Abstract
Template unwinding during DNA replication initiation requires the loading of the MCM helicase activator Cdc45 at replication origins. We show that Cdc45 interacts with the DNA unwinding element (DUE) binding protein DUE-B and that these proteins localize to the DUEs of active replication origins. DUE-B and Cdc45 are not bound at the inactive c-myc replicator in the absence of a functional DUE or at the recently identified ataxin 10 (ATX10) origin, which is silent before disease-related (ATTCT)n repeat length expansion of its DUE sequence, despite the presence of the origin recognition complex (ORC) and MCM proteins at these origins. Addition of a heterologous DUE to the ectopic c-myc origin, or expansion of the ATX10 DUE, leads to origin activation, DUE-B binding, and Cdc45 binding. DUE-B, Cdc45, and topoisomerase IIβ binding protein 1 (TopBP1) form complexes in cell extracts and when expressed from baculovirus vectors. During replication in Xenopus egg extracts, DUE-B and Cdc45 bind to chromatin with similar kinetics, and DUE-B immunodepletion blocks replication and the loading of Cdc45 and a fraction of TopBP1. The coordinated binding of DUE-B and Cdc45 to origins and the physical interactions of DUE-B, Cdc45, and TopBP1 suggest that complexes of these proteins are necessary for replication initiation.
The initiation of DNA replication requires the assembly of multiple heteromeric protein complexes at origins of replication. Binding of the origin recognition complex (ORC) to chromatin is the earliest known step in origin specification (5). ORC binding is not sequence specific but is modulated by singularities of chromatin covalent modification, transcription factor binding, DNA supercoiling, and local A-T density (2, 19, 41, 54). However, aside from the presence of regions of predicted helical instability termed DNA unwinding elements (DUEs) at origins, the precise properties of a replication origin that promote ORC binding are not known.
The binding of ORC leads to the recruitment of Cdc6, Cdt1, and the Mcm2-7 helicase to form prereplication complexes (pre-RCs) (46). Additional MCM family members facilitate pre-RC assembly (27) or subsequently bind to the pre-RC (42, 58). A fraction of the BRCA1 C-terminal (BRCT) domain protein TopBP1 (the ortholog of Mus101/Cut5) and the MCM helicase-activating protein Cdc45 (13, 53) associate with the pre-RC in a manner contingent on the activities of the S-phase-promoting cyclin-dependent kinases and Cdc7/Dbf4 (Cdc7/Drf1 in Xenopus egg extracts) to convert the pre-RC into the preinitiation complex (pre-IC) (65). Cdc45 activation of the MCM helicase leads to origin unwinding and recruitment of replication protein A (RPA), GINS (Sld4, Sld5, Psf1, and Psf2), DNA polymerases, and the replication fork progression/stabilization complex (11).
The c-myc DUE binding protein (DUE-B) was identified in a yeast one-hybrid screen, using the c-myc DUE as bait (7). The 209-amino-acid (23.4-kDa) protein has been strongly conserved during metazoan evolution (18). By chromatin immunoprecipitation (ChIP), DUE-B has been shown to localize with MCM proteins to the DUEs of the c-myc and lamin B2 replication origins, and DUE-B binding to an ectopic copy of the c-myc core replicator is eliminated by deletion of the DUE (12). Suggestive of an important role for DUE-B in DNA replication, DUE-B small interfering RNA (siRNA) slows the G1- to S-phase transition of HeLa cells. In the Xenopus egg extract cell-free replication system, DUE-B depletion inhibits DNA replication, and a dominant-negative form of DUE-B blocks replication after pre-RC formation but before origin unwinding and RPA binding (7, 18).
In Saccharomyces cerevisiae, the Sld3 protein is necessary for the loading of Cdc45 on pre-RCs (16, 36), and the origin binding of Cdc45 is coordinated with that of Sld3, i.e., Cdc45 and Sld3 bind in early S phase to early-firing origins and in late S phase to late-firing origins (3, 51, 64). Cdc45 has been shown to stimulate the activity of the MCM helicase in vitro (35) and to move with replication forks in vivo, whereas Sld3 does not appear to move with replication forks (4, 17). The recruitment of yeast Cdc45 and its binding partner Sld3 to origins requires the Cdk1/cyclin-dependent association of Sld3 with Dpb11, which is structurally similar to the amino-terminal region of TopBP1 comprising BRCT domains 1 to 5 (48, 61). This segment of TopBP1 is necessary and sufficient for the replication functions of TopBP1 in the Xenopus egg extract cell-free system, while the carboxy-terminal half of TopBP1 is required for the activation of the ATR (ataxia telangiectasia and Rad3-related)/Chk1 kinase DNA damage response pathway (14, 59). Partially mimicking the interaction between Dpb11, Cdc45, and Sld3 (16, 48), human Cdc45 and TopBP1 have been reported to bind to one another in cell extracts (45); however, a human structural homolog of Sld3 that interacts with TopBP1 or Cdc45 has not been isolated or identified by database searches.
The conformation of the amino-terminal 150-residue domain of DUE-B shows strong evolutionary conservation to the structures of orthologous proteins in yeast, eubacteria, and archaebacteria (18). Remarkably, the structurally similar proteins in lower organisms do not appear to play direct roles in DNA replication but are involved in the proofreading of tRNA aminoacylation. Indeed, human DUE-B retains the amino acid deacylase and esterase activities of these proteins (18). Carboxyl to the protease-resistant amino-terminal domain, human DUE-B also contains a 59-amino-acid extension not found in the tRNA deacylases. As shown below, this region of DUE-B shows limited amino acid similarity to the carboxy-terminal Dpb11-binding domains of Sld3 in S. cerevisiae and Schizosaccharomyces pombe.
In the current work, we show that DUE-B binds to TopBP1 and Cdc45 and that DUE-B and Cdc45 bind in parallel to replication origins in vivo, dependent on the presence of a functional DUE. The binding of DUE-B to chromatin is decreased in a cell line expressing a hypomorphic Orc2 mutant, and DUE-B is inhibited from binding to Xenopus sperm chromatin by geminin in egg extracts, suggesting that DUE-B binding to origins is ORC and pre-RC dependent. In the Xenopus cell-free replication system, DUE-B associates with sperm chromatin after MCM deposition, in parallel with the binding of Cdc45, and the binding of DUE-B and Cdc45 is similarly blocked by inhibition of S-phase-promoting kinase activity. DUE-B is released from the DNA template coincident with origin unwinding and RPA binding. Immunodepletion of DUE-B inhibits Cdc45 loading and decreases TopBP1 binding to sperm chromatin, indicating that DUE-B is necessary for Cdc45 loading. These results suggest that DUE-B may function analogously to the Sld3 proteins for the loading of Cdc45 onto prereplication complexes.
MATERIALS AND METHODS
Cell culture and chromatin preparation.
HeLa and HCT116 cells were maintained as adherent cultures in Dulbecco modified Eagle medium (DMEM) containing 10% newborn calf serum and 10 μg/ml gentamicin (Gibco). The HCT116 cell line expressing reduced levels of Orc2 (Orc2Δ/−) was provided by Anindya Dutta (10, 49). FLP-recombinase-mediated cassette exchange (FLP-RMCE) construction of the ectopic c-myc/(ATTCT)n cell lines has been described previously (23). Cross-linked chromatin was prepared for immunoprecipitation as described previously (12). Soluble chromatin was isolated according to Mendez and Stillman (32).
Immunoblotting.
Antiactin antibody was purchased from Oncogene. Anti-DUE-B antibody was raised in rabbits against a His6-tagged recombinant human DUE-B expressed in bacteria. Anti-xOrc2 antibody was raised in rabbits against baculovirus-expressed recombinant xOrc2. Anti-RPA70 and anti-p21 antibodies were purchased from Santa Cruz Biotechnology. Anti-xCdc45 antibody was generously provided by Johannes Walter (Harvard University), and anti-hsCdc45 (human) was purchased from Santa Cruz Biotechnology. The use of the anti-hsCdc45 antibody in ChIP was validated by ChIP with anti-Gal4 antibody in cells transfected with Gal4 epitope-tagged Cdc45. Anti-Cdk2 antibody was obtained from Transduction Laboratories. Mcm3 and Mcm4 antibodies were obtained from Aloys Schepers (GSF-Haematologikum) or purchased from Santa Cruz Biotechnology. Anti-His6 (C-terminal) monoclonal antibody was purchased from Invitrogen. Antiphosphoserine antibody Q5 was obtained from Qiagen. A. Vaillant (Replicor, Quebec, Canada) provided anti-Ku80 antibody. Anti-TopBP1 antibody was provided by Lawrence Karnitz (Mayo Clinic).
Indirect immunofluorescence.
Cells were grown for 36 to 48 h (30 to 50% confluence) on glass coverslips. Cells were washed with phosphate-buffered saline (PBS), then fixed with 2% paraformaldehyde, freshly dissolved in PBS for 15 min at room temperature, and washed with PBS (three times, 5 min each). In some experiments, cells were extracted prior to fixation by incubating them for 2 min on ice with 0.2% Triton X-100 in a buffer containing 10 mM HEPES, 50 mM KCl, 25 mM MgCl2, and 200 mM sucrose. After fixation, cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature and washed with 0.5% normal goat serum in PBS (blocking buffer) three times for 5 min each. Cells were incubated with the primary antibodies diluted in a volume of 30 to 50 μl of the blocking buffer in a humid chamber for 1 h, washed three times for 5 min each with the blocking buffer, and then incubated with the fluorescence-conjugated secondary antibodies (1:200; Jackson ImmunoResearch Laboratory) for 1 h in the dark. Cells on the coverslips were washed three times for 5 min each. DAPI (4′,6-diamidino-2-phenylindole; Sigma) was added in the last washing for DNA staining. The following antibody dilutions were used for immunofluorescence: anti-DUE-B antibody, 1:20; anti-His6 antibody, 1:200; and anti-SAP145 antibody (provided by R. Reed, Harvard Medical School), anti-hSm, and anti-BrUdR antibodies, 1:100 (Sigma). Immunofluorescence microscopy was performed using a DeltaVision RT apparatus (Applied Precision), running SoftWoRx 3.5.1 for image deconvolution and protein colocalization.
Xenopus egg extracts.
Low-speed Xenopus interphase egg extracts were prepared according to the procedure of Walter and Newport (55). DUE-B immunodepletion, isolation of recombinant DUE-B from HeLa cells, and reconstitution of immunodepleted extracts were performed as described previously (7). Sperm chromatin was prepared according to Chong et al. (8) and frozen in aliquots at −80°C. Supercoiled plasmid pNeo.myc-2.4 (30) was biotinylated by photocoupling with Photoprobe (S-S) biotin (Vector Laboratories), which has a cleavable disulfide bond in the linker arm. The procedure was performed according to the manufacturer's instructions as described previously (62). The biotinylated plasmid was bound to streptavidin-Dynabeads M-280 (Dynal Biotech) using a Dynabeads kilobaseBinder kit (Dynal Biotech), according to the manufacturer's instructions.
Sperm chromatin was added to extract at a final concentration of 3,000 sperm/μl or to bead-bound plasmid DNA at a final concentration of 15 ng/μl. Chromatin was purified by centrifugation at 6,000 × g for 5 min at 4°C through a 1.5-ml 0.7 M sucrose cushion in XB buffer (28). Chromatin pellets were washed once in XB buffer containing 0.15% Triton X-100, and proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting. To reduce background protein binding to the bead-bound plasmid, the plasmid chromatin was released from the beads by incubation in 100 μl of 100 mM dithiothreitol. The released chromatin was then separated from the beads using a magnet and boiled with SDS sample buffer for SDS-PAGE and immunoblot analysis.
Isolation of recombinant proteins.
Recombinant DUE-B was purified from a HeLa cell line stably expressing His6-tagged DUE-B cDNA (A1 cells). Cell pellets were lysed on ice in lysis buffer (50 mM NaH2PO4 at pH 8.0, 300 mM NaCl, 20 mM imidazole, 0.5 to 1% NP-40, 1× protease inhibitor cocktail [Sigma]). The lysate was clarified by centrifugation, and DUE-B was isolated by Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) chromatography. The resin was washed with washing buffer (50 mM NaH2PO4 at pH 8.0, 500 mM NaCl, 40 mM imidazole, 0.1% NP-40, protease inhibitor cocktail) to remove heterodimers of recombinant and endogenous DUE-B. His6-tagged DUE-B was eluted with elution buffer (50 mM NaH2PO4 at pH 8.0, 100 mM NaCl, 200 mM imidazole, protease inhibitor cocktail) and concentrated by centrifugal ultrafiltration. DUE-B was added to immunodepleted extract at a final concentration of approximately 30 ng/μl (7). Recombinant glutathione S-transferase (GST)-p27 was isolated by glutathione-Sepharose chromatography (52) and added to egg extracts at a final concentration of 1 μM (57). Alternatively, the Cdk inhibitor roscovitine (dissolved in dimethyl sulfoxide [DMSO]) was added to 0.5 mM. The final DMSO concentration was <1% (25).
Baculoviruses.
The pFastBAC-Cdc45 (a gift from I. Kukimoto, National Institute of Infectious Diseases, Tokyo) and pFastBAC-HF-XTopBP1 (a gift from A. Kumagai and W. Dunphy, California Institute of Technology) vectors were transformed into Escherichia coli DH10BAC (Invitrogen). Baculoviruses were used to infect Sf9 or High Five cells. The human DUE-B cDNA was PCR amplified and ligated into the baculovirus transfer vector pBlueBac4.5/V5-His. Plasmids were cotransfected with linearized Bac-N-Blue DNA (Invitrogen) using Cellfectin reagent into Sf9 cells to generate recombinant baculovirus, as recommended by the manufacturer.
Protein alignment.
Protein sequences were aligned using DNAMAN 6.03 (Lynnon Corp.), and amino acid similarities were assigned by contact potential, according to Tan et al. (47).
RESULTS
DUE-B and Cdc45 binding correlates with replication origin activation.
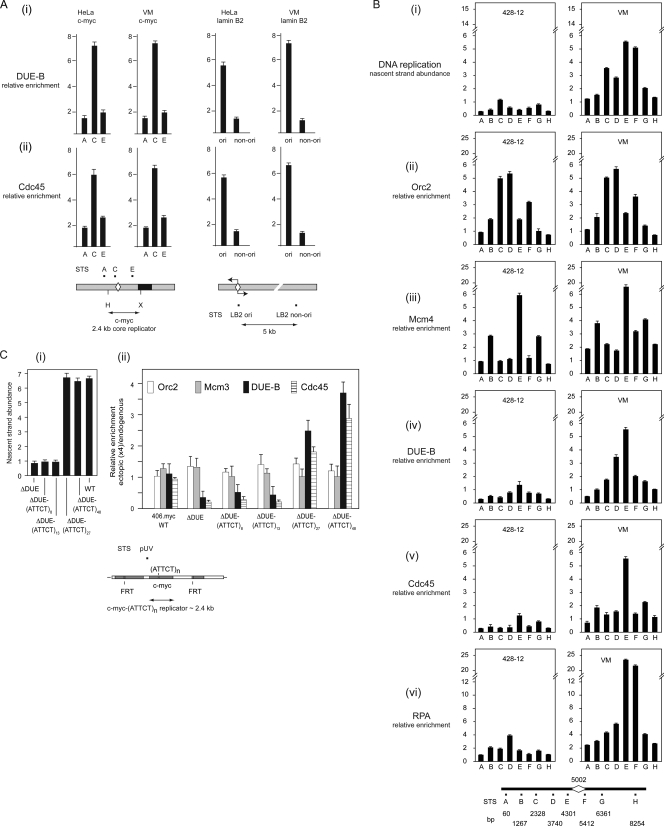
DUE-B and pre-RC proteins bind nonrandomly and with equal efficiency to the endogenous c-myc origin and an ectopic copy of the 2.4-kb c-myc core replicator in HeLa cells (12). To test whether Cdc45 localizes with DUE-B at active replication origins, the binding of these proteins to the c-myc and binding to the lamin B2 origins were compared by chromatin immunoprecipitation. As shown in Fig. 1, DUE-B (Fig. 1Ai) and Cdc45 (Fig. 1Aii) bound to the DUE regions of the c-myc and lamin B2 replication origins in HeLa and VM cells. Taken with the earlier demonstration showing that the Mcm3 and Mcm7 proteins are also enriched at these DUEs (12), these results suggest that DUE-B, Cdc45, and the MCM complex may interact at the most easily unwound DUE sites of active replication origins.
FIG. 1.
Protein binding to replication origins. (A) DUE-B (i) and Cdc45 (ii) binding to the c-myc and lamin B2 origins (ori) in HeLa cells and VM (SCA10 patient) cells. Maps show the positions of sequence-tagged sites (STS) used for quantitative ChIP within the c-myc and lamin B2 origins. Diamond shapes, DUEs predicted by the SIDD (stress-induced DNA duplex destabilization) algorithm (3, 7). Restriction sites flanking the c-myc replicator core: H, HindIII; X, XhoI. (B) Origin activity (nascent strand abundance) (i) and Orc2 (ii), Mcm4 (iii), DUE-B (iv), Cdc45 (v), and RPA (vi) binding at the ATX10/SCA10 locus (map at bottom) in non-SCA10 lymphoblastoid cells (left, 428-12) and SCA10 patient cells (right, VM). Diamond shape, position of expanded (ATTCT)n repeats in SCA10 patient cells. (C) Origin activity (i) and Orc2, Mcm3, DUE-B, and Cdc45 binding (ii) at the ectopic c-myc replicator (map at bottom, STS pUV) in FLP-RMCE cell lines containing the wild-type (WT; 406.myc) c-myc replicator, the c-myc replicator minus the DUE (ΔDUE), or the c-myc replicator in which the DUE has been replaced by the SCA10 DUE containing (ATTCT)8, (ATTCT)13, (ATTCT)27, or (ATTCT)48 repeats. The data shown in panels Bi and Ci are reproduced from reference 23 (copyright, American Society for Microbiology; reproduced with permission).
ORC is bound to origin and nonorigin sites in S. cerevisiae (33), and it has been proposed that MCM complexes are bound at dormant origins that are silent under nonstressed cell cycle conditions (11). To compare the origin binding of DUE-B with that of pre-RC proteins and markers of origin activity (Cdc45 and RPA70), replication protein binding was assayed at the inactive ATX10 replication origin in lymphoblastoid cells obtained from a myotonic dystrophy type 1 (DM1) patient (428-12 cells) and at the active ATX10/SCA10 (spinocerebellar ataxia type 10) origin in lymphoblastoid cells obtained from a SCA10 patient, in which the DUE has expanded from comprising fewer than 20 copies of the (ATTCT)n pentanucleotide to more than 4,000 copies (Fig. 1Bi) (23). The patterns of Orc2 and Mcm4 binding were similar at the inactive and active origins, confirming that pre-RC assembly is not sufficient for ATX10/SCA10 origin activity. In contrast, DUE-B, Cdc45, and the large (70-kDa) subunit of RPA (RPA70) were bound only at the DUE of the active ATX10/SCA10 origin. These results show that the binding of DUE-B, Cdc45, and RPA at the DUE is associated with origin activation.
To investigate the relationship between pre-RC formation, DUE-B binding, Cdc45 binding, and origin activity further, we used engineered cell lines in which the DUE of an ectopic copy of the c-myc origin was deleted or replaced by heterologous SCA10 DUEs, comprising easily unwound (ATTCT)n pentanucleotide repeat sequences. All c-myc replicator integrant cells were derived from the same parental acceptor cell line, and all forms of the integrated c-myc replicator were integrated at the same ectopic chromosomal site (23, 24). The endogenous c-myc DUE can be functionally replaced at the ectopic c-myc core replicator by DUEs containing (ATTCT)n repeat tracts of sufficient length (Fig. 1Ci) (23), indicating that DUE activity is not sequence specific. As shown in Fig. 1Cii, Orc2, Mcm3, DUE-B, and Cdc45 bind with equal efficiency at the endogenous and ectopic wild-type c-myc replicators. In the c-myc replicator DUE deletion mutant (23, 24) ChIP confirmed that Orc2 and Mcm3 binding was not affected but that DUE-B binding (Fig. 1Cii) mirrored the length-dependent restoration of origin activity (Fig. 1Ci) by the heterologous DUEs. The binding of Cdc45 quantitatively paralleled the binding of DUE-B to the ectopic origin. These results imply that dormant pre-RCs can be assembled at inactive origins, that DUE-B binding and Cdc45 binding are coordinated and DUE dependent, and that DUE-B binding and Cdc45 binding are involved in origin activation.
DUE-B and replication protein binding to chromatin.
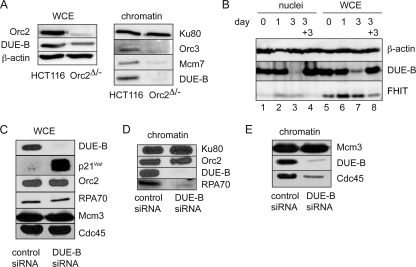
The results shown in Fig. 1 indicate that MCM binding does not require DUE-B, in agreement with previous results showing that MCM and DUE-B bind independently to the ectopic c-myc origin (12). To test whether DUE-B binding is ORC dependent, we used HCT116 Orc2Δ/− cells in which one Orc2 allele is deleted and the second allele produces a hypomorphic mutant of Orc2 at 5 to 10% of wild-type levels. Orc3 is destabilized in Orc2Δ/− cells; in contrast, total levels of Mcm7 are unaffected, but Mcm7 loading onto chromatin is significantly decreased (49). These cells are viable but show a defect in the G1- to S-phase transition; once the Orc2Δ/− hypomorphic cells enter S phase, replication is comparable to that in wild-type cells (10, 49). DUE-B levels were modestly reduced in total lysates of the Orc2Δ/− cells (Fig. 2A); however, the amounts of Orc3, Mcm7, and DUE-B that were bound to chromatin were strongly decreased. This result suggests that DUE-B binding to chromatin is ORC dependent in vivo. Consistent with the decrease in DUE-B levels in slow-growing Orc2Δ/− cells, serum deprivation of HeLa cells resulted in the loss of DUE-B protein from whole-cell lysates and permeabilized nuclei (Fig. 2B), in contrast to the accumulation of the fragile histidine triad (FHIT) protein, which is stabilized during serum deprivation (44).
FIG. 2.
Replication protein abundance and chromatin binding. (A, left) Whole-cell extract (WCE) levels of Orc2, DUE-B, and β-actin; (right) chromatin-bound Orc3, Mcm7, and DUE-B in HCT116 cells or HCT116 Orc2Δ/− cells. Ku80 was used as a loading control (20, 38). (B) DUE-B, β-actin, and FHIT protein levels were assayed in permeabilized nuclei (lanes 1 to 4) and WCE (lanes 5 to 8) after serum (0.5%) deprivation of cells for 0, 1, and 3 days (lanes 1 to 3 and 5 to 7) and after 3 days of serum deprivation, followed by 3 days of serum (10%) restoration (lanes 4 and 8). (C) The levels of the indicated proteins were compared in WCE from control (scrambled) siRNA- or DUE-B siRNA-treated HeLa cells. (D and E) Chromatin binding of the indicated proteins in control siRNA- or DUE-B siRNA-treated HeLa cells.
RNA interference (RNAi) knockdown of DUE-B in HeLa cells delays the cell cycle transition from G1 to S phase (7). As shown in Fig. 2C, siRNA knockdown of DUE-B did not result in significant changes in the total levels of Orc2, RPA70, Mcm3, or Cdc45. In contrast, DUE-B knockdown led to a large increase in the level of the cyclin-dependent kinase inhibitor p21WAF1 protein, possibly related to the inhibition of replisome assembly and p21 degradation (1, 37). Since p53 is inactivated in HeLa cells, and DUE-B siRNA also inhibited proliferation of p53-null H1299 cells (M. Kemp, unpublished data), this effect does not appear to be due to stabilization of p53. DUE-B knockdown also resulted in dramatically decreased levels of RPA (Fig. 2D) and Cdc45 (Fig. 2E) loading on chromatin, in agreement with the coordinated binding of these proteins at the activated ATX10/SCA10 origin. Based on the homology between the DUE-B and aminoacyl-tRNA deacylating enzymes, it is formally possible that the inhibition of replication by DUE-B siRNA could arise indirectly through effects on protein synthesis. However, DUE-B siRNA knockdown did not affect tRNATyr aminoacylation or [3H]leucine incorporation in HeLa cells (63), suggesting that protein synthesis is not the point at which DUE-B siRNA inhibits cell cycle progress. Nevertheless, the present results cannot distinguish between a decrease in the chromatin loading of RPA and that of Cdc45 due to a direct inhibition of pre-IC formation by DUE-B knockdown or due to an indirect effect through the inhibition of S-phase Cdk activity by elevated p21 protein levels.
Intracellular localization of DUE-B.
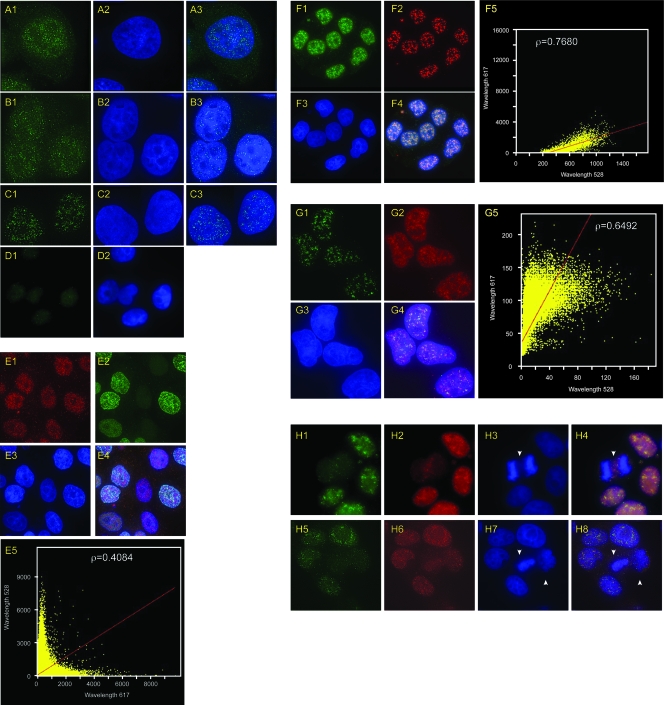
Immunoprecipitation of cross-linked chromatin indicates that DUE-B is bound to chromosomal replication origins, but fractionation of uncross-linked, detergent-permeabilized cells revealed that DUE-B can partition between soluble and insoluble nuclear fractions after cell lysis (7). HeLa cells, or HeLa cells stably expressing a hexahistidine-tagged DUE-B protein in addition to endogenous DUE-B (HeLa A1 cells), were fixed and stained with anti-DUE-B antibody (Fig. 3A) or antihexahistidine antibody (Fig. 3B). Indirect immunofluorescence of the endogenous DUE-B (Fig. 3A) or the epitope-tagged DUE-B (Fig. 3B) revealed a punctate pattern of staining over a diffuse chromatin distribution of DUE-B, whereas the hexahistidine-directed antibody showed only background staining in control untransfected HeLa cells (Fig. 3D). When cells were permeabilized with 0.2% Triton X-100 before fixation, the diffuse nuclear staining was decreased (Fig. 3C), while the punctate staining became more evident. We considered the possibility that the punctate DUE-B staining represents the presence of nuclear replication foci. However, when HeLa A1 cells were pulse-labeled for 10 min with bromodeoxyuridine (BrUdR) and stained with anti-BrUdR and antihexahistidine antibodies, the two labels did not colocalize (Fig. 3E), suggesting that neither the punctate staining nor the diffuse chromatin staining of DUE-B indicates the presence of active replication foci. The absence of DUE-B at sites of nucleotide incorporation is consistent with the observations that DUE-B binds preferentially to double-stranded DNA (18) but is released from origins upon template unwinding (see below).
FIG. 3.
Indirect immunofluorescence localization of DUE-B. (A) HeLa cells stained with anti-DUE-B antibody (A1) and DAPI (A2) (merged image [A3]). (B) HeLa A1 cells expressing epitope (His6)-tagged DUE-B stained with anti-His6 antibody (B1) and DAPI (B2) (merged image [B3]). (C) Staining done as in panel B, with 0.2% Triton X-100 preextraction. (D) Negative-control HeLa cells stained with anti-His6 antibody (D1) and DAPI (D2). (E) HeLa A1 cells pulse-labeled with BrUdR and stained with anti-His6 antibody (E1), anti-BrUdR antibody (E2), and DAPI (E3) (merged image [E4]). (E5) Plot showing degree of colocalization of DUE-B (λ = 617 nm) and BrUdR (λ = 528 nm) signals (Pearson correlation coefficient, ρ = 0.4084). (F) HeLa A1 cells stained with anti-His6 antibody (F1), anti-SAP145 antibody (F2), and DAPI (F3) (merged image [F4]). (F5) Plot showing degree of colocalization of DUE-B (λ = 528 nm) and SAP145 (λ = 617 nm) signals (Pearson correlation coefficient, ρ = 0.7680). (G) HeLa A1 cells stained with anti-His6 antibody (G1), anti-SM antibody (G2), and DAPI (G3) (merged image [G4]). (G5) Plot showing degree of colocalization of DUE-B (λ = 528 nm) and Sm (λ = 617 nm) signals (Pearson correlation coefficient, ρ = 0.6492). (H) HeLa A1 cells stained with anti-His6 antibody (H1, H5), anti-SAP145 antibody (H2, H6), and DAPI (H3, H7) (merged images [H4, H8]). Arrowheads indicate cells in mitosis.
The strong conservation of structure and function between aminoacyl-tRNA proofreading enzymes and the amino-terminal 150 amino acids of DUE-B suggested that the punctate staining bodies could be interchromatin granule clusters (IGCs), which are storage depots for pre-mRNA processing factors (43). The SAP145 subunit of the SF3b150 splicing factor and the snRNP core protein Sm are concentrated in IGCs (21, 43). As shown in Fig. 3, SAP145 (Fig. 3F) and Sm (Fig. 3G) colocalized with DUE-B in IGCs. In mammalian cell nuclei, IGCs disassemble at the beginning of mitosis and begin to reassemble toward the end of mitosis (6). Consistent with the colocalization of DUE-B and SAP145 in IGCs in HeLa A1 cells, the punctate staining of DUE-B and SAP145 was lost from cells in mitosis (Fig. 3H). The conclusion that DUE-B and SAP145 are both found in IGCs is supported by coimmunoprecipitation and mass spectrometry results, indicating that DUE-B interacts with SAP145 (N. Katrangi, unpublished data).
DUE-B binding to replicating chromatin.
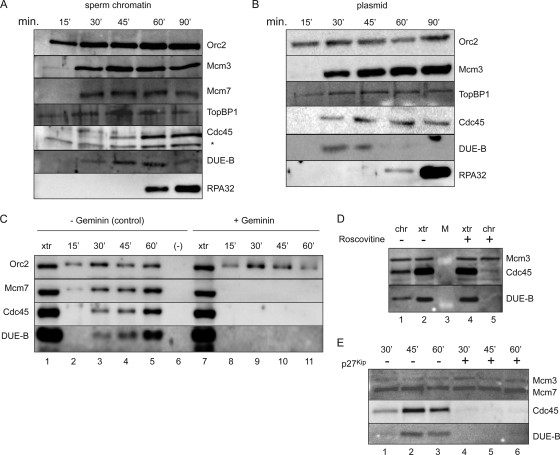
When sperm chromatin or bead-bound plasmid DNA is added to the Xenopus cell-free egg extract, ORC-dependent MCM binding and pre-RC formation on DNA are followed by Ddk (Cdc7/Drf1)- and Cdk (Cdk2/cyclin)-dependent loading of Cdc45 (53, 57, 62). Cdc45 loading activates the MCM helicase and marks the conversion of the pre-RC into the pre-IC. The unwound form of the DNA template binds the single-stranded DNA binding protein RPA, which is required to recruit DNA polymerase α/primase (Fig. 4A and B). In this system, DUE-B binds to chromatin with kinetics similar to those of Cdc45 (Fig. 4A to C), although the stoichiometry of chromatin-bound Cdc45 to DUE-B may be slightly less at the earliest time, possibly reflecting a less-stable chromatin association of Cdc45 immediately after loading. At later time points, DUE-B is released from chromatin upon RPA binding, while Cdc45 remains associated with the replicating template (Fig. 4A, 90′, and B, 60′). The BRCT domain protein TopBP1 binds to chromatin in an ORC-dependent manner and has been shown to play roles in both replication and DNA damage signaling (13, 22, 34, 39, 59). In the Xenopus egg extracts, a portion of TopBP1 binds to chromatin before MCM binding, while additional TopBP1 binds in parallel with the binding of DUE-B and Cdc45. These results agree with prior work showing Cdk-independent and Cdk-dependent modes of Xenopus Cut5 binding to sperm chromatin (13).
FIG. 4.
Protein binding to chromatin-Xenopus cell-free replication system. (A) Time course of binding of the indicated proteins to sperm chromatin. *, cross-reactive band. (B) Time course of protein binding to bead-bound plasmid DNA. (C) Geminin inhibits Mcm7, DUE-B, and Cdc45 binding to sperm chromatin. Lane 1, egg extract (xtr); lanes 2 to 5, control (no geminin); lane 6, control (sperm chromatin omitted); lane 7, egg extract (geminin added); lanes 8 to 11, geminin treated. (D) Inhibition of DUE-B and Cdc45 binding to sperm chromatin (chr) by roscovitine. Lanes 1 and 2, untreated; lanes 4 and 5, roscovitine treated. Lanes 1 and 5, protein bound to sperm chromatin (45′ time point); lanes 2 and 4, egg extract. Lane 3, protein markers. (E) Inhibition of DUE-B and Cdc45 binding to sperm chromatin by p27Kip. Lanes 1 to 3, untreated; lanes 4 to 6, p27Kip treated.
It was reported previously that the binding of DUE-B to an ectopic copy of the c-myc replicator in vivo was not diminished by a mutation that altered the chromatin structure of the origin and partially decreased MCM binding (12). To test whether DUE-B binding is pre-RC dependent in the egg extract system, geminin was added to block MCM binding (26, 29). As shown in Fig. 4C, complete inhibition of MCM loading blocked both DUE-B and Cdc45 binding, indicating that the loading of these proteins is pre-RC dependent. Lesser inhibition of MCM loading by geminin did not result in a decrease in DUE-B loading (A. Chowdhury, unpublished results), suggesting that the persistence of DUE-B binding to the mutant c-myc origin in vivo may have been due to the incomplete loss of MCM binding or to additional interactions between DUE-B and the origin DUE.
The membrane fraction of egg extract contributes to the formation of pseudonuclei around sperm chromatin that concentrates the cytosolic S-phase-promoting kinases required for activation of the pre-RC (56). Since DUE-B and Cdc45 do not bind efficiently to sperm chromatin in a high-speed egg cytosol devoid of membrane components (7, 9), we tested the effects of the Cdk2 inhibitors roscovitine (31) (Fig. 4D) and p27Kip (Fig. 4E) on DUE-B and Cdc45 loading. As reported previously, the binding of Cdc45 to chromatin was blocked by Cdk inhibition (53). The loading of DUE-B was also blocked in extracts containing these inhibitors, suggesting that Cdk2 activity is necessary for the loading of both DUE-B and Cdc45.
Interaction of DUE-B, TopBP1, and Cdc45.
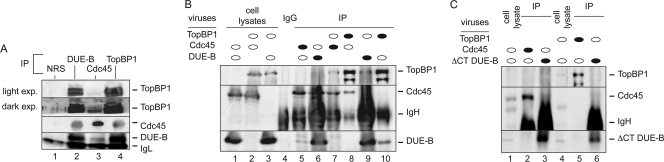
TopBP1 and Cdc45 have been reported to interact in vivo and in cell extracts (45). In view of the similar loading kinetics of DUE-B and Cdc45, we asked whether DUE-B can also interact with TopBP1 and Cdc45. Immunoprecipitation with antibodies directed against DUE-B, TopBP1, or Cdc45 coprecipitated each of the other two proteins from HeLa cell lysates (Fig. 5A). Recombinant DUE-B can also interact with TopBP1 and Cdc45 in Xenopus egg extracts (see below). To gain further evidence for direct interactions among DUE-B, TopBP1, and Cdc45, these proteins were expressed pairwise from baculovirus vectors in insect Sf9 cells (Fig. 5B). We observed that each of these proteins could be coprecipitated with either of the other two proteins in appropriately infected cells in the absence of the appearance of a cross-reactive band of the third, nonexpressed protein. These results suggest that DUE-B, TopBP1, and Cdc45 interact in vivo and may form a ternary complex.
FIG. 5.
Interaction of DUE-B, Cdc45, and TopBP1. (A) Immunoprecipitation (IP) of HeLa cell extracts using normal rabbit serum (NRS, lane 1) or antibody against DUE-B (lane 2), Cdc45 (lane 3), or TopBP1 (lane 4). Proteins indicated to the right of the figure were visualized by Western blotting (WB) with the cognate antibodies. (B) Western blot of lysates (input) of Sf9 cells infected pairwise with baculoviruses expressing DUE-B, TopBP1, and Cdc45 (lanes 1 to 3, unfilled ovals). Lane 4, IP with preimmune IgG; lanes 5 to 10, IP with antibody (filled ovals) against one of each pair of baculovirus-expressed proteins. (C) Western blot of immunoprecipitates of lysates (input) of Sf9 cells infected pairwise with baculoviruses expressing ΔCT DUE-B, TopBP1, and Cdc45 (lanes 1 and 4, unfilled ovals). Lanes 2, 3, 5, and 6, IP with antibody (filled ovals) against one of each pair of baculovirus-expressed proteins.
The amino-terminal 150-residue region of DUE-B shows strong structural and enzymatic similarities to those of tRNA proofreading enzymes in lower organisms, while the carboxy-terminal 59-amino-acid region of DUE-B absent from these enzymes is necessary for DUE-B binding to DNA in vitro (18) and shows nominal similarity to the carboxy-terminal regions of Sld3 in S. pombe and S. cerevisiae (Fig. 6). Immunoprecipitation of lysates of Sf9 cells coinfected with baculoviruses expressing the 1- to 150-amino-acid region of DUE-B (DUE-B ΔCT mutant) and either TopBP1 or Cdc45 showed that neither TopBP1 nor Cdc45 bound efficiently to the truncated DUE-B ΔCT mutant (Fig. 5C). These results suggest that the ability to bind TopBP1 and Cdc45 was acquired along with the carboxy-terminal extension of DUE-B.
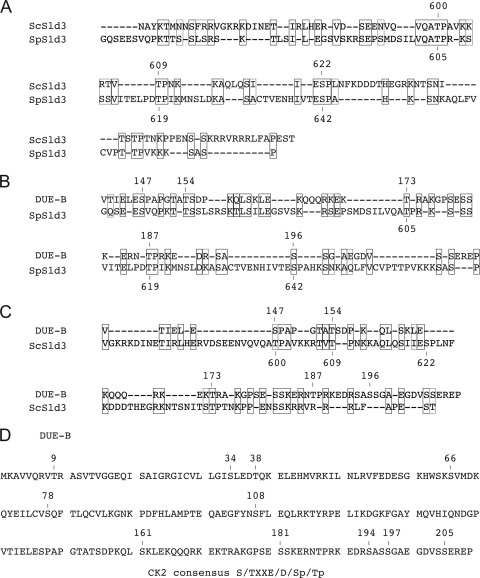
FIG. 6.
Alignment of yeast Sld3 and DUE-B. (A to C) The carboxy-terminal regions of Sld3 from S. cerevisiae Sld3 (ScSld3), S. pombe Sld3 (SpSld3), and human DUE-B were aligned as indicated, using DNAMAN. Amino acid similarity (47) and identity are indicated by light and dark outlines, respectively. (D) CK2 consensus target sites (numbered) in DUE-B.
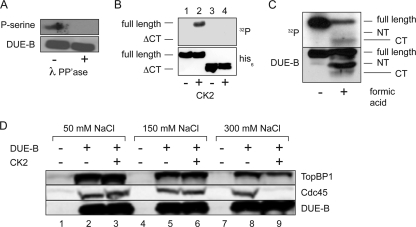
Cdk phosphorylation of the carboxyl terminus of S. cerevisiae Sld3 (T600, T609, and S622) has been implicated in Sld3 binding to Dpb11 and the loading of Cdc45 onto pre-RCs (48, 61). Allowing for gaps, this region of S. pombe and S. cerevisiae Sld3 shows ∼45% amino acid conservation, including the Cdk consensus S/T-P sites implicated in pre-IC formation (Fig. 6A). Alignment of the 69-amino-acid carboxyl domain of DUE-B with the carboxy-terminal regions of S. cerevisiae Sld3 and S. pombe Sld3 revealed limited similarity, allowing for gaps (Fig. 6B and C). However, except for the DUE-B T187P188 dipeptide, the Cdk consensus sites found in yeast Sld3 were not conserved, and this dipeptide is not conserved in other metazoan DUE-B proteins (18). Since mass spectrometric analysis indicated that three serines in the carboxy-terminal region of HeLa DUE-B are phosphorylated in vivo but were not phosphorylated in baculovirus-expressed DUE-B (M. Kemp and J. Yao, unpublished data), we considered the possibility that DUE-B might be phosphorylated by another kinase in vivo. Indeed, 11 matches to the casein kinase 2 (CK2) consensus target sequence S/TXXE/D/Sp/Tp are present in DUE-B, of which 9 are highly conserved (S34, T38, S66, S78, S161, S181, S194, S197, and S205) in mice, chickens, frogs, and fish (18). Treatment of HeLa DUE-B with λ phosphatase removed the phosphorylation of the protein (Fig. 7A), and treatment of baculovirus-expressed DUE-B with CK2 resulted in selective phosphorylation of the carboxy-terminal tail of the protein (Fig. 7B and C). To determine whether CK2 phosphorylation of DUE-B affected its interaction with TopBP1 or Cdc45, baculovirus-expressed DUE-B was added to Xenopus egg extract, pulled down through its hexahistidine tag by Ni-NTA beads, and washed with increasing concentrations of NaCl. At low and intermediate concentrations of salt, Xenopus TopBP1 and Cdc45 were coisolated with the added DUE-B. However, at the highest salt concentration tested, the affinity between Cdc45 and phosphorylated DUE-B was lost. These results imply that the carboxyl terminus of DUE-B is important for Cdc45 binding and that phosphorylation of the DUE-B carboxyl terminus weakens its binding to Cdc45. These data also suggest that TopBP1 can interact with DUE-B independently of Cdc45 binding.
FIG. 7.
Phosphorylation of DUE-B. (A) Recombinant DUE-B was purified by Ni-NTA chromatography from HeLa A1 cells and visualized by antiphosphoserine antibody before (−) or after (+) treatment with λ phosphatase (PP'ase). (B) Epitope-tagged (His6) full-length DUE-B or the ΔCT-DUE-B fragment was expressed in baculovirus-infected Sf9 cells and purified. Proteins were untreated (lanes 1 and 3) or treated (lanes 2 and 4) with casein kinase 2 (CK2) and [γ-32P]ATP. Top, autoradiogram; bottom, anti-His6 Western blot. (C) Baculovirus-expressed DUE-B was phosphorylated by CK2 in the presence of [γ-32P]ATP and electrophoresed before or after treatment with formic acid, which cleaves preferentially between D156 and P157 (18). Top, autoradiogram; bottom, anti-DUE-B Western blot. NT, N-terminal fragment; CT, C-terminal fragment. (D) Egg extracts were either untreated (lanes 1, 4, and 7), added to baculovirus-expressed His6-tagged DUE-B that was untreated (lanes 2, 5, and 8), or phosphorylated by CK2 (lanes 3, 6, and 9). Ni-NTA beads were added to each extract and washed with buffer of the indicated NaCl concentration, and the proteins pulled down were visualized by Western blotting with anti-DUE-B, anti-Cdc45, or anti-TopBP1 antibodies.
DUE-B is required for preinitiation complex formation.
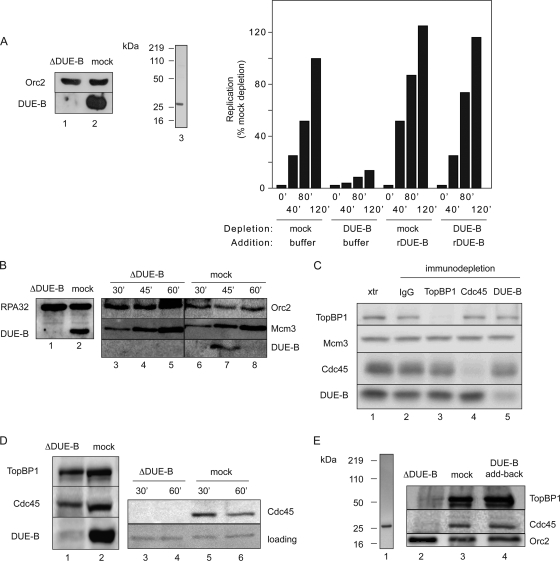
Baculovirus-expressed DUE-B cannot complement the replication activity of DUE-B-depleted extracts, and indeed, preincubation of sperm chromatin in egg cytosol containing baculovirus-expressed DUE-B inhibits subsequent replication at a step after MCM binding but before RPA-stabilized origin unwinding (7). In contrast, recombinant DUE-B purified from HeLa cells restored the end point (7) and time course of sperm chromatin replication (Fig. 8A). DUE-B immunodepletion does not block ORC or MCM binding to sperm chromatin (Fig. 8B) and does not appreciably deplete TopBP1 or Cdc45 from the extract, and vice versa (Fig. 8C). Considering the dominant-negative effect of baculovirus-expressed DUE-B on replication initiation, these results suggest that inhibition of DUE-B function interferes with a step after pre-RC formation but before pre-IC formation. To test directly whether DUE-B plays a role in the loading of pre-IC proteins, egg extracts were immunodepleted of DUE-B, and the loading of Cdc45 was assayed. When sperm chromatin was added to DUE-B-immunodepleted extract, we observed a significant decrease in the loading of Cdc45 (Fig. 8D, lanes 3 to 6). In a repeat experiment, DUE-B immunodepletion was observed to decrease the chromatin binding of Cdc45 and TopBP1; this effect was reversed by the addition of HeLa-expressed DUE-B (Fig. 8E) (7), confirming that the depletion of DUE-B is specifically responsible for the inhibition of Cdc45 and TopBP1 loading. The inability of DUE-B-immunodepleted extracts to load the essential initiation factor Cdc45 provides an explanation for the inhibition of replication observed when DUE-B levels are depleted.
FIG. 8.
DUE-B-dependent loading of Cdc45. (A) Xenopus egg extract was immunodepleted (left) with anti-DUE-B antibody (lane 1, ΔDUE-B) or preimmune serum (lane 2, mock), and buffer or recombinant His6-DUE-B purified from HeLa A1 cells (lane 3, Coomassie-stained gel) was added to the extracts for time course analysis of sperm chromatin replication (right). (B) Sperm chromatin was added to extract immunodepleted with anti-DUE-B antibody (lane 1) or preimmune serum (lane 2, mock) for analysis of the time course of Orc2 and Mcm3 binding (lanes 3 to 8). (C) Xenopus egg extract was undepleted (lane 1) or immunodepleted with preimmune serum (lane 2) or antiserum against TopBP1 (lane 3), Cdc45 (lane 4), or DUE-B (lane 5). (D) Extract was immunodepleted with anti-DUE-B antibody (lane 1) or preimmune serum (lane 2), and sperm chromatin was added for analysis of the time course of Cdc45 binding (lanes 3 to 6). The loading control is a nonspecific cross-reacting band. (E) Lane 1, His6-DUE-B purified from HeLa A1 cells for add back (silver-stained gel). Lanes 2 to 4, TopBP1, Cdc45, and Orc2 binding to sperm chromatin (45′ time point) in DUE-B-immunodepleted extract (lane 2), mock-depleted extract (lane 3), or DUE-B-immunodepleted extract containing His6-DUE-B purified from HeLa A1 cells (lane 4).
DISCUSSION
The c-myc DNA unwinding element binding protein DUE-B has a bipartite nature, in which the amino-terminal 150-residue domain bears strong structural similarity to aminoacyl-tRNA proofreading enzymes in lower organisms, while the carboxy-terminal 59 residues are involved in binding to DNA and the pre-IC proteins TopBP1 and Cdc45. The in vivo distribution of DUE-B between nuclear speckles and chromatin may reflect this bipartite structure. The interaction of DUE-B and SAP145 may also be relevant to a possible role for DUE-B in activation of the TopBP1/ATR DNA damage response, in view of the report that disruption of SAP145 function by the HIV viral protein R activates the ATR kinase and results in a late-S/G2-phase cell cycle arrest (50).
Cdc45 and TopBP1 have been shown to interact directly (45), which has been confirmed in the present work. We have also shown that DUE-B coprecipitates individually with Cdc45 and TopBP1. Indeed, sequential immunoprecipitation results are consistent with the formation of a ternary complex when purified, baculovirus-expressed TopBP1, DUE-B, and Cdc45 are mixed (J. Yao, unpublished data). The notion that DUE-B, Cdc45, and TopBP1 interact in vivo is supported by the coimmunoprecipitation of these proteins from human and insect cell lysates. The observations that DUE-B and Cdc45 are enriched at the DUEs of the c-myc, lamin B2, and ATX10/SCA10 origins and that binding of DUE-B and Cdc45 at the ATX10/SCA10 and ectopic c-myc origins show similarly strong dependence on the presence of a functional DUE suggest that the interaction between DUE-B and Cdc45 at the origin is important for replication initiation.
In egg extracts, we find that DUE-B and Cdc45 bind to the sperm chromatin template with comparable kinetics and dependencies on MCM binding and Cdk activity. In close agreement with previous reports of bimodal, Cdk-dependent, and Cdk-independent binding of Cut5 (13, 14), we observe that a fraction of TopBP1 binds to the template before DUE-B and Cdc45 in a Cdk-independent fashion, and binding increases in parallel with the Cdk-dependent loading of DUE-B and Cdc45. The dual phases of TopBP1 loading may reflect the roles in DNA replication initiation and the DNA damage checkpoint response (14, 60). The demonstration showing that the Cdk-independent binding of TopBP1 is sufficient for replication initiation suggests that the Cdk- and DUE-B-dependent loading of TopBP1 may be necessary for the response to DNA damage during S phase.
In addition to the ORC dependence of TopBP1 and DUE-B binding, siRNA knockdown of TopBP1 or DUE-B each inhibits the G1- to S-phase cell cycle transition and increases the levels of the Cdk inhibitor p21Waf1. In the case of TopBP1 knockdown, the inhibition of replication was independent of increased p21Waf1 (or p27Kip) expression (15). Jeon et al. concluded that TopBP1 is necessary for a step in replication initiation that occurs after pre-RC formation and Cdc7/Dbf4 activity but before pre-IC formation (15); the data in the present report indicate that DUE-B acts in the same temporal window as TopBP1.
DUE-B expressed in HeLa cells is phosphorylated at three sites in its carboxy-terminal tail and potentially at other sites yet to be mapped, whereas baculovirus-expressed DUE-B lacks detectable phosphorylation (M. Kemp and J. Yao, unpublished data). Although both purified forms of DUE-B chromatograph as homodimers, only baculovirus-expressed DUE-B assembles into high-molecular-weight complexes in HeLa cell or Xenopus egg extracts and is inhibitory to replication (7). The binding of baculovirus-expressed DUE-B, TopBP1, and Cdc45 implies that DUE-B phosphorylation is not essential for this interaction. This represents a significant difference from the Cdk1-dependent phosphorylation required for the binding of yeast Sld3 and Dpb11 (48, 61) and suggests that the phosphorylation of DUE-B is important at a step coming after the assembly of a TopBP1-DUE-B-Cdc45 complex.
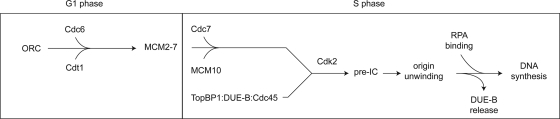
Van Hatten et al. have proposed a branched model for ORC-dependent Cdc45 loading and pre-IC formation in which one branch of the pathway comprises pre-RC assembly, while the second branch involves the Xenopus TopBP1 ortholog Mus101 (53) in Cdc45 loading and pre-IC formation. We wish to offer a modification of their model, as shown in Fig. 9, in which the fraction of TopBP1 that binds to DUE-B and Cdc45 is involved in Cdk2-dependent pre-IC formation; subsequent origin unwinding by the activated MCM helicase leads to the release of DUE-B and the binding of RPA. Our data do not address the loading of that fraction of TopBP1 that is Cdk independent. This model emphasizes the similarity in function of DUE-B and Sld3 and implies that DUE-B and Cdc45 will coordinately bind early in S phase to early-firing origins and late in S phase to late-firing origins. We speculate further that a TopBP1-DUE-B-Cdc45 complex may interact weakly with the pre-RC and that Cdk2 phosphorylation of the carboxy-terminal tail of DUE-B may enable the transfer and tight binding of Cdc45 to the pre-RC. The phosphorylation of DUE-B is significantly decreased by the treatment of HeLa cells with purvalanol A at levels which selectively inhibit Cdk activity and is enhanced when cells are treated with the PP2A inhibitor okadaic acid (7). Thus, if this speculation is correct, recycling of DUE-B into a new TopBP1-DUE-B-Cdc45 complex might involve dephosphorylation by PP2A, as presciently suggested by Petersen et al. (40). Experiments are currently under way to test these hypotheses.
FIG. 9.
Model for pre-IC formation. In this model, interacting DUE-B, Cdc45, and TopBP1 associate with the pre-RC. Cdk2 phosphorylation triggers Cdc45 activation of the pre-IC. Upon origin unwinding, DUE-B is released, and RPA binds to the origin, followed by replisome assembly and DNA synthesis. See the text for further details.
Acknowledgments
We sincerely thank the many investigators who generously provided reagents used in this work.
X.C. was supported by the WSU Biomedical Sciences Ph.D. Program. This work was funded by a WSU Research Challenge award and NIGMS grants GM53819 and GM82995 to M.L.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Abbas, T., U. Sivaprasad, K. Terai, V. Amador, M. Pagano, and A. Dutta. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal, B. D., and B. R. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372-376. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., A. M. Stout, and S. P. Bell. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. U. S. A. 96:9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 6.Bubulya, P. A., K. V. Prasanth, T. J. Deerinck, D. Gerlich, J. Beaudouin, M. H. Ellisman, J. Ellenberg, and D. L. Spector. 2004. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 167:51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casper, J. M., M. G. Kemp, M. Ghosh, G. M. Randall, A. Vaillant, and M. Leffak. 2005. The c-myc DNA-unwinding element-binding protein modulates the assembly of DNA replication complexes in vitro. J. Biol. Chem. 280:13071-13083. [DOI] [PubMed] [Google Scholar]
- 8.Chong, J. P., P. Thommes, A. Rowles, H. M. Mahbubani, and J. J. Blow. 1997. Characterization of the Xenopus replication licensing system. Methods Enzymol. 283:549-564. [DOI] [PubMed] [Google Scholar]
- 9.Chou, D. M., P. Petersen, J. C. Walter, and G. Walter. 2002. Protein phosphatase 2A regulates binding of Cdc45 to the prereplication complex. J. Biol. Chem. 277:40520-40527. [DOI] [PubMed] [Google Scholar]
- 10.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 11.Gambus, A., R. C. Jones, A. Sanchez-Diaz, M. Kanemaki, F. van Deursen, R. D. Edmondson, and K. Labib. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8:358-366. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh, M., M. Kemp, G. Liu, M. Ritzi, A. Schepers, and M. Leffak. 2006. Differential binding of replication proteins across the human c-myc replicator. Mol. Cell. Biol. 26:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto, Y., and H. Takisawa. 2003. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 22:2526-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto, Y., T. Tsujimura, A. Sugino, and H. Takisawa. 2006. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells 11:993-1007. [DOI] [PubMed] [Google Scholar]
- 15.Jeon, Y., K. Y. Lee, M. J. Ko, Y. S. Lee, S. Kang, and D. S. Hwang. 2007. Human TopBP1 participates in cyclin E/CDK2 activation and preinitiation complex assembly during G1/S transition. J. Biol. Chem. 282:14882-14890. [DOI] [PubMed] [Google Scholar]
- 16.Kamimura, Y., Y. S. Tak, A. Sugino, and H. Araki. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanemaki, M., and K. Labib. 2006. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 25:1753-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp, M., B. Bae, J. P. Yu, M. Ghosh, M. Leffak, and S. K. Nair. 2007. Structure and function of the c-myc DNA-unwinding element-binding protein DUE-B. J. Biol. Chem. 282:10441-10448. [DOI] [PubMed] [Google Scholar]
- 19.Kohzaki, H., and Y. Murakami. 2005. Transcription factors and DNA replication origin selection. Bioessays 27:1107-1116. [DOI] [PubMed] [Google Scholar]
- 20.Koike, M., T. Ikuta, T. Miyasaka, and T. Shiomi. 1999. Ku80 can translocate to the nucleus independent of the translocation of Ku70 using its own nuclear localization signal. Oncogene 18:7495-7505. [DOI] [PubMed] [Google Scholar]
- 21.Kramer, A., P. Gruter, K. Groning, and B. Kastner. 1999. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 145:1355-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumagai, A., J. Lee, H. Y. Yoo, and W. G. Dunphy. 2006. TopBP1 activates the ATR-ATRIP complex. Cell 124:943-955. [DOI] [PubMed] [Google Scholar]
- 23.Liu, G., J. J. Bissler, R. R. Sinden, and M. Leffak. 2007. Unstable spinocerebellar ataxia type 10 (ATTCT)*(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol. Cell. Biol. 27:7828-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, G., M. Malott, and M. Leffak. 2003. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 23:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciani, M. G., M. Oehlmann, and J. J. Blow. 2004. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J. Cell Sci. 117:6019-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutzmann, M., D. Maiorano, and M. Mechali. 2006. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during S phase in Xenopus. EMBO J. 25:5764-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutzmann, M., and M. Mechali. 2008. MCM9 binds Cdt1 and is required for the assembly of prereplication complexes. Mol. Cell 31:190-200. [DOI] [PubMed] [Google Scholar]
- 28.Maiorano, D., J. M. Lemaitre, and M. Mechali. 2000. Stepwise regulated chromatin assembly of MCM2-7 proteins. J. Biol. Chem. 275:8426-8431. [DOI] [PubMed] [Google Scholar]
- 29.McGarry, T. J., and M. W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043-1053. [DOI] [PubMed] [Google Scholar]
- 30.McWhinney, C., and M. Leffak. 1988. Episomal persistence of a plasmid containing human c-myc DNA, p. 467-471. In B. Stillman and T. Kelly (ed.), Cancer cells, vol. 6. CSH Laboratory Press, New York, NY. [Google Scholar]
- 31.Meijer, L., A. Borgne, O. Mulner, J. P. Chong, J. J. Blow, N. Inagaki, M. Inagaki, J. G. Delcros, and J. P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527-536. [DOI] [PubMed] [Google Scholar]
- 32.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Micklem, G., A. Rowley, J. Harwood, K. Nasmyth, and J. F. Diffley. 1993. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature 366:87-89. [DOI] [PubMed] [Google Scholar]
- 34.Mordes, D. A., G. G. Glick, R. Zhao, and D. Cortez. 2008. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 22:1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer, S. E., P. W. Lewis, and M. R. Botchan. 2006. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U. S. A. 103:10236-10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima, R., and H. Masukata. 2002. SpSld3 is required for loading and maintenance of SpCdc45 on chromatin in DNA replication in fission yeast. Mol. Biol. Cell 13:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishitani, H., Y. Shiomi, H. Iida, M. Michishita, T. Takami, and T. Tsurimoto. 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283:29045-29052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novac, O., D. Matheos, F. D. Araujo, G. B. Price, and M. Zannis-Hadjopoulos. 2001. In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell 12:3386-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parrilla-Castellar, E. R., and L. M. Karnitz. 2003. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J. Biol. Chem. 278:45507-45511. [DOI] [PubMed] [Google Scholar]
- 40.Petersen, P., D. M. Chou, Z. You, T. Hunter, J. C. Walter, and G. Walter. 2006. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol. Cell. Biol. 26:1997-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remus, D., E. L. Beall, and M. R. Botchan. 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23:897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricke, R. M., and A. K. Bielinsky. 2004. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol. Cell 16:173-185. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh, N., C. S. Spahr, S. D. Patterson, P. Bubulya, A. F. Neuwald, and D. L. Spector. 2004. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15:3876-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sard, L., P. Accornero, S. Tornielli, D. Delia, G. Bunone, M. Campiglio, M. P. Colombo, M. Gramegna, C. M. Croce, M. A. Pierotti, and G. Sozzi. 1999. The tumor-suppressor gene FHIT is involved in the regulation of apoptosis and in cell cycle control. Proc. Natl. Acad. Sci. U. S. A. 96:8489-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, U., Y. Wollmann, C. Franke, F. Grosse, H. P. Saluz, and F. Hanel. 2008. Characterization of the interaction between the human DNA topoisomerase IIbeta-binding protein 1 (TopBP1) and the cell division cycle 45 (Cdc45) protein. Biochem. J. 409:169-177. [DOI] [PubMed] [Google Scholar]
- 46.Sclafani, R. A., and T. M. Holzen. 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41:237-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, Y. H., H. Huang, and D. Kihara. 2006. Statistical potential-based amino acid similarity matrices for aligning distantly related protein sequences. Proteins 64:587-600. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, S., T. Umemori, K. Hirai, S. Muramatsu, Y. Kamimura, and H. Araki. 2007. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445:328-332. [DOI] [PubMed] [Google Scholar]
- 49.Teer, J. K., Y. J. Machida, H. Labit, O. Novac, O. Hyrien, K. Marheineke, M. Zannis-Hadjopoulos, and A. Dutta. 2006. Proliferating human cells hypomorphic for origin recognition complex 2 and pre-replicative complex formation have a defect in p53 activation and Cdk2 kinase activation. J. Biol. Chem. 281:6253-6260. [DOI] [PubMed] [Google Scholar]
- 50.Terada, Y., and Y. Yasuda. 2006. Human immunodeficiency virus type 1 Vpr induces G2 checkpoint activation by interacting with the splicing factor SAP145. Mol. Cell. Biol. 26:8149-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tercero, J. A., K. Labib, and J. F. Diffley. 2000. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 19:2082-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyoshima, H., and T. Hunter. 1994. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78:67-74. [DOI] [PubMed] [Google Scholar]
- 53.Van Hatten, R. A., A. V. Tutter, A. H. Holway, A. M. Khederian, J. C. Walter, and W. M. Michael. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 56.Walter, J., L. Sun, and J. Newport. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1:519-529. [DOI] [PubMed] [Google Scholar]
- 57.Walter, J. C. 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275:39773-39778. [DOI] [PubMed] [Google Scholar]
- 58.Wohlschlegel, J. A., S. K. Dhar, T. A. Prokhorova, A. Dutta, and J. C. Walter. 2002. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol. Cell 9:233-240. [DOI] [PubMed] [Google Scholar]
- 59.Yan, S., H. D. Lindsay, and W. M. Michael. 2006. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 173:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan, S., and W. M. Michael. 2009. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle 8:2877-2884. [DOI] [PubMed] [Google Scholar]
- 61.Zegerman, P., and J. F. Diffley. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445:281-285. [DOI] [PubMed] [Google Scholar]
- 62.Zembutsu, A., and S. Waga. 2006. De novo assembly of genuine replication forks on an immobilized circular plasmid in Xenopus egg extracts. Nucleic Acids Res. 34:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng, G., W. Liu, Y. Gong, H. Yang, B. Yin, J. Zhu, Y. Xie, X. Peng, B. Qiang, and J. Yuan. 2009. Human D-Tyr-tRNA(Tyr) deacylase contributes to the resistance of the cell to D-amino acids. Biochem. J. 417:85-94. [DOI] [PubMed] [Google Scholar]
- 64.Zou, L., and B. Stillman. 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20:3086-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou, L., and B. Stillman. 1998. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280:593-596. [DOI] [PubMed] [Google Scholar]