial suspension was transferred to an Oxygraph-2K for respiratory analysis in Mir05 buffer (Oroboros Instruments). Complex I respiration was obtained by combined supplement of pyruvate (5 M), malate (2 mM), and glutamate (10 mM). Respiration capacity was determined after the addition of ADP (1 mM), followed by saturating amounts of FCCP to completely uncouple the mitochondria. The coupling numbers in mutUNG1-expressing mice and wild-type littermates were statistically indifferent, and varied: 6 (cerebellum), 2 (hippocampus), and 4 (caudate putamen). Complex II-based respiration was quantified after inhibition of complex I with rotenone (50 uM) and the addition of succinate (10 mM) to the uncoupled mitochondria.
Paraffin sections.
Whole brains were fixed in 10% (vol/vol) neutral buffered formalin (Richard-Allan Scientific) for at least 48 h and paraffin embedded. Brains were cut by using a Rotary microtome 355 S (Microm) into 4-μm-thick sections and dyed with hematoxylin and eosin (HE; Richard-Allan Scientific). HE-stained sections were studied with a Zeiss Axioplan 2 microscope, and pictures were taken and analyzed with an AxioCamHRc and AxioVision Rel. 4.6 software.
Cryosections.
Whole brains were frozen on a razorblade on dry ice. Frozen brains were immersed in Neg50 frozen section medium (Richard-Allan Scientific). An HM 550 Cryostat was used to cut the brains into 8-μm-thick sections.
ARP/streptavidin staining.
Cryosections were treated with biotin blocking kit (Invitrogen) and probed with 20 mM aldehyde-reactive probe (N-(aminooxyacetyl)-N′-(d-biotinoyl) hydrazine, trifluoroacetic acid salt (ARP; Invitrogen) for 1 h at 37°C. Then, the sections were treated with 40 μg/ml of streptavidin-Alexa Fluor 488 conjugate (Invitrogen), which recognizes the biotin tag of the ARP, for 1 h at 37°C and stained with 1 μg/ml of DAPI (4′,6′-diamidino-2-phenylindole; Sigma). Sections were studied with a Zeiss Axiovert 200M microscope, and pictures were taken and analyzed with an AxioCamMRm and AxioVision Rel 4.6 software.
Immunostaining and immunohistochemistry.
Cryosections were fixed in 4% paraformaldehyde and permeabilized in 0,1% Triton X-100 (Sigma) in PBS. The sections were blocked in 5% bovine serum albumin (BSA; Sigma), 5% goat serum (Sigma), and 0.1% Triton X-100 (Sigma). Sections were incubated with anti-MAP2 (Chemicon), diluted 1:500, and monoclonal anti-GFAP (Sigma), diluted 1:400. Both antibodies were diluted in 0.5% BSA (Sigma), 0.5% goat serum, and 0.1% Tween 20 (Sigma) in PBS. The secondary antibodies utilized were Alexa Fluor 594-conjugated goat anti-mouse IgG (H+L; Invitrogen catalog no. A11005) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L; Invitrogen catalog no. A11008). In addition, sections were incubated with 1 μg/ml of DAPI. Mowiol 4-88 reagent (Merck Biosciences, Ltd.) mounting solution and a coverslip were applied, and the sections were studied with a Zeiss Axiovert 200M microscope. Pictures were taken and analyzed with an AxioCamMRm and AxioVision Rel 4.6 software.
TUNEL.
Apoptosis was investigated in cryosections by using an in situ cell death detection kit, TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling), and TMR Red staining (Roche) according to the manufacturer's recommendations. In addition, sections were stained with 1 μg/ml DAPI. Sections were studied with a Zeiss Axiovert 200M microscope, and pictures were taken and analyzed with an AxioCamMRm and AxioVision Rel 4.6 software.
Tissue preparation for electron microscopy.
Hippocampus and cerebellum in mice (sections were typically 0.5 by 0.5 by 1 mm) were cryoprotected by immersion in graded concentrations of glycerol (10, 20, and 30%) in PBS. The samples were then plunged into liquid propane cooled to −190°C by liquid nitrogen in a Universal Cryofixation System KF80 (Reichert-Jung). For freeze substitution, the tissue samples were immersed in a solution of anhydrous methanol and 0.5% uranyl acetate overnight at −90°C. The temperature was then raised stepwise in 4°C increments per hour from −90°C to −45°C, at which temperature it was kept for the subsequent steps. The tissue samples were then washed several times with anhydrous methanol to remove residual water and uranyl acetate. The infiltration in Lowicryl resin HM20 went stepwise from Lowicryl-methanol 1:2, 1:1, and 2:1 (1 h each) to pure Lowicryl (overnight). For polymerization, the tissue was placed in a precooled embedding mall, and the polymerization was catalyzed by UV light of 360 nm wavelength for 2 days at −45°C, followed by 1 day at room temperature. Ultrathin sections (90 nm) were cut by a diamond knife on a Reichert-Jung ultramicrotome and mounted on nickel grids using an adhesive pen (Electron Microscopy Sciences).
The ultrathin sections were contrasted in uranyl acetate (5%) and lead citrate (30%). Sections were observed in a Philips CM100 electron microscope. Pictures were taken at primary magnifications of ×25,000.
Quantitative electron microscopic analysis.
Electron micrographs with clearly visible synapses were taken randomly in the stratum radiatum of CA1, ∼10 μm apical to the pyramidal cell layer. The excitatory synapses were identified as small terminals forming asymmetric synaptic specializations on dendritic spines (Schaffer collateral synapses). The lengths (in nanometers) of the postsynaptic membranes of excitatory synapses were recorded (6) in mutUNG1-expressing mice and wild-type littermates. The lengths of postsynaptic membranes were also recorded at synapses between excitatory parallel fiber and Purkinje cell dendritic spines in the molecular layer of the cerebellum in mutUNG1-expressing mice and wild-type littermates. Mice were induced for 7 to 8 months.
Behavior.
Open field tests were conducted using a VersaMax animal activity monitoring system (AccuScan Instruments, Inc.). Two mice were measured simultaneously for 10 min, once a month. The tester was blind to the genotypes, and the mice were measured in random order. Twenty mice (eleven mutUNG1-expressing mice and nine wild-type littermates) were used in this experiment. The data were analyzed by using VersaDat software (AccuScan Instruments, Inc.).
The Morris water maze task was conducted in a white circular tank, 120 cm in diameter and 30 cm deep, filled with 20 cm of white opaque water (SikaLatex liquid) that was maintained at 24 ± 1°C. A pneumatically controlled escape platform 11 cm in diameter was located at a fixed position midway between the center and the wall. The platform was usually 1.0 cm below the water surface, but on Atlantis and probe trials the platform was submerged to the bottom; during the Atlantis tests, this was only for the first 40 s. The experimenter sat in a fixed position 2 m from the pool and was visible to the animals during the task.
Training in the Morris water maze consisted of eight blocks of four trials. The mice were given two blocks of trials per day with an interblock interval of 3 h. The first trial of the first block each day was an Atlantis trial. The start position for each trial was randomly drawn from four possible positions. The mice were released into the water with their heads facing the maze wall and were given a maximum of 60 s to locate the hidden platform. The mice remained on the platform for 15 s after each trial. Retention tests, or probe trials, with the escape platform submerged to the bottom of the pool, were conducted on days 5 and 11. The test order was randomized (50). For thigmotaxis measurements, the thigmotaxis zone was extended 20 cm form the wall into the pool. Twelve mice (six mutUNG1-expressing mice and six wild-type littermates) were used in this experiment. The position of the mice was tracked and stored at 10 Hz by a video tracking system (Any-Maze, Stoelting, IL).
The zero-maze task (44) was conducted in a custom-made apparatus consisting of a white 5-cm-wide circular runway with an inner diameter of 50 cm and elevated 65 cm above the floor. The runway had alternating open and closed quadrants. The closed quadrants had a metal outer wall and a transparent inner wall of 20-cm height. The open quadrants had a 1-cm-high lip along the inner and outer edges to prevent the mice from falling off the runway.
Testing took place under 110 lx of illumination recorded at the level of runway in both open quadrants. The test order was randomized. The mice were placed on the maze facing a closed quadrant and allowed 5 min for exploration of the apparatus. The position of the mice was tracked and stored at 10 Hz by a video tracking system, and the number and duration of head dips was manually recorded in the tracking software. An open area entry was defined as 85% of the mouse inside an open area. Twelve mice (six mutUNG1-expressing mice and six wild-type littermates) were used in this experiment.
Statistical analysis.
All quantitative date are represented as means ± the standard error of the mean. Unpaired, two-tailed t tests were performed unless otherwise stated. The null hypothesis was rejected at the 0.05 level.
RESULTS
Characterization of the CaMKIIα-inducible mutUNG1 mouse model.
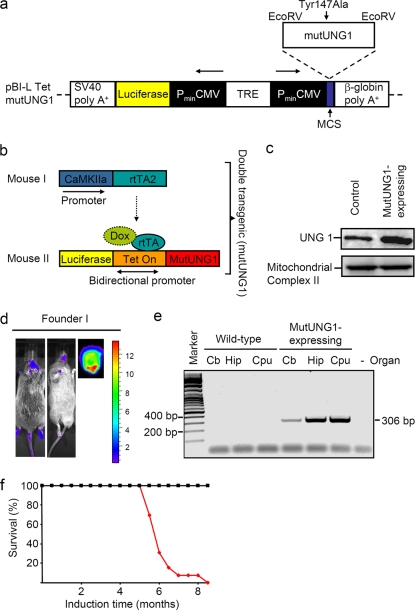
The present study was initiated to explore the influence of mtDNA damage in the forebrain neurons of mice, particularly in association with behavioral defects, aging, and associated diseases. We wanted to investigate the impact of mtDNA damage and have addressed this by producing transgenic mice expressing a mutated mitochondrion-targeted DNA repair enzyme. The enzyme is an engineered modification of human UNG1 (Y147A; mutUNG1) that catalyzes the removal of thymine from DNA in addition to uracil (23). Thus, mutUNG1 induces DNA damage in the form of AP sites. In contrast to the endogenous murine Ung1 protein, which is transported both to the nucleus and to the mitochondria (36), human UNG1 used in the present study is sorted exclusively to the mitochondria (39). We used the Tet-on system combined with the CaMKIIα promoter to restrict mutUNG1 expression to forebrain neurons in the adult mouse brain (5, 34). The mutUNG1 gene was cloned into a vector containing a bidirectional Tet on promoter (4). In this vector, luciferase was coexpressed with mutUNG1 and used as a reporter gene (Fig. 1a and b). To test the inducible potential of mutUNG1 and luciferase, the construct was transfected into Tet-on stable transfected cells and induced by the addition of doxycycline. Reporter expression was confirmed by luminometer analysis (data not shown), and increased levels of UNG1 (consisting of the endogenous murine Ung1 and the transgene expressed mutUNG1) were confirmed by Western analysis (Fig. 1c). Twelve transgenic founder mice were made by using pronuclear injection of the Tet-on luciferase/mutUNG1 DNA construct, and these were bred with rtTA2-mice (34). Founder I was picked for continual studies based on its high and specific luciferase activity (Fig. 1d). Expression of the mutUNG1 was also confirmed by RT-PCR (Fig. 1e). The CaMKIIα promoter is forebrain specific, and the expression in the hippocampus and caudate putamen is severalfold higher than that in the cerebellum (34). The presence of the genes for luciferase, mutUNG1 and the transactivator from the Tet-on system, rtTA2, was also confirmed by genotyping (data not shown).
FIG. 1.
Verification of the CaMKIIα-inducible mutUNG1 mouse model. (a) Construct design for the generation of mutUNG1 inducible mice. (b) Working model of the transgenic mouse. (c) Western analysis showing increased expression of UNG1 in stable transfected cells. Antibodies against mitochondrial complex II were used as a loading control. (d) Luciferase activity measured with a charge-coupled device camera in transgenic founder mouse. Bioluminescence is expressed as photons/s cm2 steradian. (e) Expression of mutUNG1-transgene analyzed by using reverse transcriptase PCR visualized on an agarose gel. Cb, cerebellum; Hip, hippocampus; Cpu, caudate putamen. (f) Survival of wild-type (black) and mutUNG1-expressing mice (red).
The mutUNG1 mice started to show severe signs of sickness between 5.5 and 8.5 months of doxycycline induction (Fig. 1f): they were generally very restless and showed erratic behavior with exaggerated scratching that eventually led to open wounds, especially in areas of the body that they could reach easily, such as the backs of their heads and behind the ears. Therefore, it was no longer ethically correct to keep the mice alive.
Wild-type mice that were on a diet containing doxycycline and mutUNG1 mice that were fed with normal food without doxycycline (and thereby not expressing mutUNG1) resembled the wild-type littermates and did not exhibit the phenotype of the mutUNG1-expressing mice.
Activity of mutUNG1 in transgenic mice.
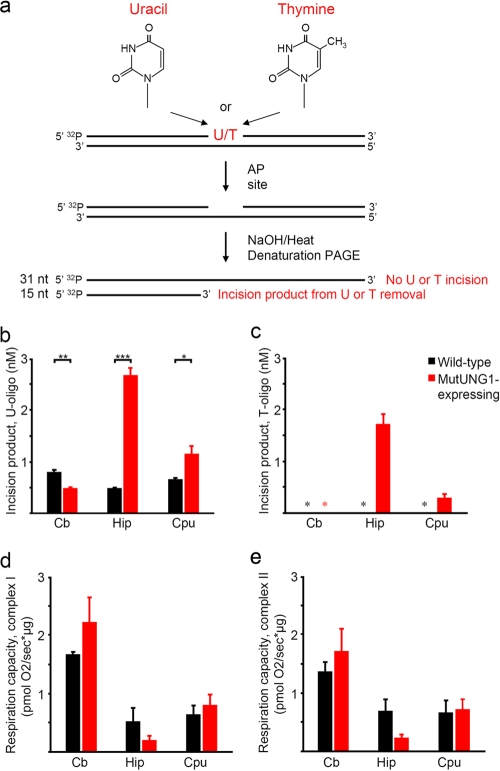
The activity of mutUNG1 was analyzed in protein extracts from hippocampus, caudate putamen, and cerebellum from induced mutUNG1 mice and wild-type littermates. Enzymatic activities were analyzed from the amount of incised thymine or uracil residues after cleavage of the resulting intermediate AP site with NaOH and heat (Fig. 2a to c) (24). A fivefold increase in UDG activity was revealed in hippocampal extracts in mutUNG1-expressing mice compared to wild-type littermates (P ≤ 0.0001). There was a more moderate increase, twofold, in the caudate putamen (P = 0.0133). Whereas significant thymine removal activity was registered for induced hippocampus and, to a lesser extent, in the caudate putamen in mutUNG1-expressing mice, no activity was observed in extracts from wild-type littermates or in induced cerebellum (Fig. 2c). Thus, all detectable thymine removal appears to be associated with expression of mutUNG1. This is expected, since no endogenous protein is capable of removing thymine from normal T:A base pairs.
FIG. 2.
Activity and consequence of mutUNG1expression in transgenic mice. (a) Uracil and thymine removal activity was measured using a 5′-end radioactively labeled 31-mer oligonucleotide containing either a single uracil or a single thymine residue placed at position 16 from the 5′ terminus of the substrate and hybridized to a complementary strand. After incubation with tissue extracts, enzymatic activities were analyzed from the amount of incised thymine or uracil residues after cleavage of the resulting intermediate AP site with NaOH and heat and strand separation by denaturating PAGE. Bands were visualized and quantified by using a Typhoon 9410 and ImageQuant TL v2003.02 software (Amersham/GE Healthcare). (b) Uracil removal activity is highest in induced brain areas in mutUNG1 mice. (c) Thymine removal activity is only present in induced brain areas in mutUNG1 mice. *, no activity was identified. (d) Oxygraph measurements indicating reduced mitochondrial respiration capacity in complex I in the hippocampus of mutUNG1-expressing mice compared to wild-type littermates. (e) Oxygraph measurements indicating reduced mitochondrial respiration capacity in complex II in the hippocampus of mutUNG1 expressing mice compared to wild-type littermates. Cb, cerebellum; Hip, hippocampus; Cpu, caudate putamen.
Consequences of mutUNG1 expression for mitochondrial function.
In order to analyze the functional consequence of mutUNG1 expression, the respiratory activity of mitochondria from mutUNG1-expressing mice and wild-type littermates was measured using an oxygraph. We aimed to quantify the respiratory capacity of individual mice, omitting pooling. Mitochondria from different brain subregions from single mice were isolated and analyzed by high-resolution respirometry. The low amounts of tissue, especially for the hippocampus and caudate putamen, means that we were on the threshold for the oxygraph to register data and measure respiratory activity. As seen (Fig. 2d and e), the hippocampal mitochondria displayed lower activity, during both complex I- and II-supported respiration. This indicates reduced electron transport chain activity in the hippocampal region, whereas no change was observed for the cerebellum or caudate putamen.
Previously, targeting of the proofreading domain of the mtDNA polymerase gamma was reported to increase the frequency of the mtDNA point mutation up to 2,500-fold (48). However, our mice (expressing mutUNG1) have mutation frequencies that are undistinguishable from wild-type littermates. In all tissues tested—hippocampus, caudate putamen, and cerebellum—the frequencies of mutations were always less than 1.2 × 10−5 (7.4 × 104 to 8.5 × 104 bases were sequenced for each tissue sample with three mice in each group).
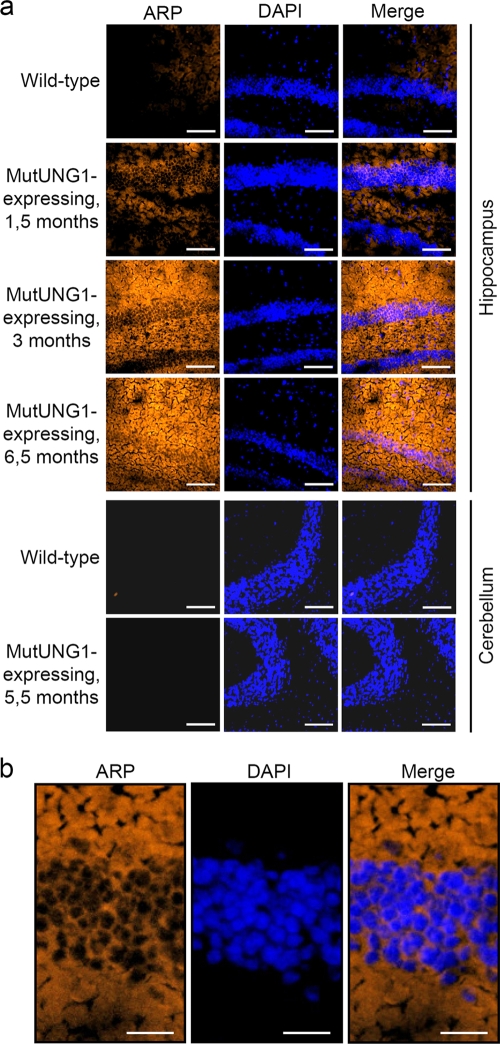
However, mtDNA from the hippocampus of mutUNG1-expressing mice was characterized by highly elevated levels of AP sites compared to wild-type littermates. This was obtained by utilizing an aldehyde-reactive probe, which reacts with the exposed aldehyde group formed at AP sites (Fig. 3a). In the cerebellum, in which mutUNG1 was not expressed, we did not detect an increased level of AP sites. It is worth noticing that AP sites in the hippocampus were completely excluded from the nuclei, highlighting the specific export of mutUNG1 to the mitochondria (Fig. 3b). This corresponds well with the elevated UDG and thymine removal activity (Fig. 2b and c). The lack of mutations is likely due to the existence of enzymes that accurately complete the later stages of BER in mitochondria or, alternatively, the lack of mammalian polymerases for translesion synthesis in mitochondria (42, 52). Thus, our model visualizes the effect of mtDNA damage rather than mtDNA mutations.
FIG. 3.
AP site appearances in mutUNG1-expressing mice. (a) Coronal cryosections of the hippocampus and cerebellum probed with an aldehyde-reactive probe and stained with DAPI demonstrate a high level of AP sites, but only in the hippocampus of induced mutUNG1 mice. Scale bar, 100 μm. (b) AP sites are only seen in parts of the hippocampus containing mitochondria, not nuclei, as demonstrated here in a mutUNG1 mouse induced for 3 months. Scale bar, 25 μm.
Apoptosis and progressive atrophy in the hippocampus of mutUNG1-expressing mice.
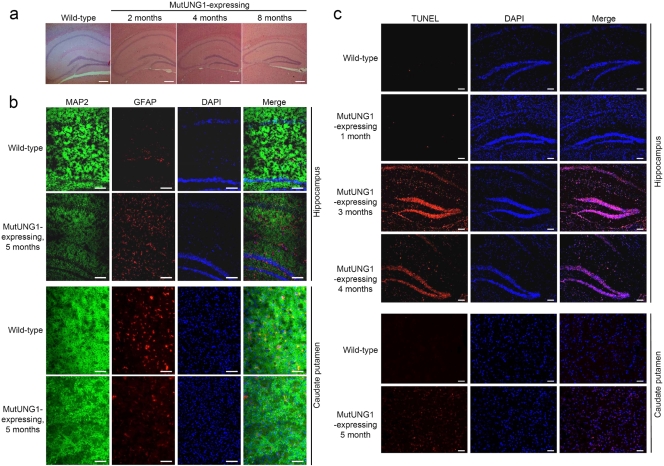
Investigation of HE-stained coronal brain sections revealed progressive atrophy and severe shrinkage of the hippocampus in mutUNG1-expressing mice (illustrated at 2, 4, and 8 months of age in Fig. 4a). Atrophy of the human hippocampus has been seen in a variety of psychiatric and neurological disorders, such as Cushing's syndrome, recurrent depression, schizophrenia, posttraumatic stress disorder, epilepsy, head injury, and normal aging preceding dementia and Alzheimer's disease (13, 33). Rough estimates, by measuring the distance from the back of the dentate gyrus over the stratum radiatum until CA1, show a shrinkage of the hippocampus of ∼25% in mice induced for 2 months, ∼40% in mice induced for 4 months, and ∼45% in mice induced for 8 months, compared to the wild-type littermates.
FIG. 4.
Induced mutUNG1 mice show progressive atrophy, neuronal loss, and apoptosis in the hippocampus. (a) HE-stained coronal paraffin sections of hippocampus of wild-type and mutUNG1-expressing mice at different time points of induction show progressive atrophy in induced mutUNG1 mice. (b) Immunostained coronal cryosections of hippocampus and caudate putamen incubated with antibodies against neuronal marker MAP2 and astrocyte marker GFAP in addition to DAPI stain reveal neuronal loss in hippocampus of induced mutUNG1 mice. (c) TUNEL (and DAPI) staining of cryosections of wild-type and mutUNG1-expressing mice at different time points of induction reveal a high degree of apoptosis in the hippocampus, but not in caudate putamen, in induced mutUNG1 mice. Scale bars, 100 μm.
To investigate the atrophic effect in the hippocampus further, cryosections were immunostained. Microtubule-associated protein 2 (MAP2) was used as a neuronal marker and glial fibrillary acidic protein (GFAP) as a glial cell/astrocyte marker (Fig. 4b). An apparent mutUNG1-dependent progressive reduction in nerve cells was identified in the hippocampus but not in caudate putamen (Fig. 4b). Using a TUNEL assay on cryosections, we found a high degree of apoptosis in mutUNG1-expressing hippocampus. The level of apoptosis peaked at ∼3 months of induction and might reflect the reduced number of cells in mutUNG1-expressing hippocampus at later stages. A more modest level of apoptosis was observed at later time points, at 5 months postinduction, in caudate putamen (Fig. 4c).
Electron microscopy reveals neurodegeneration and reduced membrane length of the postsynaptic density on dendritic spines from hippocampal CA1 pyramidal cells.
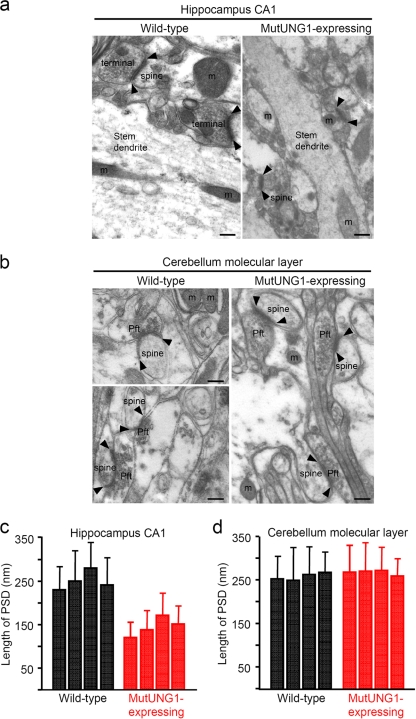
Analysis of hippocampal sections from mutUNG1-expressing mice by electron microscopy revealed signs of neurodegeneration (Fig. 5a), while in the cerebellum—a tissue in which the CaMKIIα promoter is not active—we found no morphological differences when comparing mutUNG1-expressing mice and wild-type littermates (Fig. 5b). Closer inspection of the Schaffer collateral synapses and measurements of the membrane length of the postsynaptic densities (PSD) on dendritic spines of CA1 pyramidal cells revealed a reduction in the PSD size in mutUNG1-expressing mice (P ≤ 0.02, Mann-Whitney test) (Fig. 5c). When the same measurements of the membrane length of the PSD at parallel fiber-Purkinje cell synapses were made in the cerebellar molecular layer, no significant difference was observed between mutUNG1-expressing mice and the wild-type littermates (Fig. 5d). The PSD, a highly complex protein network consisting of hundreds of different proteins, is believed to be an important part of the structural basis for learning and memory formation (7). Thus, a reduction in PSD size in CA1 pyramidal cells is likely to influence these processes in mutUNG1-expressing mice and may ultimately affect the ability to learn and form new memories.
FIG. 5.
Neurodegeneration and reduced membrane length of the postsynaptic density on dendritic spines from mutUNG1-expressing CA1 pyramidal cells. Electron micrographs showing examples of terminals making asymmetric (i.e., excitatory) synapses with dendritic spines in wild-type and mutUNG1-expressing mice in CA1 stratum radiatum of hippocampus (a) and in the cerebellar molecular layer (b). Note the small size of the tissue profiles, including a short length of the PSD (arrowheads), in the mutUNG1-expressing hippocampus. The following structures are indicated: axon terminal (terminal), postsynaptic spine (spine), ends of PSD (arrowheads), mitochondria (m), parallel fiber-Purkinje cell synapses (Pft), and stem dendrite. Scale bars, 150 nm. (c) Histograms showing the mean PSD length ± the standard deviation of excitatory synapses in and four mutUNG1-expressing mice and four wild-type littermates in CA1 stratum radiatum of hippocampus (c) and the cerebellar molecular layer (d). *, the PSD values in all hippocampal mutUNG1-expressing mice were significantly lower than those in wild-type littermates (P < 0.02).
Electron microscopy showed no significant difference in the number of mitochondria between mutUNG1-expressing mice and wild-type littermates, leading us to speculate that the observed phenotype is due to poorly functioning mitochondria rather than to reduced numbers of the organelle.
Behavioral tests reveal increased locomotor activity, but decreased cognitive ability and anxietylike behavior, in mutUNG1-expressing mice.
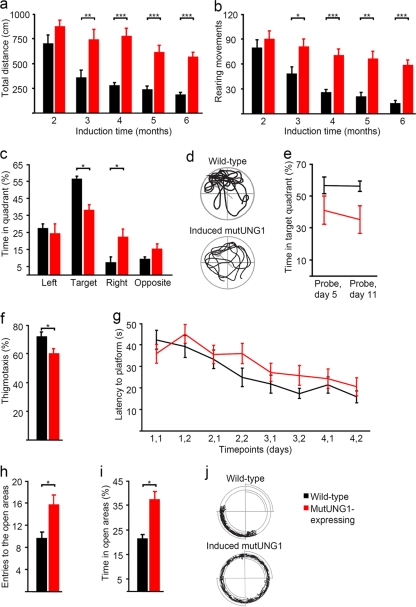
Locomotion and exploratory activity of mutUNG1-expressing mice were tested in an open field. We observed a general decline in exploration with time for both mutUNG1-expressing mice and controls but a relative increase in exploration in mutUNG1-expressing mice compared to controls. Thus, there were significant differences between the mouse groups in the total distance traveled and number of rearings (Fig. 6a and b). Compared to littermate controls, mutUNG1-expressing mice showed a threefold increase in the total distance traveled after 6 months of doxycycline administration (mice were induced from 2 months of age so they are 8 months old, P ≤ 0.0001). However, repeated measurements between 2 and 6 months of induction showed a long-term 1.5-fold decrease in the total distance traveled for mutUNG1-expressing mice (P = 0.0015) and a 3.8-fold decrease for wild-type littermates (P ≤ 0.0001) (Fig. 6a). Compared to littermate controls, analysis of rearing movements showed a 4.5-fold increase in mutUNG1-expressing mice after 6 months of doxycycline administration (8 months old, P ≤ 0.0001). However, between the second and sixth months of induction, there was a 1.4-fold decrease in rearing movements for the mutUNG1-expressing mice (P = 0.0161) and a 3.7-fold decrease for the control mice (P ≤ 0.0001) (Fig. 6b).
FIG. 6.
Increased locomotor activity, but decreased cognitive ability and anxietylike behavior, in mutUNG1-expressing mice. (a) Mice expressing mutUNG1 show higher locomotor activity in the form of total distance traveled in open field tests compared to wild-type littermates. (b) MutUNG1-expressing mice show higher activity in the form of rearing movements in open field tests compared to wild-type littermates. (c) Percentage of time spent in each of the four quadrants of the water maze during probe trials. The mutUNG1 mice spend less time in the target quadrant than the wild-type littermates. The results from both probe trials have been pooled. (d) Representative swimming paths in the water maze during the probe trial on day 5. (e) Percentage of time spent in the target quadrant of the water maze for both probe trials. (f) Percentage of time in thigmotaxis (that is, the tendency of rodents to hug the wall) during the water maze test. (g) Latency to locate platform position during the learning phase of the water maze task. (h) Entries to the open areas of the elevated zero maze. Induced mutUNG1 mice show reduced anxiety responses by a higher entry frequency into the open areas. (i) Percentage of time in the open areas of the elevated zero maze. Induced mutUNG1 mice show reduced anxiety responses by spending more time in the open areas. (j) Representative paths of the head during the elevated zero maze task. Closed quadrants are marked with double lines.
Given the observed morphological and anatomical changes in the hippocampus, we investigated whether mutUNG1 expression affected spatial learning and memory performance in the Morris water maze. This was of particular interest since the Morris water maze measures activities that strongly correlate with hippocampal plasticity and long-term potentiation, and it has previously been shown that memory loss in old rats is associated with brain mitochondrial decay and DNA/RNA oxidation (29, 50). Compared to littermate controls, mutUNG1-expressing mice tended to spend less time searching inside the target quadrant for the submerged platform during the probe trials (P = 0.036, repeated-measures analysis of variance, and P = 0.043, Mann-Whitney test, for both probe trials pooled) (Fig. 6c) were and inclined to spend less time close to the pool wall (P = 0.005, Mann-Whitney test) (Fig. 6f). Furthermore, although no significant effect of treatment was observed (P = 0.079, repeated-measures analysis of variance), analysis of the escape latencies during the learning trials also suggested that the mutUNG1-expressing mice were poorer learners than the wild-type littermates (Fig. 6g). These results indicate that mutUNG1-expressing mice have a learning or memory deficit and a reduced anxietylike response.
Based on the observed thigmotaxis in the water maze (Fig. 6f), we wanted to investigate anxietylike behavior in mutUNG1 expressing mice in the elevated zero maze. Compared to littermate controls, mutUNG1-expressing mice made more entries into (P = 0.024, Mann-Whitney test) and spent a greater percentage of the time (P = 0.005, Mann-Whitney test) in the open areas (Fig. 6h, i, and j). These data indicate that the mutUNG1-expressing mice have reduced anxietylike behavior compared to wild-type littermates.
DISCUSSION
mutUNG1-expressing mice are characterized by a dramatic increase in the steady-state level of cytotoxic AP sites in mtDNA. However, no increase in the mutation frequency could be detected. Possibly, AP sites, which are common intermediates in BER of all types of base lesions, are eventually processed in an error-free manner by the multistep mitochondrial BER pathway (12) or, alternatively, AP sites containing mtDNA are disfavored during replication. DNA damage that is generally reparable by BER is efficiently repaired in mtDNA, whereas mtDNA damage that requires another sort of repair machinery is not (8). AP sites that remain in DNA present a serious threat to the genome by presenting a block to the replicative polymerases. In order to overcome the toxicity of such replication blocks, a set of translesion synthesis polymerases have evolved. Some of these DNA polymerases replicate through the AP site in an error-prone manner. AP sites are noninstructional DNA lesions, and any replication through them are highly mutagenic. In prokaryotes and yeast and mammalian cells an adenine is predominantly inserted opposite an AP site, probably because this confers the least amount of helix distortion, and this has been termed the “A rule” (3, 40, 46). Even though mitochondrial transport of the translesion synthesis components polymerase zeta and Rev1p in Saccharomyces cerevisiae has been demonstrated (52), the only known polymerase in mammalian mitochondria is the replicative polymerase gamma (42). Thus, we propose that the phenotypes observed here are caused entirely by the increase in cytotoxic AP sites. Previous studies in cell cultures have provided evidence that the capacity of the BER pathway for handling AP sites after oxidative stress correlates strongly with mitochondrial initiated apoptosis and neurodegeneration (20). Targeted damage of the DNA structure is considered to be a key approach in the induction of cell death and, among the plethora of death execution programs, the mitochondrial events are of the utmost importance (17, 21). Thus, a significant increase in the number of mitochondrial AP sites could explain the severe apoptosis in our aging colony of doxycycline-induced mice. Similarly, endogenously generated AP sites cause cell death in repair-deficient S. cerevisiae (18). A reduction in mitochondrial respiration could also explain why mutUNG1-expressing cells are more prone to undergo apoptosis (51). Similar to our data, caloric restriction in mice, plus the overexpression of catalase targeted to mouse mitochondria—two treatments that extend the life span of mice—specifically decrease mitochondrial DNA damage and inhibit stress-induced apoptotic cell death (10, 43).
Neurodegenerative disease progression, caused by mitochondrial dysfunction, are often associated with apoptosis and a progressive loss of neurons (41). In addition, defects in energy consumption have been found in Alzheimer's disease, Parkinson's disease and Huntington's disease (14, 15, 32). We observed gross abnormalities and loss of neuronal tissue and atrophy in the hippocampus of mutUNG1-expressing mice. The integrity of the mtDNA is believed to be particularly important for postmitotic tissue such as neurons, since mitochondria have a central role in synapses in that they provide the energy required for synaptic activity, and dysfunction can cause aberrant nerve function. The mutUNG1-expressing mice also show differences in behavior compared to wild-type littermates, such as increased locomotor activity and rearing in open-field tests and reduced anxietylike responses in elevated zero maze tests. These results demonstrate the severity and toxicity of damage to mtDNA in the form of AP sites. In contrast, mice expressing polymerase gamma carrying a deleted proofreading domain in forebrain neurons were characterized by elevated levels of mtDNA point mutations and deletions, and such mice displayed a mood disorder. However, degeneration in the brain or behavioral abnormalities were not identified in these mice (22).
The PSD contains proteins that are essential for synaptic transmission, and dynamic rearrangements of the PSD are thought to contribute to synaptic regulation and plasticity, underlying learning and memory formation (7). Therefore, the reduced PSD size in CA1 pyramidal cells of mutUNG1-expressing mice is likely to influence synaptic transmission and plasticity and may thus ultimately affect the ability to learn and form new memories (9, 27). Hence, the memory deficits displayed by the mutUNG1-expressing mice is probably related to both the observed hippocampal atrophy and the reduced PSD size.
Finally, the mouse model created for the present study, which carries a bidirectional doxycycline-responsive promoter for simultaneous induction of mutUNG1 and luciferase, should allow us to address the role of the mtDNA damage and mitochondrial dysfunction in more or less any organs of the mouse by using a different tissue specific promoter. In particular, mitochondria in cardiac cells are important for generating energy, and mitochondria have been implicated in the loss of cells in various cardiac pathologies, including ischemia/reperfusion and congestive heart failure (19).
Acknowledgments
This study was supported by the Norwegian Cancer Society and by the Research Council of Norway (funding from Centre of Excellence programme SFF to the CMBN).
We thank David Kunke, Hege Wiksèn, and Cesilie Granum Castellanos for work with microscopy, histology, and sectioning and Hans Krokan and Geir Slupphaug for providing plasmid and antibodies. We also thank Christian Johannes van den Bout and the Norwegian Transgenic Centre for excellent service.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Anderson, C. T., and E. C. Friedberg. 1980. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic Acids Res. 8:875-888. [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. F., and N. R. Sims. 2000. Improved recovery of highly enriched mitochondrial fractions from small brain tissue samples. Brain Res. Brain Res. Protoc. 5:95-101. [DOI] [PubMed] [Google Scholar]
- 3.Avkin, S., S. Adar, G. Blander, and Z. Livneh. 2002. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 99:3764-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, U., S. Freundlieb, M. Gossen, and H. Bujard. 1995. Coregulation of two gene activities by tetracycline via a bidirectional promoter. Nucleic Acids Res. 23:3605-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumgartel, K., D. Genoux, H. Welzl, R. Y. Tweedie-Cullen, K. Koshibu, M. Livingstone-Zatchej, C. Mamie, and I. M. Mansuy. 2008. Control of the establishment of aversive memory by calcineurin and Zif268. Nat. Neurosci. 11:572-578. [DOI] [PubMed] [Google Scholar]
- 6.Bergersen, L. H., J. Storm-Mathisen, and V. Gundersen. 2008. Immunogold quantification of amino acids and proteins in complex subcellular compartments. Nat. Protoc. 3:144-152. [DOI] [PubMed] [Google Scholar]
- 7.Boeckers, T. M. 2006. The postsynaptic density. Cell Tissue Res. 326:409-422. [DOI] [PubMed] [Google Scholar]
- 8.Bogenhagen, D. F., K. G. Pinz, and R. M. Perez-Jannotti. 2001. Enzymology of mitochondrial base excision repair. Prog. Nucleic Acids Res. Mol. Biol. 68:257-271. [DOI] [PubMed] [Google Scholar]
- 9.Brun, V. H., S. Leutgeb, H. Q. Wu, R. Schwarcz, M. P. Witter, E. I. Moser, and M. B. Moser. 2008. Impaired spatial representation in CA1 after lesion of direct input from entorhinal cortex. Neuron 57:290-302. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, H. Y., C. Miller, K. J. Bitterman, N. R. Wall, B. Hekking, B. Kessler, K. T. Howitz, M. Gorospe, C. R. de, and D. A. Sinclair. 2004. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305:390-392. [DOI] [PubMed] [Google Scholar]
- 11.de Souza-Pinto, N. C., L. Eide, B. A. Hogue, T. Thybo, T. Stevnsner, E. Seeberg, A. Klungland, and V. A. Bohr. 2001. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on the oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 61:5378-5381. [PubMed] [Google Scholar]
- 12.de Souza-Pinto, N. C., D. M. Wilson, III, T. V. Stevnsner, and V. A. Bohr. 2008. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair 7:1098-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhikav, V., and K. S. Anand. 2007. Is hippocampal atrophy a future drug target? Med. Hypotheses 68:1300-1306. [DOI] [PubMed] [Google Scholar]
- 14.DiMauro, S., and E. A. Schon. 2003. Mitochondrial respiratory chain diseases. N. Engl. J. Med. 348:2656-2668. [DOI] [PubMed] [Google Scholar]
- 15.Druzhyna, N. M., G. L. Wilson, and S. P. LeDoux. 2008. Mitochondrial DNA repair in aging and disease. Mech. Ageing Dev. 129:383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ekstrand, M. I., M. Terzioglu, D. Galter, S. Zhu, C. Hofstetter, E. Lindqvist, S. Thams, A. Bergstrand, F. S. Hansson, A. Trifunovic, B. Hoffer, S. Cullheim, A. H. Mohammed, L. Olson, and N. G. Larsson. 2007. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U. S. A. 104:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, D. R., and G. Kroemer. 2004. The pathophysiology of mitochondrial cell death. Science 305:626-629. [DOI] [PubMed] [Google Scholar]
- 18.Guillet, M., and S. Boiteux. 2003. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:8386-8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustafsson, A. B., and R. A. Gottlieb. 2008. Heart mitochondria: gates of life and death. Cardiovasc. Res. 77:334-343. [DOI] [PubMed] [Google Scholar]
- 20.Harrison, J. F., S. B. Hollensworth, D. R. Spitz, W. C. Copeland, G. L. Wilson, and S. P. LeDoux. 2005. Oxidative stress-induced apoptosis in neurons correlates with mitochondrial DNA base excision repair pathway imbalance. Nucleic Acids Res. 33:4660-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara, T., M. Kubota, T. Miyauchi, Y. Noda, A. Mouri, T. Nabeshima, and T. Kato. 2006. Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol. Psychiatry 11:577-593. [DOI] [PubMed] [Google Scholar]
- 23.Kavli, B., G. Slupphaug, C. D. Mol, A. S. Arvai, S. B. Peterson, J. A. Tainer, and H. E. Krokan. 1996. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 15:3442-3447. [PMC free article] [PubMed] [Google Scholar]
- 24.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 25.Kujoth, G. C., A. Hiona, T. D. Pugh, S. Someya, K. Panzer, S. E. Wohlgemuth, T. Hofer, A. Y. Seo, R. Sullivan, W. A. Jobling, J. D. Morrow, R. H. Van, J. M. Sedivy, T. Yamasoba, M. Tanokura, R. Weindruch, C. Leeuwenburgh, and T. A. Prolla. 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309:481-484. [DOI] [PubMed] [Google Scholar]
- 26.LeDoux, S. P., G. L. Wilson, E. J. Beecham, T. Stevnsner, K. Wassermann, and V. A. Bohr. 1992. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis 13:1967-1973. [DOI] [PubMed] [Google Scholar]
- 27.Leutgeb, S., J. K. Leutgeb, A. Treves, M. B. Moser, and E. I. Moser. 2004. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science 305:1295-1298. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., E. Head, A. M. Gharib, W. Yuan, R. T. Ingersoll, T. M. Hagen, C. W. Cotman, and B. N. Ames. 2002. Memory loss in old rats is associated with brain mitochondrial decay and RNA/DNA oxidation: partial reversal by feeding acetyl-l-carnitine and/or R-alpha-lipoic acid. Proc. Natl. Acad. Sci. U. S. A. 99:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb, L. A., D. C. Wallace, and G. M. Martin. 2005. The mitochondrial theory of aging and its relationship to reactive oxygen species damage and somatic mtDNA mutations. Proc. Natl. Acad. Sci. U. S. A. 102:18769-18770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longley, M. J., R. Prasad, D. K. Srivastava, S. H. Wilson, and W. C. Copeland. 1998. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc. Natl. Acad. Sci. U. S. A. 95:12244-12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustbader, J. W., M. Cirilli, C. Lin, H. W. Xu, K. Takuma, N. Wang, C. Caspersen, X. Chen, S. Pollak, M. Chaney, F. Trinchese, S. Liu, F. Gunn-Moore, L. F. Lue, D. G. Walker, P. Kuppusamy, Z. L. Zewier, O. Arancio, D. Stern, S. S. Yan, and H. Wu. 2004. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science 304:448-452. [DOI] [PubMed] [Google Scholar]
- 33.McEwen, B. S. 1997. Possible mechanisms for atrophy of the human hippocampus. Mol. Psychiatry 2:255-262. [DOI] [PubMed] [Google Scholar]
- 34.Michalon, A., K. Koshibu, K. Baumgartel, D. H. Spirig, and I. M. Mansuy. 2005. Inducible and neuron-specific gene expression in the adult mouse brain with the rtTA2S-M2 system. Genesis 43:205-212. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls, D. G. 2008. Oxidative stress and energy crises in neuronal dysfunction. Ann. N. Y. Acad. Sci. 1147:53-60. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, H., K. S. Steinsbekk, M. Otterlei, G. Slupphaug, P. A. Aas, and H. E. Krokan. 2000. Analysis of uracil-DNA glycosylases from the murine Ung gene reveals differential expression in tissues and in embryonic development and a subcellular sorting pattern that differs from the human homologues. Nucleic Acids Res. 28:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishioka, K., T. Ohtsubo, H. Oda, T. Fujiwara, D. Kang, K. Sugimachi, and Y. Nakabeppu. 1999. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen, L. C., R. Aasland, C. U. Wittwer, H. E. Krokan, and D. E. Helland. 1989. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 8:3121-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otterlei, M., T. Haug, T. A. Nagelhus, G. Slupphaug, T. Lindmo, and H. E. Krokan. 1998. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 26:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pages, V., R. E. Johnson, L. Prakash, and S. Prakash. 2008. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl. Acad. Sci. U. S. A. 105:1170-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reeve, A. K., K. J. Krishnan, and D. Turnbull. 2008. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann. N. Y. Acad. Sci. 1147:21-29. [DOI] [PubMed] [Google Scholar]
- 42.Ropp, P. A., and W. C. Copeland. 1996. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics 36:449-458. [DOI] [PubMed] [Google Scholar]
- 43.Schriner, S. E., N. J. Linford, G. M. Martin, P. Treuting, C. E. Ogburn, M. Emond, P. E. Coskun, W. Ladiges, N. Wolf, R. H. Van, D. C. Wallace, and P. S. Rabinovitch. 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308:1909-1911. [DOI] [PubMed] [Google Scholar]
- 44.Shepherd, J. K., S. S. Grewal, A. Fletcher, D. J. Bill, and C. T. Dourish. 1994. Behavioural and pharmacological characterisation of the elevated “zero-maze” as an animal model of anxiety. Psychopharmacology 116:56-64. [DOI] [PubMed] [Google Scholar]
- 45.Slupphaug, G., F. H. Markussen, L. C. Olsen, R. Aasland, N. Aarsaether, O. Bakke, H. E. Krokan, and D. E. Helland. 1993. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 21:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, M., X. Shen, E. G. Frank, M. O'Donnell, R. Woodgate, and M. F. Goodman. 1999. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. U. S. A. 96:8919-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trifunovic, A., A. Wredenberg, M. Falkenberg, J. N. Spelbrink, A. T. Rovio, C. E. Bruder, Y. Bohlooly, S. Gidlof, A. Oldfors, R. Wibom, J. Tornell, H. T. Jacobs, and N. G. Larsson. 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429:417-423. [DOI] [PubMed] [Google Scholar]
- 48.Vermulst, M., J. H. Bielas, G. C. Kujoth, W. C. Ladiges, P. S. Rabinovitch, T. A. Prolla, and L. A. Loeb. 2007. Mitochondrial point mutations do not limit the natural life span of mice. Nat. Genet. 39:540-543. [DOI] [PubMed] [Google Scholar]
- 49.Vermulst, M., J. Wanagat, G. C. Kujoth, J. H. Bielas, P. S. Rabinovitch, T. A. Prolla, and L. A. Loeb. 2008. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 40:392-394. [DOI] [PubMed] [Google Scholar]
- 50.Vorhees, C. V., and M. T. Williams. 2006. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc. 1:848-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J., J. P. Silva, C. M. Gustafsson, P. Rustin, and N. G. Larsson. 2001. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc. Natl. Acad. Sci. U. S. A. 98:4038-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, H., A. Chatterjee, and K. K. Singh. 2006. Saccharomyces cerevisiae polymerase zeta functions in mitochondria. Genetics 172:2683-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]