Abstract
ACK (activated Cdc42-associated tyrosine kinase) (also Tnk2) is an ubiquitin-binding protein and plays an important role in ligand-induced and ubiquitination-mediated degradation of epidermal growth factor receptor (EGFR). Here we report that ACK is ubiquitinated by HECT E3 ubiquitin ligase Nedd4-1 and degraded along with EGFR in response to EGF stimulation. ACK interacts with Nedd4-1 through a conserved PPXY WW-binding motif. The WW3 domain in Nedd4-1 is critical for binding to ACK. Although ACK binds to both Nedd4-1 and Nedd4-2 (also Nedd4L), Nedd4-1 is the E3 ubiquitin ligase for ubiquitination of ACK in cells. Interestingly, deletion of the sterile alpha motif (SAM) domain at the N terminus dramatically reduced the ubiquitination of ACK by Nedd4-1, while deletion of the Uba domain dramatically enhanced the ubiquitination. Use of proteasomal and lysosomal inhibitors demonstrated that EGF-induced ACK degradation is processed by lysosomes, not proteasomes. RNA interference (RNAi) knockdown of Nedd4-1, not Nedd4-2, inhibited degradation of both EGFR and ACK, and overexpression of ACK mutants that are deficient in either binding to or ubiquitination by Nedd4-1 blocked EGF-induced degradation of EGFR. Our findings suggest an essential role of Nedd4-1 in regulation of EGFR degradation through interaction with and ubiquitination of ACK.
Activated Cdc42-associated tyrosine kinase (ACK) (also Tnk2) is a member of the type VIII tyrosine kinase family. Activation of ACK, including both ACK1 and ACK2, occurs in response to signaling of epidermal growth factor receptor (EGFR), platelet-derived growth factor (PDGF) receptor, insulin receptor, Gas-6 receptor (Mer), M3 muscarinic receptor, integrins, or proteoglycan (3, 7, 11, 23, 26, 30, 44, 47). In Drosophila, D-ACK mediates the function of Cdc42 in dorsal closure during embryonic development (31). The ACK homologue, Ark-1, in Caenorhabditis elegans negatively regulates EGF signaling (15).
A number of studies suggest a role for ACK in EGFR degradation. ACK1 and ACK2, two alternatively spliced isoforms, possess a highly conserved clathrin-binding motif and interact with clathrin (37, 45). Overexpression of ACK2 severely impairs transferrin receptor endocytosis, causes aberrant localization of AP-2, and induces changes in clathrin assembly. Furthermore, ACK2 interacts with sorting nexin 9 (SNX9, also named SH3PX1), a member of the sorting nexin family, via its proline-rich domain 1 and phosphorylates SNX9 to facilitate the degradation of EGF receptors (22). In C. elegans, Ark-1 genetically interacts with UNC101, the homologue of mammalian clathrin-associated protein AP47, and SLI-1, the homologue of mammalian Cbl that is an E3 ubiquitin ligase for ubiquitination of EGFR, and negatively regulates EGFR signaling (15).
Our previous studies showed that ACK1 interacts with EGFR upon EGF stimulation via a region at the carboxyl terminus, designated the EGFR-binding domain (EBD), which is highly homologous to the EGFR/ErbB2-binding domain of Gene-33/Mig-6/RALT (32, 43). The interaction of ACK1 with EGFR is dependent on kinase activity and tyrosine phosphorylation of EGFR. Immunofluorescent staining using anti-EGFR and GFP-ACK1 indicates that ACK1 is colocalized with EGFR on large vacuolar structures upon EGF stimulation. Suppression of the expression of ACK1 by ACK-RNA interference (RNAi) inhibits ligand-induced degradation of EGFR, suggesting that ACK1 plays an important role in the regulation of EGFR degradation in cells. Furthermore, we identified ACK1 as an ubiquitin-binding protein. Through an ubiquitin association (Uba) domain at the carboxyl terminus, ACK1 is capable of interacting with both poly- and monoubiquitin. Overexpression of an Uba domain deletion mutant of ACK1 blocked the ligand-dependent degradation of EGFR, suggesting that ACK1 regulates EGFR degradation via its Uba domain. Thus, ACK1 senses EGF signaling and regulates degradation of EGFR.
EGF-induced degradation of EGFR is mediated by ubiquitination (16). The ubiquitination of EGFR is activated upon EGF stimulation by recruiting the RING family E3 ubiquitin ligase Cbl to pY1045 (20, 21). This ubiquitination functions as a sorting signal for transporting EGFR to lysosomes for degradation (14). Nedd4, the HECT domain-containing E3 ubiquitin ligase, is also involved in the regulation of EGFR trafficking by ubiquitination of endocytic or vesicle sorting proteins (28). For example, it has been observed that Nedd4 ubiquitinates Cbl, Eps15, Tsg101, Hrs, and secretory carrier membrane proteins (SCAMPs) and participates in the processes of EGFR endocytosis and degradation (1, 18, 25, 42). However, exactly how Nedd4 engages in the EGFR degradation process in response to EGF stimulation is not known.
In this report, we show that EGF stimulation induces ACK degradation. This degradation is associated with ubiquitination of ACK. Nedd4-1, but not Nedd4-2, is identified as the E3 ubiquitin ligase for ubiquitination of ACK. Furthermore, EGF-induced degradation of ACK is EGFR activation dependent and processed by lysosomes. RNAi knockdown and mutational analysis demonstrated that Nedd4-1 and Nedd4-1-catalyzed ubiquitination of ACK are required for EGF-induced degradation of EGFR and ACK. Our findings suggest a new mechanism in regulation of EGFR degradation.
MATERIALS AND METHODS
Reagents (i) Biochemicals.
EGF was purchased from Invitrogen (GIBCO); MG-132, PSI, lactacystin, SB203850, and bafilomycin were from CalBiochem; NH4Cl was from Fisher. Velcade (bortezomib) was from LC Laboratories.
(ii) Antibodies.
Anti-EGFR (Mab528) was obtained from hybridoma Mab528 cell culture medium and used for immunoprecipitation; anti-EGFR (1005) and anti-ACK (A11) were from Santa Cruz Biotech; anti-ACKPCC is a polyclonal rabbit anti-ACK antibody against the N-terminal 100-amino-acid residues of ACK2, as described previously (32); antiubiquitin and antihemagglutinin (anti-HA) (12CA5) were from Covance; anti-Myc (9E10) was from the Developmental Studies Hybridoma Bank (DSHB, University of Iowa); anti-PY (4G10) and anti-Nedd4-1 were from Chemicon (Upstate Biotech); anti-β-actin was from Sigma. cDNAs of MGC (Mammalian Gene Collection) and IMAGE (the Integrated Molecular Analysis of Genomes and their Expression) clones were purchased from Open Biosystems. RNAi oligonucleotides were synthesized by Invitrogen. Cell lines were purchased from ATCC.
Construction of cDNA plasmids and mutagenesis.
(i) Nedd4 constructs. pcDNA3-HA-human Nedd4-1 and pcDNA3 HA-human Nedd4-2b were constructed by subcloning human Nedd4-1 cDNA (IMAGE no. 8862584) and human Nedd4-2b (IMAGE no. 5528964), respectively. The pcDNA3-HA-human Nedd4-2 was constructed by ligation of the partial human Nedd4-2 cDNA (IMAGE no. 3604024) with the 5′ end of human Nedd4-2b cDNA. Nedd4-2b is a splicing variant of Nedd4-2 without exons 13 and 14.
(ii) Construction of GST fusion proteins.
The Uba domain or the WW-binding domain (WWBD) of ACK, or the WW domains of human Nedd4-1, was subcloned into the glutathione S-transferase (GST) fusion protein expression vector pGEX4T3 by PCR. The constructs were confirmed by DNA sequencing.
(iii) Mutagenesis.
Mutations of ACK and Nedd4s were generated by site-directed mutagenesis using a mutagenesis kit manufactured by Stratagene.
Cell culture, transfection, and preparation of cell lysates.
COS7, HEK293T, HeLa, T47D, and A549 cells were maintained in Dulbecco modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS) at 37°C and 5% CO2. The cells were cultured to 90% confluence before transfection. For transfection, the plasmid DNA (1 to 2 μg/60 mm dish) was premixed with ESCORT V transfection reagent (SIGMA) in ESCORT V transfection reagent buffer (0.2 ml per 60-mm dish) or Lipofectamine 2000 (Invitrogen) and incubated at room temperature for 15 min. The cells were incubated in the plasmid DNA/ESCORT V or Lipofectamine 2000 mixture for 5 h. After 48 h of transfection, the cells were lysed with precooled mammalian cell lysis buffer (40 mM HEPES [pH 7.4], 100 mM NaCl, 1% Triton X-100, 25 mM glycerol phosphate, 1 mM sodium orthovanadate, 1 mM EDTA, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) or Radioimmunoprecipitation assay (RIPA) buffer (40 mM HEPES [pH 7.4], 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS, 100 mM NaCl, 1 mM EDTA, 25 mM β-glycerolphosphate, 1 mM Na orthovanadate, 10 μg/ml each leupeptin and aprotinin) (for GST-ACK1Uba pulldown assays) by rocking the plates at 4°C for 30 min. The cell lysates were cleared by centrifugation at 14,000 rpm in a Microfuge (Eppendorf) for 10 min at 4°C before use.
Immunoprecipitation and immunoblotting.
For immunoprecipitation, the precleared cell lysate was incubated with primary antibody on ice for 30 min; then protein A beads were added, and the mixture was incubated at 4°C for 2 h with rotation. The beads were washed with lysis buffer three times, and the immunoprecipitation complexes were either ready for enzymatic assays or directly dissolved in SDS-PAGE sample buffer for SDS-PAGE. Immunoblotting was performed as instructed by Western Lightning immunoblot kits (Perkin Elmer).
Endogenous Nedd4 was detected by immunoblotting with an anti-Nedd4-1 antibody that was against the WW2 domain (amino acid residues 395 to 462) of rat Nedd4-1 (UBI). Because this antibody also recognizes Nedd4-2, we did not specify detected endogenous Nedd4 as Nedd4-1, except in Nedd4-1 RNAi knockdown experiments.
Expression and purification of GST fusion protein.
GST fusion proteins were expressed in Escherichia coli (JM109) and purified by affinity purification with glutathione-agarose beads as described previously (45).
Expression and purification of His-tagged Nedd4-1 proteins.
His-tagged E3 ligase Nedd4-1 was expressed in E. coli BL21 cells by isopropyl-β-d-thiogalactopyranoside (IPTG) induction. The cells were lysed in His-lysis buffer (25 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.5% Triton X-100, 10% glycerol, 10 mM imidazole). The cell lysates were cleared by 14,000 rpm centrifugation at 4°C for 20 min, and the cleared lysates was incubated with Ni-nitrilotriacetic acid (Ni-NTA) agarose beads at 4°C with rotation for 2 h. The Ni beads were washed three times with His-lysis buffer containing 20 mM imidazole. The bead-bound Nedd4-1 was eluted by incubation of the beads with elution buffer (25 mM Tris-HCl [pH 8.0], 50 mM NaCl, 10% glycerol, 250 mM imidazole) at 4°C with rotation for 15 min. The eluted Nedd4-1 was dialyzed overnight in Nedd4 dialysis buffer (25 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM MgCl2) and stored at −80°C for use.
Pulldown assays with GST fusion proteins.
The GST fusion protein beads containing 15 to 30 μg of GST fusion protein were incubated with the mammalian cell lysates (about 1 mg) at 4°C for 3 h with rotation. The beads were washed three times with the mammalian cell lysis buffer and resuspended in 2× SDS-PAGE sample buffer. After SDS-PAGE, the precipitated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane for immunoblotting.
Precipitation of denatured ACK by Ni-NTA-bead pulldown.
His-tagged mouse ACK1 in pcDNA3 vector was transfected into HEK293 cells for 36 h followed by 12 h of serum starvation. The cells were treated with MG-132 for 30 min followed by EGF (50 ng/ml) for the indicated time and lysed in mammalian cell lysis buffer plus 6 M urea for 30 min at 4°C. To precipitate denatured His-tagged ACK, 20 μl of Ni-NTA beads (Qiagen) was added to 500 μl of the cell lysates, and the mixture was incubated for 3 h with rotation at 4°C. The beads were precipitated, washed three times with mammalian cell lysis buffer, and used for SDS-polyacrylamide gel electrophoresis and immunoblotting.
In vitro ubiquitination of ACK1 by Nedd4-1.
To examine in vitro ubiquitination of ACK1 by Nedd4-1, Myc-tagged ACK1 was transiently expressed in HEK293 cells and immunoprecipitated by protein A-bead-bound anti-Myc antibody. In vitro ubiquitination of ACK1 was performed by adding 15 μl reaction mixture containing a final concentration of 100 nM E1, 0.5 μM UbcH7 (E2), 5 μM monoubiquitin, and 2 mM ATP in ULR buffer (25 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2.0 mM MgCl2, and 1 mM dithiothreitol) to 10 μl of the ligase/substrate fraction containing protein A bead-bound immunoprecipitated ACK1 and/or purified His-Nedd4-1 (100 ng). The reaction was initiated by adding the reaction mixture and was carried out at 22°C for 20 min. The protein A bead-bound ACK1 was washed three times by mammalian cell lysis buffer to remove non-ACK-conjugated ubiquitin and resuspended in 25 μl 2× SDS-PAGE sample buffer for SDS-PAGE.
RNA interference.
Nedd4-1, Nedd4-2, and ACK-RNAi experiments were performed according to the method described by Elbashir et al. (9). Two 21-nucleotide small interference RNA (siRNA) sequences (h-Nedd4-1 RNAi-A, UUGAGAUGAUUUCCUAGGUCA; h-Nedd4-1 RNAi-B, UUCCAUGAAUCUAGAAGAACA; h-Nedd4-2 RNAi, AACCACAACACAAAGUCACAC; h-ACK RNAi, AAGAUGGUGACAGAGCUGGCA) corresponding to the coding region of human Nedd4-1, Nedd4-2, or ACK were selected. The siRNA oligonucleotides were chemically synthesized by Invitrogen. The negative control was set up using the 21-nucleotide RNA oligonucleotide that corresponded to the coding sequence of luciferase. Transfection of the siRNA (final concentration, 40 nM) into HEK293 or A549 cells was carried out using Lipofectamine 2000 (Invitrogen). Suppression of expression of endogenous Nedd4-1, Nedd4-2, or ACK by the siRNAs was determined by immunoblotting.
RESULTS
EGF signaling induces ACK degradation.
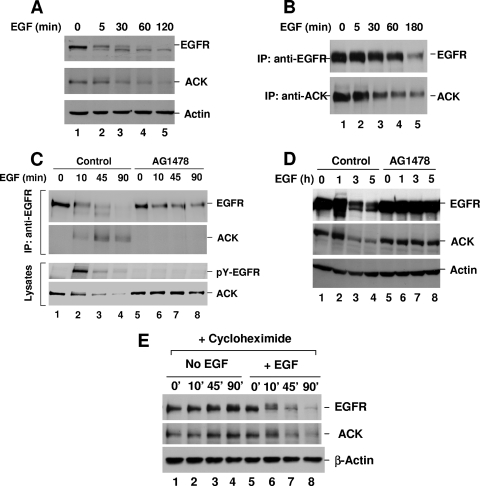
Our previous studies observed that interaction of endogenous ACK with EGFR in response to EGF stimulation occurred at a late stage (30 to 60 min after the addition of EGF) of the stimulation, while exogenously expressed ACK interacted with EGFR immediately upon EGF stimulation (as fast as 1 min) (32). We speculated that EGF stimulation might affect the expression of ACK as it affects that of Mig-6 (13). Thus, we examined the expression of endogenous ACK in response to EGF stimulation in HeLa and COS7 cells. As shown in Fig. 1A and B, unexpectedly, EGF treatment significantly reduced the expression of ACK accompanied by degradation of EGFR. This EGF-induced reduction of ACK was dependent on EGFR kinase activity. When the kinase activity of EGFR was inhibited by treatment with AG1478, a specific kinase inhibitor of EGFR, the reduction of expression of ACK in HeLa or COS7 cells upon EGF stimulation was eliminated (Fig. 1C and D), accompanied by inhibition of EGFR degradation (Fig. 1C and D, lanes 5 to 8).
FIG. 1.
EGF induces degradation of ACK. HeLa or COS7 cells were cultured to 90% confluence and starved overnight (12 h) in either 0.1% FBS medium (A, C, and E, HeLa cells) or serum-free medium (B and D, COS7 cells). The cells were stimulated with EGF (50 ng/ml) alone (A and B) or EGF (50 ng/ml) plus EGFR inhibitor AG1478 (1 μM) (C and D) or translational inhibitor cycloheximide (10 μg/ml) (E). AG1478 or cycloheximide was added to culture medium 30 min before EGF stimulation. (A) EGFR (top panel) and ACK (middle panel) in the cell lysates were detected by immunoblotting with anti-EGFR (1005) or anti-ACK (A11) antibody. (B) EGFR or ACK was immunoprecipitated (IP) with anti-EGFR (Mab528) or anti-ACK (anti-ACKPCC) and detected by immunoblotting with anti-EGFR (1005) (top panel) or anti-ACK (A11) (bottom panel). (C) EGFR was immunoprecipitated and immunoblotted with anti-EGFR antibody (top panel). The coprecipitated ACK was detected by immunoblotting with anti-ACK (A11) antibody (second top panel). The tyrosine phosphorylation of EGFR was determined by immunoblotting the lysates with anti-PY(4G10) (second bottom panel). The amount of ACK in the cell lysates was detected by immunoblotting with anti-ACK (A11) (bottom panel). (D and E) EGFR (top panel) and ACK (middle panel) in cell lysates were detected by immunoblotting with anti-EGFR (1005) and anti-ACK (A11) antibodies. The amount of actin was used for indication of the lysate loading (bottom panels in panels A, D, and E).
There are two possible causes for EGF-induced reduction of ACK expression in HeLa or COS7 cells. One is suppression of biosynthesis of ACK; the other is induction of degradation of ACK. To determine the cause, we pretreated HeLa cells before EGF stimulation with the protein translation inhibitor cycloheximide to eliminate protein biosynthesis and then examined the effect of EGF stimulation on ACK expression. As shown in Fig. 1E, cycloheximide pretreatment did not prevent EGF-induced reduction of ACK level and EGFR degradation (lanes 6 to 8), indicating that EGF-induced reduction of ACK is protein synthesis independent. As a control, cycloheximide alone did not cause significant change in ACK expression level (lanes 1 to 4). Thus, we conclude that EGF induces ACK degradation and that this EGF-induced degradation of ACK is EGFR kinase activity dependent.
ACK binds to Nedd4 and is ubiquitinated upon EGF stimulation.
To elucidate the molecular mechanism underlying EGF-induced degradation of ACK, we first examined the ubiquitination of ACK in response to EGF stimulation. However, we found it difficult to detect the ubiquitination of endogenous ACK by immunoblotting the immunoprecipitated ACK with antiubiquitin antibody because of the low level of endogenous ACK. To overcome this difficulty, we developed a GST-ACK-Uba pulldown assay for detection of ubiquitinated ACK, based on our previous studies showing that the ACK-Uba domain has a high binding affinity for ubiquitinated proteins (32). The GST-ACK-Uba precipitates only ubiquitinated proteins from cell lysates; thus it is able to enrich ubiquitinated ACK for detection.
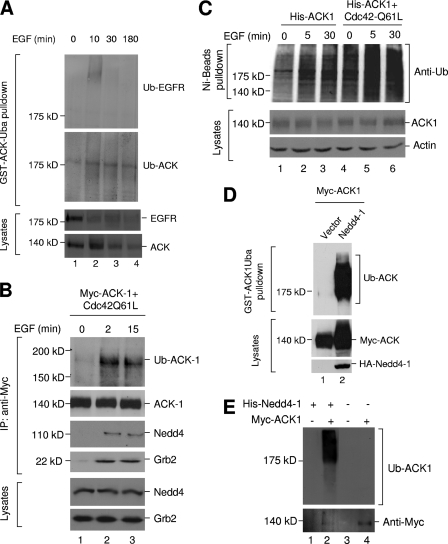
With the GST-ACK-Uba pulldown assay in HeLa cells, EGFR ubiquitination was detected after 10 min stimulation with EGF (Fig. 2A, top panel, lane 2). The EGFR ubiquitination declined rapidly, due to either degradation or dissociation of Cbl, the E3 ubiquitin ligase for EGFR. ACK was persistently ubiquitinated in response to EGF stimulation (Fig. 2A, second panel from top, lanes 2 to 4), suggesting that ubiquitination of ACK might be catalyzed by a different E3 ubiquitin ligase from that for EGFR ubiquitination, which is consistent with our previous data that ACK was not able to complex with Cbl (32). To further characterize EGF-induced ubiquitination of ACK, we overexpressed Myc-tagged ACK1 (Myc-ACK1) along with Cdc42Q61L, a GTPase-defective mutant of Cdc42 that activates ACK (44), in HEK293 cells and examined the ubiquitination of Myc-ACK1 immunoprecipitation complex upon EGF stimulation (Fig. 2B). We found that the HECT family E3 ubiquitin ligase Nedd4 was coprecipitated with ACK upon EGF stimulation (Fig. 2B, third panel, lanes 2 and 3), suggesting that ACK1 and Nedd4 are in complex in response to EGFR signaling. We also observed that Grb2 was associated with ACK1 upon EGF stimulation (Fig. 2B, fourth panel). EGF stimulation rapidly induced ubiquitination of the ACK1 immunoprecipitation complex at 180 kDa (Fig. 2B, top panel), similar to the ubiquitinated ACK band in Fig. 2A. To confirm that this EGF-induced ubiquitination is on ACK1, not the coprecipitated proteins, we transiently expressed His-tagged ACK1 alone or with Cdc42Q61L in HEK293 cells, stimulated the cells with EGF, and denatured the cell lysates with 6 M urea to dissemble any protein complex. Then we precipitated denatured ACK1 with nickel-NTA beads and detected ubiquitination of precipitated ACK1 by immunoblotting with anti-ubiquitin antibody. As shown in Fig. 2C, EGF stimulation induced ubiquitination of ACK (lanes 2 and 3). Coexpression with Cdc42Q61L enormously enhanced EGF-induced ubiquitination of ACK1 (lanes 5 and 6). Taken together, these data indicate that EGF stimulation promotes the association of ACK with E3 ubiquitin ligase Nedd4 and therefore the ubiquitination of ACK.
FIG. 2.
ACK is ubiquitinated in response to EGF stimulation. (A) HeLa cells were starved overnight (12 h) in 0.1% FBS medium and treated with MG-132 (10 μM) 30 min before EGF stimulation. After EGF stimulation for the indicated time, the cells were lysed. The ubiquitinated proteins in cell lysates (1 mg) were precipitated by glutathione-bead-bound GST-ACK-Uba (15 to 20 μg). Ubiquitinated EGFR (top panel) or ACK (second panel from top) was detected by immunoblotting with anti-EGFR (1005) or anti-ACK (A11) antibodies. EGF-induced degradation of EGFR and ACK is shown in the bottom two panels by immunoblotting the cell lysates with anti-EGFR (1005) and anti-ACK (A11) antibodies. (B) Myc-tagged mouse ACK1 was transfected into HEK293 cells for 36 h. The cells were serum starved for 12 h and stimulated with EGF for the indicated time. ACK1 was immunoprecipitated with anti-Myc antibody (second panel from top), and ubiquitinated ACK1 was determined by immunoblotting with antiubiquitin antibody (top panel). Coprecipitated endogenous Nedd4 (third panel from top) and Grb2 (fourth panel from top) were detected by immunoblotting with anti-Nedd4-1 and anti-Grb2 antibodies. The amount of Nedd4 or Grb2 in the lysates is shown in the two bottom panels by immunoblotting. (C) His-tagged mouse ACK1 was transfected into HEK293 cells for 36 h. The cells were serum starved for 12 h, treated with MG-132 (10 μM) for 30 min, and then stimulated with EGF (50 ng/ml) for the indicated time. The cells were lysed in mammalian cell lysis buffer plus 6 M urea. The denatured His-tagged ACK1 was precipitated by Ni-NTA beads. The ubiquitination of His-tagged ACK1 was detected by immunoblotting with antiubiquitin antibody (top panel). Expression of ACK1 was determined by immunoblotting of cell lysates with anti-ACK antibody (A11) (middle panel). (D) Myc-tagged ACK1 was cotransfected with HA-tagged Nedd4-1 (lane 2) or vector (lane 1) into HEK293 cells for 48 h. Ubiquitinated ACK1 was precipitated by glutathione-bead-bound GST-ACK1Uba and detected by immunoblotting with antiubiquitin antibody (top panel). The expression level of ACK1 or Nedd4 was determined by immunoblotting the lysates with anti-Myc or anti-HA antibodies (bottom two panels). (E) Myc-tagged ACK1 was immunoprecipitated from Myc-ACK1-transfected HEK293 cell lysates and used for an in vitro ubiquitination assay. For the ubiquitination assay, 100 ng of purified Nedd4-1 was used in each sample. After the ubiquitination reaction, Myc-ACK-bound beads were washed with mammalian lysis buffer three times to remove non-ACK-conjugated polyubiquitin. Ubiquitination of Myc-ACK1 was detected by immunoblotting with antiubiquitin antibody (top panel). The amount of Myc-ACK1 was determined by immunoblotting with anti-Myc antibody (bottom panel). Controls that had no Myc-ACK (but with the same amount of protein A beads that were incubated with Myc-ACK-transfected HEK293 cell lysates) (lanes 1 and 3) or Nedd4-1 (lane 4) were set up to exclude endogenous E3 ligase activity and Nedd4-1 autoubiquitination.
To confirm that Nedd4 could ubiquitinate ACK in cells, we cotransfected human Nedd4-1 with ACK in HEK293 cells and examined the ubiquitination of ACK (Fig. 2D). As expected, overexpression of Nedd4-1 caused extensive ubiquitination of ACK (Fig. 2D, top panel, lane 2). We also examined in vitro ubiquitination of ACK by Nedd4-1. Incubation of purified Nedd4-1 with ACK1 immunoprecipitated from HEK293 cells caused strong ubiquitination of ACK1 (Fig. 2E, top panel, lane 2). No ubiquitination was detected in samples that had no addition of Nedd4-1 or/and ACK1 (Fig. 2E, top panel, lanes 1, 3, and 4), indicating that the ubiquitination is both ACK1 and Nedd4-1 dependent. In addition, nonubiquitinated ACK1 protein was significantly reduced by incubation with Nedd4-1 (Fig. 2E, bottom panel, compare lane 2 with lane 4), presumably resulting from an ubiquitination-caused gel shift, implicating that ACK1 is ubiquitinated by Nedd4-1 in vitro. Taken together, these data suggest that Nedd4 is the E3 ubiquitin ligase ubiquitinating ACK in response to EGF signaling.
ACK interacts with Nedd4 via a conserved WW domain-binding motif.
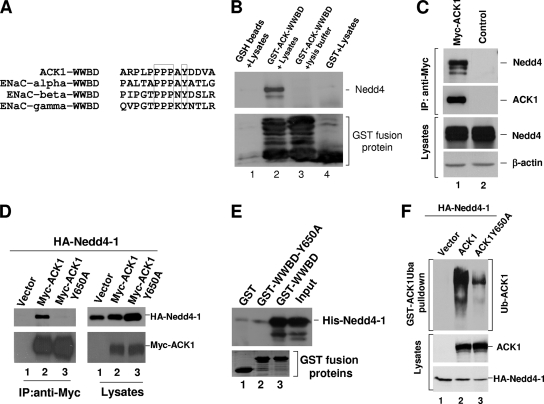
We next characterized the interaction of ACK with Nedd4. Nedd4 belongs to the WW domain-containing family of E3 ubiquitin ligases (17, 29) and has three or four type I WW domains that interact with the PPXY motif in its ubiquitination substrates or regulatory proteins (36). The peptide sequence of ACK contains a PPXY motif located in a proline-rich region from amino acids 647 to 650 in mouse ACK1 (Fig. 3A). This PPXY motif is also present in epithelial sodium channel subunits, which are known Nedd4 substrates (12, 35) (Fig. 3A). We designated this region the WW-binding domain (WWBD). To confirm that WWBD is the Nedd4 binding site, we constructed a GST-ACK-WWBD fusion protein for a Nedd4 pulldown assay. As shown in lane 2 of Fig. 3B, GST-ACK-WWBD precipitated Nedd4 from COS7 cell lysates, whereas GST (lane 4) or GSH beads (lane 1) did not, indicating that Nedd4 interacts with the WWBD. To confirm interaction of ACK with Nedd4 in cells, Myc-tagged ACK1 was expressed in HEK293 cells. Endogenous Nedd4 was coimmunoprecipitated with ACK after immunoprecipitation of ACK with anti-Myc antibody (Fig. 3C). To determine whether the PPXY motif is the site in ACK interacting with Nedd4, we mutated Y650 to alanine, changing PPAY to PPAA, and then examined coimmunoprecipitation with Nedd4-1. As shown in Fig. 3D, while wild-type ACK coprecipitated Nedd4-1 (lane 2 of the right panel), the Y650A mutation eliminated the interaction of ACK with Nedd4-1 (lane 3 of the right panel), indicating that Y650 is required for ACK to bind to Nedd4-1. To confirm that interaction between ACK and Nedd4-1 is direct and no other protein is needed, we performed an in vitro binding assay with purified Nedd4-1 and ACK-WWBD proteins. As shown in Fig. 3E, purified His-tagged Nedd4-1 was coprecipitated with purified GST-WWBD (lane 3) but not with GST-WWBD-Y650A, the Nedd4-binding-defective mutant (lane 2), or GST (control) (lane 1), demonstrating a direct interaction between the ACK-WWBD and Nedd4-1. Furthermore, in a GST-ACK-Uba pulldown assay, abolishing the binding of ACK to Nedd4-1 by mutation of Y650 reduced the ubiquitination of ACK by 90% (Fig. 3E, lane 3), suggesting that the binding is required for the ubiquitination.
FIG. 3.
ACK interacts with Nedd4 through a conserved PPXY WW-binding motif. (A) Alignment of the PPXY WW-binding motif of ACK with the known Nedd4-binding site of ENaC subunits. The boxed residues are the conserved WW-binding motif. (B) Glutathione (GSH)-conjugated beads, the bead-bound GST-ACK-WWBD, or GST was incubated with HEK293 cell lysates (lanes 1, 2, and 4). As a control, the bead-bound GST-ACK-WWBD was also incubated with the lysis buffer (lane 3). Coprecipitated Nedd4 was detected by immunoblotting with anti-Nedd4-1 antibody (top panel). The GST fusion proteins were visualized by Coomassie blue staining (bottom panel). (C) Myc-tagged ACK1 (lane 1) or the vector (lane 2) was transfected into HEK293 cells. Myc-ACK1 was immunoprecipitated and immunoblotted with anti-Myc antibody (the second top panel). Coprecipitated endogenous Nedd4 was detected by immunoblotting with anti-Nedd4-1 antibody (top panel). The amount of endogenous Nedd4 in the lysates is shown in the second panel from the bottom. Actin was used as an indication of the lysate loading (bottom panel). (D) Myc-tagged ACK1, the WW-binding-defective mutant ACK1Y650A, or the vector was cotransfected with HA-tagged Nedd4-1 into HEK293 cells. ACK1 or ACK1Y650A was immunoprecipitated with anti-Myc antibody (the left bottom panel). Coprecipitated Nedd4-1 was detected by immunoblotting with anti-HA antibody (left top panel). The expression levels of Myc-ACK and HA-Nedd4-1 in the cell lysates are shown in the right panels. (E) Purified GST-ACK-WWBD or GST-ACK-WWBD-Y650A was incubated with purified His-Nedd4-1. The bead-bound GST fusion proteins were precipitated by centrifugation. The coprecipitated Nedd4-1 was detected by immunoblotting with anti-Nedd4-1 (top panel). The GST fusion proteins were visualized by Coomassie blue staining (bottom panel). (F) Myc-tagged ACK1, the WW-binding-defective mutant ACK1Y650A, or the vector was cotransfected with HA-tagged Nedd4-1 into HEK293 cells. The ubiquitinated ACK1 was precipitated by GST-ACK1Uba and detected by immunoblotting with anti-ACK (A11) (top panel). Expression of Myc-ACK1 and Nedd4-1 is shown in the two bottom panels.
The WW3 domain of human Nedd4-1 interacts with ACK and is required for ubiquitination of ACK.
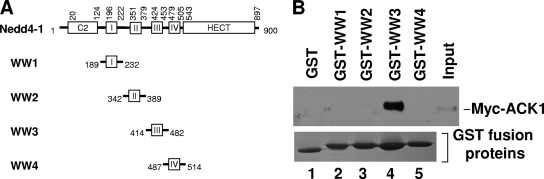
To determine the region in Nedd4 that interacts with ACK, we subcloned each of four WW domains of human Nedd4-1 into a GST fusion protein expression vector (Fig. 4A) and performed the GST fusion protein pulldown assay by incubating purified bead-bound GST-WW domain fusion proteins with ACK-overexpressed HEK293 cell lysates. As shown in Fig. 4B, the bead-bound GST-WW3 precipitated Myc-ACK1 from the cell lysates (lane 4, top panel), while the GST-WW1, GST-WW2, and GST-WW4 did not (lanes 2, 3, and 5, top panel), indicating that WW3 is the domain binding to ACK1.
FIG. 4.
ACK1 interacts with the WW3 domain of Nedd4-1. (A) Schematic representation of GST-human Nedd4-1 WW domain constructs. (B) The bead-bound GST or GST-human Nedd4-1 WW domain was incubated with Myc-ACK1-expressed HEK293 cell lysates and precipitated by centrifugation. Coprecipitated Myc-ACK1 was detected by immunoblotting with anti-Myc antibody (top panel). Loaded GST fusion proteins were visualized by Coomassie blue staining (bottom panel).
Nedd4-1 is the E3 ubiquitin ligase for ubiquitination of ACK.
There are two members in the Nedd4 family, Nedd4-1 and Nedd4-2 (also Nedd4L), in mammalian cells (5). Both Nedd4-1 and Nedd4-2 have a similar domain structure, with a C2 domain at the N terminus, followed by four WW domains and the HECT domain at the C terminus. Human Nedd4-1 and Nedd4-2 are about 65% identical in primary sequence. The most divergent region is the segment between the WW1 and the WW3 domains. Despite their structural similarity, Nedd4-1 and Nedd4-2 recognize distinct substrates, for example, ion channels and membrane transporters for Nedd4-2 (8, 33, 34, 40, 49) and endocytic proteins for Nedd4-1 (1, 18, 25, 42).
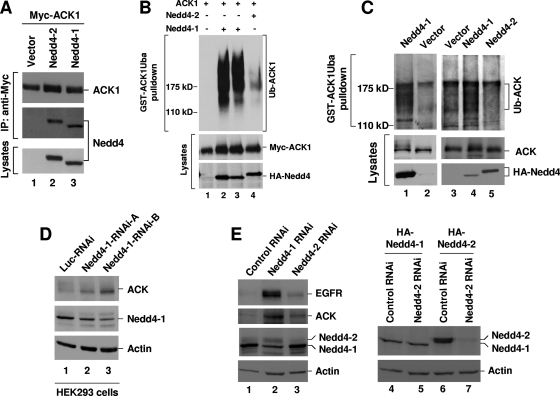
To distinguish the specificity of Nedd4-1 and Nedd4-2 in ubiquitination of ACK, we first examined the binding of Nedd4-1 and Nedd4-2 to ACK1. HA-tagged Nedd4-1 or Nedd4-2 was cotransfected with Myc-ACK1 into HEK293 cells. ACK1 was immunoprecipitated with anti-Myc antibody, and coimmunoprecipitated Nedd4-1 or Nedd4-2 was detected by immunoblotting with anti-HA antibody. As shown in Fig. 5A, both Nedd4-1 and Nedd4-2 were coprecipitated with ACK1 (lanes 2 and 3, middle panel).
FIG. 5.
Nedd4-1 is the E3 ubiquitin ligase for ACK ubiquitination. (A and B) HA-tagged human Nedd4-1, Nedd4-2, or vector was coexpressed with Myc-ACK1 in HEK293 cells. In panel A, Myc-ACK1 was immunoprecipitated with anti-Myc antibody (top panel). Coprecipitated Nedd4 was detected by immunoblotting with anti-HA antibody (middle panel). The expression level of Nedd4 was determined by immunoblotting the cell lysates with anti-HA antibody (bottom panel). In panel B, ubiquitinated ACK1 was precipitated by GST-ACK1Uba pulldown and detected by immunoblotting with anti-ACK (A11) (top panel). The expression level of ACK1 or Nedd4 is shown in the two bottom panels. Lane 2, 2 μg Nedd4-1 for transfection; lane 3, 1 μg Nedd4-1; lane 4, 2 μg Nedd4-2. (C) HA-tagged Nedd4-1, Nedd4-2, or the vector was transfected into HEK293 cells for 36 h. The cells were treated with MG-132 for 12 h to accumulate ubiquitinated ACK. The cells were lysed, and the endogenous ubiquitinated ACK was precipitated by GST-ACK1Uba pulldown and detected by immunoblotting with anti-ACK (A11) (top panels). The expression level of endogenous ACK or HA-tagged Nedd4-1 and Nedd4-2 is shown in the middle and bottom panels. (D) Nedd4-1 RNAi-A, Nedd4-1 RNAi-B, or luciferase RNAi (control) was transfected into HEK293 cells for 48 h. (E) Nedd4-1 RNAi-B, Nedd4-2 RNAi, or luciferase RNAi (control) was transfected into A549 cells (lanes 1 to 3) for 72 h or cotransfected with HA-Nedd4-1 or Nedd4-2 into HEK293 cells for 48 h (lanes 4 to 7). For both panels D and E, the cells were lysed and expression of ACK and Nedd4-1 was detected by immunoblotting with anti-ACK (A11) or anti-Nedd4-1 antibodies. The actin amounts shown in the bottom panels indicate the lysate loading.
Although both Nedd4-1 and Nedd4-2 bind to ACK, a significant difference in ubiquitination of ACK1 was observed between Nedd4-1 and Nedd4-2. Using a GST-ACK-Uba pulldown assay, we found that coexpression with Nedd4-1 resulted in ubiquitination of ACK1 that was more than 10-fold higher than coexpression with Nedd4-2 (Fig. 5B, lanes 2 to 4). A difference in ubiquitination of endogenous ACK between Nedd4-1 and Nedd4-2 was also observed (Fig. 5C). Upon overexpression of Nedd4-1 or Nedd4-2 followed by treatment with the proteasomal inhibitor MG-132 for accumulation of ubiquitinated ACK, we detected that overexpression of Nedd4-1 significantly enhanced ubiquitination of endogenous ACK (Fig. 5C, lanes 1 to 4). However, overexpression of Nedd4-2 did not yield any increase in ubiquitination of ACK; rather, it reduced ubiquitination of ACK, which might result from competing with endogenous Nedd4-1 (compare lane 5 with lane 3). We sometimes observed a significant gel shift of endogenous ACK in SDS-PAGE with expression of Nedd4-1 (Fig. 5C, compare lane 1 with lane 2 in middle panel). These data suggest that Nedd4-1, not Nedd4-2, is the E3 ubiquitin ligase for ubiquitination of ACK in cells.
To confirm that Nedd4-1 is the E3 ubiquitin ligase for ACK, we also performed Nedd4-1 RNAi knockdown experiments. We transfected the Nedd4-1 RNAi oligonucleotides into either HEK293 cells or human non-small cell lung cancer (NSCLC) A549 cells for 48 to 72 h and examined the knockdown effect of Nedd4-1 on ACK protein level by immunoblotting of the cell lysates (Fig. 5D and E). The Nedd4-1 protein level was reduced more than 50% in both HEK293 and A549 cells by Nedd4-1 RNAi knockdown over that of the control RNAi (Fig. 5D, lanes 1 to 3 in the middle panel, and E, lanes 1 and 2 in the third panel). Knockdown of Nedd4-1 dramatically increased ACK protein levels in both cell lines compared to those of the controls (Fig. 5D and E, top panel). A significant increase of EGFR expression by Nedd4-1 knockdown was also observed in A549 cells (Fig. 5E, top panel, lane 2).
We further examined the effect of Nedd4-2 knockdown on the ACK expression level in A549 cells. We could not find a suitable commercial anti-Nedd4-2 antibody for detection of endogenous Nedd4-2 in A549 cells. However, the anti-Nedd4-1 antibody (Upstate Biotech) used in our studies was raised against an antigen region in Nedd4-1 that is partially conserved in Nedd4-2. We tested the anti-Nedd4-1 in detection of exogenously expressed HA-tagged Nedd4-2 (HA-Nedd4-2) and estimated the difference in detection sensitivity between HA-Nedd4-1 and HA-Nedd4-2 by comparison with anti-HA antibody. We found that the anti-Nedd4-1 was about 10-fold less sensitive in detection of HA-Nedd4-2 than detection of HA-Nedd4-1 (data not shown). As expected, we detected endogenous Nedd4-2 expression with the anti-Nedd4-1 in A549 cells (Fig. 5E, third panel, lanes 1 to 3) and estimated that expression of Nedd4-2 is approximately 2-fold higher than that of Nedd4-1 in A549 cells after rectifying the detection sensitivity. We observed that more than 80% of endogenous Nedd4-2 was diminished by Nedd4-2 RNAi knockdown (Fig. 5E, lane 3). To confirm this knockdown, we examined the effect of Nedd4-2 RNAi oligonucleotide on exogenous expression of HA-Nedd4-2 in HEK293 cells and demonstrated that the Nedd4-2 RNAi depleted more than 80% of exogenously expressed Nedd4-2 (Fig. 5E, lane 7), which is consistent with the effect on knockdown of endogenous Nedd4-2. By knockdown of Nedd4-2, expression level of both ACK and EGFR was only slightly enhanced (Fig. 5E, two top panels, lanes 2 and 3), probably due to more Nedd4-1 occupied by dual substrates upon the depletion of Nedd4-2. In summary, the RNAi knockdown data confirm that Nedd4-1, not Nedd4-2, is the E3 ubiquitin ligase regulating ACK and EGFR degradation in vivo.
The effects of the SAM and the Uba domains on the ubiquitination of ACK.
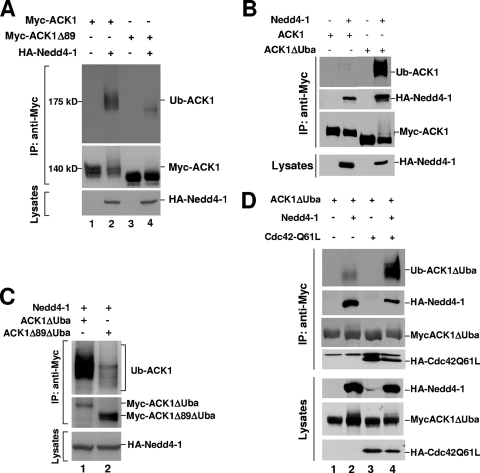
ACK contains a sterile alpha motif (SAM) domain at the N terminus and an ubiquitin association (Uba) domain at the carboxyl terminus (11, 32). Deletion of the SAM domain by truncating the first 89 amino acid residues (ACK1Δ89) resulted in a dramatic reduction of ubiquitination of ACK1 when coexpressed with Nedd4-1 (Fig. 6A), suggesting that the SAM domain is required for Nedd4-1-catalyzed ubiquitination of ACK1. How deletion of the SAM domain eliminates the ubiquitination is not known. Our speculation is that the SAM domain may contain major ubiquitination sites of ACK, as the SAM domain has a lysine-rich region (46). On the other hand, deletion of the Uba domain (ACK1ΔUba) remarkably enhanced ubiquitination (compare lane 4 with lane 2 in the top panel of Fig. 6B). This enhancement of ubiquitination seems to result from increased binding to Nedd4-1, since substantially more Nedd4-1 was coimmunoprecipitated with ACK1ΔUba than with ACK1 (Fig. 6B, compare lane 4 with lane 2 in the second panel from the top).
FIG. 6.
The effects of the SAM and the Uba domains on the ubiquitination of ACK. (A) Myc-tagged ACK1 or ACK1Δ89 was cotransfected with HA-tagged Nedd4-1 or the vector into HEK293 cells. Myc-tagged ACK1 or ACK1Δ89 was immunoprecipitated and immunoblotted with anti-Myc antibody (middle panel), and the ubiquitination of ACK was detected by immunoblotting with antiubiquitin antibody (top panel). The expression level of Nedd4-1 was determined by immunoblotting with anti-HA (bottom panel). (B and C) Myc-tagged ACK1, ACK1ΔUba, or ACK1Δ89ΔUba was cotransfected with HA-tagged Nedd4-1 into HEK293 cells and immunoprecipitated with anti-Myc. The ubiquitination of ACK1 was determined by immunoblotting with antiubiquitin antibody (top panel). Immunoprecipitated ACK1 and its mutants were detected by immunoblotting with anti-Myc. The expression level of HA-Nedd4-1 in cells was determined by immunoblotting the cell lysates with anti-HA (bottom panel). In panel B, coimmunoprecipitated Nedd4-1 was detected by immunoblotting with anti-HA (the second top panel). (D) Myc-ACK1ΔUba was cotransfected with HA-Nedd4-1 and/or HA-Cdc42Q61L into HEK293 cells and immunoprecipitated with anti-Myc antibody. Immunoprecipitated ACK (the third panel from top), coimmunoprecipitated Nedd4-1 (the second panel from top), and coimmunoprecipitated Cdc42Q61L (the fourth panel from top) were detected by immunoblotting with anti-Myc or anti-HA. The expression levels of ACK1ΔUba, Nedd4-1, and Cdc42Q61L in cells are shown in the three bottom panels.
To test whether the enhanced ubiquitination caused by deletion of the Uba domain occurs in the lysine-rich region of the SAM domain, we compared the ubiquitination catalyzed by Nedd4-1 between ACK1 ΔUba and ACK1Δ89ΔUba. As shown in Fig. 6C, deletion of the SAM domain in ACK1ΔUba significantly reduced the ubiquitination catalyzed by Nedd4-1 (lane 2), supporting the hypothesis that the lysine-rich region in the SAM domain may contain the majority of the ubiquitination sites of ACK.
Interestingly, when Cdc42 Q61L, a GTPase-defective mutant of Cdc42, was cotransfected with ACK1ΔUba and Nedd4-1, the ubiquitination of ACK was further enhanced (Fig. 6D, compare lane 4 with lane 2 in the top panel). This is consistent with the effect of Cdc42 on EGF-stimulated ubiquitination of ACK (Fig. 2C). Cdc42 is a member of the Rho family GTPase and an activator of ACK. These data suggest that activated ACK may be a preferential ubiquitination substrate for Nedd4-1.
EGF-induced degradation of ACK is processed by lysosomes, not proteasomes.
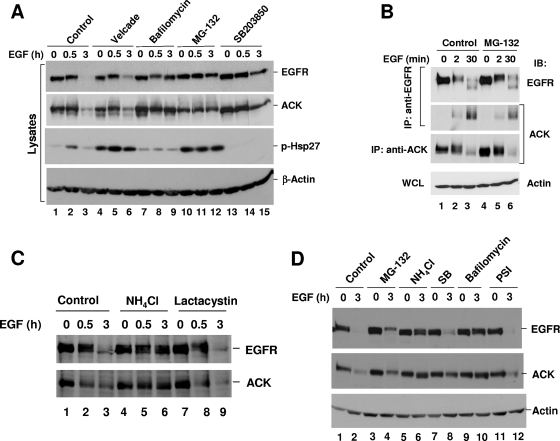
Ubiquitination-mediated protein degradation can occur via proteasomes or lysosomes. It has been demonstrated that ubiquitination-mediated EGFR degradation is mediated by the lysosomal pathway (21). To determine the mechanism of EGF-induced ACK degradation, we used inhibitors of proteasomal (MG-132, PSI, Velcade [bortezomib], lactacystin) or lysosomal (bafilomycin and NH4Cl) pathways to assess the degradation route for ACK in COS7, HeLa, and A549 cells. As shown in Fig. 7A, in COS7 cells, treatment with bafilomycin (lanes 7 to 9) or MG-132 (lanes 10 to 12) significantly inhibited EGF-induced degradation of both EGFR and ACK (top and middle panels). However, MG-132 is known to inhibit calpain in addition to proteasomes, and thus it is not a specific proteasomal inhibitor (6, 39). The specific proteasome inhibitor Velcade (bortezomib) only slightly blocked the degradation of EGFR and ACK (Fig. 7A, top and middle panels, compare lane 6 with lane 3). Both MG-132 and Velcade activated p38 kinase (phosphorylation of Hsp27) to the same extent (Fig. 7A, second panel from the bottom, lanes 4 to 6 and 10 to 12), suggesting that both effectively inhibited proteasomes. Therefore, inhibition of EGFR and ACK degradation by MG-132 in COS7 cells might result from a nonproteasomal inhibitory activity.
FIG. 7.
EGF-induced degradation of ACK is mediated by lysosomes. COS7 (A), HeLa (B and C) or A549 (D) cells were cultured to 90% confluence, followed by serum starvation (0.1% FBS for HeLa cells, serum-free for COS7 and A549 cells) for 12 h. The proteasomal, lysosomal, or p38 inhibitor (10 μM Velcade [bortezomib], 1 μM bafilomycin, 10 μM MG-132, 10 μM SB203850, 20 mM NH4Cl, 10 μM lactacystin, or 10 μM PSI) was added to the culture medium 30 min before EGF stimulation. After EGF (50 ng/ml) stimulation for the indicated time, the cells were lysed. EGF-induced degradation of EGFR or ACK1 was determined either by directly immunoblotting the cell lysates with anti-EGFR (1005) or anti-ACK (A11) (top two panels in panels A and D) or by immunoprecipitation of EGFR followed by immunoblotting with anti-EGFR (1005) and combined with immunoblotting the cell lysates with anti-ACK (A11) (top and second bottom panel in panel B, top and bottom panels in panel C). Coimmunoprecipitated ACK1 with EGFR was detected by immunoblotting with anti-ACK (A11) antibody (second top panel in panel B). Detection of phosphorylation of Hsp27 was performed by immunoblotting with anti-phospho-Hsp27 to indicate activation of p38 kinase by proteasomal inhibition treatments in COS7 cells (second bottom panel in panel A). The actin amount was used to show the cell lysate loading (bottom panels in panels A, B, and D). WCL, whole-cell lysate.
As shown in Fig. 7B, MG-132 treatment did not significantly affect EGF-induced degradation of either EGFR or ACK (top and bottom panels) or the binding of ACK to EGFR (middle panel) in HeLa cells. Similar results were observed with the proteasomal inhibitor lactacystin (Fig. 7C, top and bottom panels, compare lane 9 with lane 3). Treatment with NH4Cl, an inhibitor of lysosomes, significantly blocked EGF-induced degradation of EGFR and ACK (Fig. 7C, top and bottom panels, lane 6). These data demonstrate that EGF-induced degradation of ACK is processed by lysosomes.
Similar results were observed in A549 cells. The proteasomal inhibitors MG-132 and PSI did not inhibit degradation of EGFR or ACK (Fig. 7D, two top panels, lanes 3 and 4 and lanes 11 and 12). The lysosomal inhibitors, NH4Cl and bafilomycin, blocked 70 to 90% of the EGF-induced degradation of EGFR and ACK (Fig. 7D, two top panels, lanes 5 and 6 and lanes 9 and 10).
It has been reported that p38 kinase activity was required for ligand-induced EGFR degradation via regulation of tyrosine phosphorylation of Y1045 of EGFR, which is the binding site for Cbl, the E3 ubiquitin ligase for EGFR (10). The p38 kinase inhibitor SB203850 blocked EGF-induced degradation of EGFR (Fig. 7A, top panel, compare lanes 13 to 15 with lanes 1 to 3) and ACK (Fig. 7A, second panel from top, compare lanes 13 to 15 with lanes 1 to 3). This result suggests that EGF-induced ACK degradation is dependent on EGFR degradation, likely via cotransporting with EGFR to lysosomes.
Nedd4-1, not Nedd4-2, is required for EGF-induced ACK and EGFR degradation.
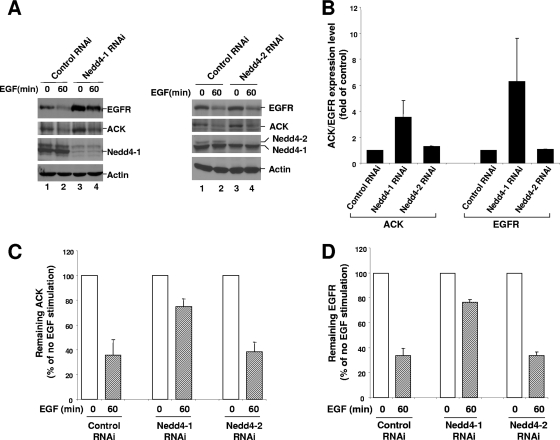
To address the role of Nedd4-1 and Nedd4-1-catalyzed ubiquitination of ACK in regulation of EGF-induced degradation of EGFR and ACK, we compared the degradation of EGFR and ACK in Nedd4-1 knockdown cells with that in Nedd4-2 knockdown and control cells (Fig. 8). Knockdown of Nedd4-1 significantly increased both EGFR and ACK expression levels (Fig. 8A, two top panels, lanes 1 and 3), while knockdown of Nedd4-2 caused little change in expression levels of ACK and EGFR (Fig. 8A, two top panels, lanes 5 and 7). Quantification of the data from three independent experiments shows that knockdown of 70 to 80% of Nedd4-1 caused a 2- to 5-fold increase in ACK1 expression and a 3- to 10-fold increase in EGFR expression in A549 cells, whereas knockdown of more than 80% of Nedd4-2 had little effect on expression of both ACK and EGFR (Fig. 8B). EGF-induced degradation of ACK and EGFR was significantly inhibited by Nedd4-1 knockdown. While about 65% of EGFR and ACK was degraded upon 60 min EGF stimulation in control RNAi-transfected cells, only about 20% of EGFR or ACK was degraded upon 60 min EGF stimulation in Nedd4-1 knockdown cells (Fig. 8C and D). Thus, Nedd4-1 knockdown yielded inhibition of about 70% of the degradation of both EGFR and ACK. Again, knockdown of Nedd4-2 caused little effect on EGF-induced degradation of ACK and EGFR (Fig. 8C and D). These data indicate that Nedd4-1, not Nedd4-2, plays an important role in regulation of EGF-induced degradation of EGFR and ACK.
FIG. 8.
Knockdown of Nedd4-1 by RNAi enhances the expression level of EGFR and ACK and inhibits EGF-induced degradation of EGFR and ACK. (A) Nedd4-1 RNAi-B, Nedd4-2 RNAi, or the luciferase RNAi (control RNAi) was transfected into A549 cells for 60 h, followed by serum starvation for 12 h. The cells were stimulated with EGF for 60 min. The amounts of EGFR, ACK1, Nedd4-1, and Nedd4-2 were determined by immunoblotting with anti-EGFR (1005) (top panel), anti-ACK (A11) (the second top panel), and anti-Nedd4-1 (the second bottom panel). The amount of actin was used as an indication of the lysate loading (bottom panel). (B to D) Expression of EGFR and ACK1 and degradation of EGFR and ACK1 induced by 60 min EGF (50 ng/ml) treatment in A549 cells with or without knockdown of Nedd4-1 or Nedd4-2 by RNAi were determined by immunoblotting and quantified by the Gel Logic 100 Image system (Kodak) from three independent experiments.
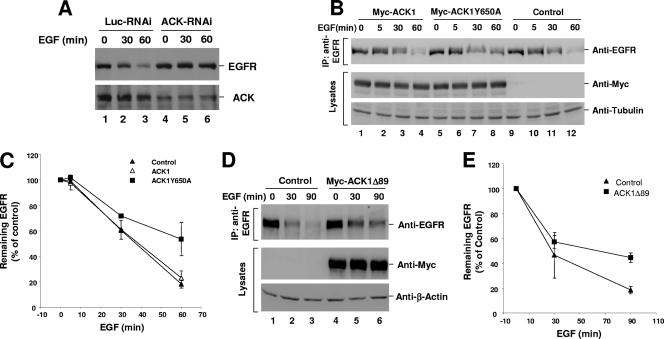
We further assessed the role of Nedd4-1-catalyzed ubiquitination of ACK in regulation of EGFR degradation. We first confirmed the role of endogenous ACK in regulation of EGFR degradation by depletion of ACK with RNAi. As shown in Fig. 9A, knockdown of about 70% of ACK1 in A549 cells inhibited EGF-induced degradation of EGFR (top panel, lanes 5 to 6). We next examined the effects of overexpression of the Nedd4-1-binding-defective mutant ACK1Y650A and the ubiquitination-defective mutant ACK1Δ89 on ligand-induced EGFR degradation in HEK293 cells. With overexpression of the Nedd4-1-binding-defective mutant ACK1Y650A, EGFR degradation upon 60 min EGF stimulation was reduced to 45% from 80% in wild-type ACK1-overexpressed or vector-transfected cells (Fig. 9B and C), indicating that about 45% of EGF-induced degradation of EGFR was inhibited by overexpression of ACK1Y650A. Similar inhibition of ligand-induced EGFR degradation was observed with overexpression of the ubiquitination-defective mutant ACK1Δ89 in HEK293 cells (Fig. 9D and E). Overexpression of ACK1Δ89 caused inhibition of EGF-induced degradation by about 40% compared with that in the vector-transfected cells (Fig. 9E). These data suggest that Nedd4-1-catalyzed ubiquitination of ACK is required for ACK in the regulation of ligand-induced degradation of EGFR.
FIG. 9.
Overexpression of the Nedd4-1-binding-defective mutant ACK1Y650A and the ubiquitination-defective mutant ACK1Δ89 inhibits EGF-induced degradation of EGFR. (A) ACK1 RNAi or luciferase RNAi (control RNAi) was transfected into A549 cells for 60 h followed by 12 h serum starvation. The cells were stimulated with EGF (50 ng/ml) for 0, 30, and 60 min. The amounts of EGFR and ACK1 were detected by immunoblotting with anti-EGFR (1005) and anti-ACK (A11). (B to E) pcDNA3 (vector control), pcDNA3-Myc-ACK1, pcDNA3-Myc-ACK1Y650A, or pcDNA3-Myc-ACK1Δ89 was transfected into HEK293 cells for 36 h followed by 12 h serum starvation. The cells were stimulated with EGF (50 ng/ml) for 0, 5, 30, and 60 min (B and C) or 0, 30, and 90 min (D and E). EGFR was immunoprecipitated with anti-EGFR (Mab528) and immunoblotted with anti-EGFR (1005). The amount of tubulin (B) or actin (D) was used to indicate lysate loading. In panels C and E, the amount of EGFR determined by immunoblotting was quantified from two independent experiments by FUJIFILM Multi Gauge V3.0.
DISCUSSION
Our previous studies have shown that ACK interacts with EGFR in response to EGF stimulation and regulates EGF-induced EGFR degradation (32). We also observed that the Uba domain is required for ACK to facilitate EGFR degradation, suggesting that ACK might be involved in ubiquitination-mediated EGFR degradation via Uba-ubiquitin interaction (32). In this report, we found that EGF signaling induced ACK degradation along with EGFR degradation. This degradation is mediated by the E3 ubiquitin ligase Nedd4-1 and processed by lysosomes. Interestingly, the degradation of ACK induced by EGF requires EGFR kinase activity, since EGFR kinase inhibitor AG1478 completely blocked the degradation (Fig. 1). Because EGFR tyrosine kinase activity is also required for ACK to bind to EGFR (32), association of ACK with activated EGFR might be necessary for EGF-induced degradation of ACK. Furthermore, EGF-induced degradation of ACK is tightly correlated with EGF-induced degradation of EGFR (Fig. 8), suggesting that ACK might be cotransported with EGFR to lysosomes for degradation. This cotransportation may be regulated by Nedd4-1-mediated ubiquitination of ACK, which may serve as a sorting signal for transporting the EGFR/ACK complex from early endosomes to late endosomes/lysosomes. In fact, knockdown of Nedd4-1 by RNAi and overexpression of the Nedd4-1-binding-defective and ubiquitination-defective mutants of ACK inhibited EGFR degradation, suggesting that Nedd4-1 and Nedd4-1-catalyzed ubiquitination of ACK are required for EGF-induced degradation of EGFR.
We observed a specificity of ACK ubiquitination, which was preferentially catalyzed by Nedd4-1, not Nedd4-2 (Fig. 5). Nedd4-2 is known to ubiquitinate ion channels, transporters, and other membrane proteins (8, 33, 34, 40, 49), while Nedd4-1 ubiquitinates endocytic proteins (1, 18, 25, 42), most of which are nonmembrane proteins. For example, previous studies observed that Nedd4-1 ubiquitinated several proteins that play critical roles in EGFR endocytosis and degradation, such as Cbl and Eps15 (25, 42). It is not known how substrate specificity is determined. The WW domains are highly conserved between Nedd4-1 and Nedd4-2. The unconserved regions are located mainly between WW1 and WW3 domains. We observed an insignificant difference between Nedd4-1 and Nedd4-2 in binding to ACK (Fig. 5). However, the difference in ubiquitination of ACK between Nedd4-1 and Nedd4-2 was more than 10-fold (Fig. 5). We speculate that there may be additional Nedd4-binding proteins interacting with the unconserved region between WW1 and WW3 that help Nedd4 to establish the specificity in recognizing substrates. During preparation of the manuscript, Chan et al. reported that ACK1 is ubiquitinated by E3 ubiquitin ligase Nedd4-2 (4), which is not consistent with our findings. We speculate that the discrepancy between their conclusion and ours may result from differences in experimental system and approaches. They assayed the ubiquitination of ACK1 by Nedd4-2 and the colocalization of ACK1 with Nedd4-2 mainly with exogenously expressed proteins. We also observed ubiquitination of ACK1 by overexpression of Nedd4-2 in HEK293 cells (Fig. 5B), although the ubiquitination was much less efficient than that by Nedd4-1.
Our data in Fig. 8 and 9 suggest an important role of Nedd4-1 but not Nedd4-2 in regulation of EGF-induced EGFR and ACK degradation. It has been proposed that ubiquitination of membrane proteins by Nedd4 generates a sorting signal for transporting these proteins to lysosomes for degradation (19, 29). The ubiquitination in these proteins may function as a protein-protein interactive module for the assembly of transporting complexes. ACK is ubiquitinated by Nedd4-1 (Fig. 5). Although treatment of MG-132, a proteasomal inhibitor, caused accumulation of ACK ubiquitination, it did not significantly affect the protein amount of ACK (Fig. 5C), indicating that inhibition of proteasomes did not affect the protein degradation of ACK. However, Nedd4-1 is required for both EGFR and ACK degradation, because knockdown of Nedd4-1 by RNAi significantly enhanced the protein expression level of EGFR and ACK and suppressed EGF-induced degradation of EGFR and ACK (Fig. 8). Thus, ubiquitination of ACK by Nedd4-1 may function as a sorting signal for transporting the EGFR/ACK complex to lysosomes for degradation. This hypothesis explains why degradation of ACK always accompanies degradation of EGFR (Fig. 7) and requires binding to EGFR (Fig. 1). Because the optimal interaction of ACK with EGFR occurs at the late stage of EGFR internalization (30 to 60 min after EGF stimulation) (32), we speculate that the EGFR/ACK complex formation would take place on endosomes and the ubiquitination of ACK by Nedd4-1 might be critical for sorting the complex to late endosomes or lysosomes. However, whether Nedd4-1-mediated ubiquitination of ACK is required for EGF-induced EGFR and ACK degradation needs to be further determined by examining the dominant negative effects of the ubiquitination site mutants of ACK and the ligase-dead mutant of Nedd4-1 on EGFR and ACK degradation.
In COS7 cells, MG-132 treatment inhibited both EGF-induced EGFR and ACK degradation (Fig. 7A). However, the specific proteasome inhibitor Velcade did not have such an effect (Fig. 7A), indicating that the inhibitory effect of MG-132 was through a nonproteasomal mechanism. An inhibitory effect on EGFR degradation by MG-132 was also observed in hepatoma Hep2 cells (24). It was proposed that inhibition of cathepsin B, a lysosomal protease, by MG-132 might block degradation of EGFR (2). We observed little effect of MG-132 on inhibition of EGFR degradation in HeLa and A549 cells (Fig. 7), arguing that inhibition of the lysosomal protease cathepsin B may not be the cause for MG-132 in blocking the degradation of EGFR in COS7 cells. MG-132 is also a known inhibitor of calpain, a calcium-activated protease (39). We speculate that inhibition of calpain in COS7 cells might contribute to the inhibitory effect by impairing an unidentified process in EGFR trafficking that is unique in COS7 or Hep2 cells.
Our data suggest that the SAM domain may contain the majority of the ubiquitination sites in ACK (Fig. 6A and C). The SAM domain is found in many proteins and functions as a protein-binding module for formation of homo- or heterooligomers (38). The SAM domain of ACK may also play a role in homodimerization or interaction with other SAM domain proteins, and the ubiquitination of the SAM domain might disrupt the homodimerization or the interaction with other proteins. We observed that deletion of the Uba domain significantly enhanced ubiquitination of ACK, probably by increasing binding to Nedd4-1 (Fig. 6B). The Uba domain may interact with the ubiquitinated SAM domain to change the conformation of ACK, resulting in reduction of the binding affinity of ACK to Nedd4-1, thus dissociating Nedd4-1 and preventing ACK from excessive ubiquitination. Such regulation may be necessary to control overubiquitination, which may interfere with the structure and function of the kinase or other domains of ACK. Interestingly, coexpression of active Cdc42 enhanced the ubiquitination of ACK even though the binding of ACK to Nedd4-1 was decreased (Fig. 6D), implying that the ACK/Cdc42 complex may activate Nedd4-1 E3 ligase activity.
Overexpression of ACK is correlated with aggressiveness of tumors (41). Amplification of the ACK gene was found in primary prostate tumor samples from patients with recurrent tumors by analysis of chromosomal alterations (41). In prostate cancer, ACK overexpression is associated with metastasis and androgen independency of the tumors (41). It has been proposed that ACK promotes tumor cell metastasis through the integrin-mediated signaling pathway and possibly by interaction with and phosphorylation of the p130Cas complex (41). Other mechanisms were also proposed in different studies for the role of ACK in prostate tumorigenesis (26, 27). It was observed that the activated ACK phosphorylated WWOX, a tumor suppressor protein, and androgen receptor in prostate cancer cells (26, 27). The phosphorylation promoted ubiquitination and degradation of WWOX (26) and activated androgen receptor (27). However, connection of the role of ACK in regulation of EGFR signaling to tumor progression has not been established. EGFR signaling is known to initiate tumorigenesis and promote tumor progression. Deregulation of interaction of ACK with EGFR and ubiquitination of ACK by Nedd4-1 might result in deregulation of EGFR signaling. In addition, the tumor suppressor Mig-6 (also known as Gene-33 or RALT) has the same EGFR/ErbB2-interactive domain as ACK that directly binds to the kinase domain of EGFR/ErbB2 (13, 48). Cross talk between ACK and Mig-6 to modulate EGFR signaling might also be an important avenue to regulate tumorigenesis and tumor progression.
Acknowledgments
We thank Daniela Rotin at University of Toronto for kindly sending us the rat Nedd4-1 cDNA plasmid, which was used in our initial research, and Mark T. Bedford at M. D. Anderson Cancer Center for confirming the interaction of the ACK-PPXY motif with the WW domain by GST-WW domain dot blotting.
This work was supported by a grant to W.Y. from the American Cancer Society (ACS-RSG TBE-110602).
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Aoh, Q. L., A. M. Castle, C. H. Hubbard, O. Katsumata, and J. D. Castle. 2009. SCAMP3 negatively regulates EGFR degradation and promotes receptor recycling. Mol. Biol. Cell 20:1816-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Authier, F., M. Métioui, A. W. Bell, and J. S. Mort. 1999. Negative regulation of epidermal growth factor signaling by selective proteolytic mechanisms in the endosome mediated by cathepsin B. J. Biol. Chem. 274:33723-33731. [DOI] [PubMed] [Google Scholar]
- 3.Bourdoulous, S., G. Orend, D. A. MacKenna, R. Pasqualini, and E. Ruoslahti. 1998. Fibronectin matrix regulates activation of RHO and CDC42 GTPases and cell cycle progression. J. Cell Biol. 143:267-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, W., R. Tian, Y. F. Lee, S. T. Sit, L. Lim, and E. Manser. 2009. Down-regulation of active ACK1 is mediated by association with the E3 ubiquitin ligase Nedd4-2. J. Biol. Chem. 284:8185-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H., C. A. Ross, N. Wang, Y. Huo, D. F. MacKinnon, J. B. Potash, S. G. Simpson, F. J. McMahon, J. R. DePaulo, Jr., and M. G. McInnis. 2001. NEDD4L on human chromosome 18q21 has multiple forms of transcripts and is a homologue of the mouse Nedd4-2 gene. Eur. J. Hum. Genet. 9:922-930. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, L. J., B. Walker, H. Ovaa, D. Chauhan, K. C. Anderson, T. C. Morris, and A. E. Irvine. 2006. Comparative selectivity and specificity of the proteasome inhibitors BzLLLCOCHO, PS-341, and MG-132. Cancer Res. 66:6379-6386. [DOI] [PubMed] [Google Scholar]
- 7.Eisenmann, K. M., J. B. McCarthy, M. A. Simpson, P. J. Keely, J. L. Guan, K. Tachibana, L. Lim, E. Manser, L. T. Furcht, and J. Iida. 1999. Melanoma chondroitin sulphate proteoglycan regulates cell spreading through Cdc42, Ack-1 and p130cas. Nat. Cell Biol. 1:507-513. [DOI] [PubMed] [Google Scholar]
- 8.Ekberg, J., F. Schuetz, N. A. Boase, S. J. Conroy, J. Manning, S. Kumar, P. Poronnik, and D. J. Adams. 2007. Regulation of the voltage-gated K(+) channels KCNQ2/3 and KCNQ3/5 by ubiquitination. Novel role for Nedd4-2. J. Biol. Chem. 282:12135-12142. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Frey, M. R., R. S. Dise, K. L. Edelblum, and D. B. Polk. 2006. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 25:5683-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galisteo, M. L., Y. Yang, J. Urena, and J. Schlessinger. 2006. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc. Natl. Acad. Sci. U. S. A. 103:9796-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulet, C. C., K. A. Volk, C. M. Adams, L. S. Prince, J. B. Stokes, and P. M. Snyder. 1998. Inhibition of the epithelial Na+ channel by interaction of Nedd4 with a PY motif deleted in Liddle's syndrome. J. Biol. Chem. 273:30012-30017. [DOI] [PubMed] [Google Scholar]
- 13.Hackel, P. O., M. Gishizky, and A. Ullrich. 2001. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol. Chem. 382:1649-1662. [DOI] [PubMed] [Google Scholar]
- 14.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 15.Hopper, N. A., J. Lee, and P. W. Sternberg. 2000. ARK-1 inhibits EGFR signaling in C. elegans. Mol. Cell 6:65-75. [PubMed] [Google Scholar]
- 16.Huang, F., D. Kirkpatrick, X. Jiang, S. Gygi, and A. Sorkin. 2006. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell 21:737-748. [DOI] [PubMed] [Google Scholar]
- 17.Ingham, R. J., G. Gish, and T. Pawson. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene 23:1972-1984. [DOI] [PubMed] [Google Scholar]
- 18.Katz, M., K. Shtiegman, P. Tal-Or, L. Yakir, Y. Mosesson, D. Harari, Y. Machluf, H. Asao, T. Jovin, K. Sugamura, and Y. Yarden. 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3:740-751. [DOI] [PubMed] [Google Scholar]
- 19.Leithe, E., and E. Rivedal. 2007. Ubiquitination of gap junction proteins. J. Membr. Biol. 217:43-51. [DOI] [PubMed] [Google Scholar]
- 20.Levkowitz, G., H. Waterman, E. Zamir, Z. Kam, S. Oved, W. Y. Langdon, L. Beguinot, B. Geiger, and Y. Yarden. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12:3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, S. Lipkowitz, and Y. Yarden. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 4:1029-1040. [DOI] [PubMed] [Google Scholar]
- 22.Lin, Q., C. G. Lo, R. A. Cerione, and W. Yang. 2002. The Cdc42-target ACK2 interacts with SH3PX1 (sorting nexin 9) to regulate EGFR degradation. J. Biol. Chem. 277:10134-10138. [DOI] [PubMed] [Google Scholar]
- 23.Linseman, D. A., K. A. Heidenreich, and S. K. Fisher. 2001. Stimulation of M3 muscarinic receptors induces phosphorylation of the Cdc42 effector activated Cdc42Hs-associated kinase-1 via a Fyn tyrosine kinase signaling pathway. J. Biol. Chem. 276:5622-5628. [DOI] [PubMed] [Google Scholar]
- 24.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnifico, A., S. Ettenberg, C. Yang, J. Mariano, S. Tiwari, S. Fang, S. Lipkowitz, and A. M. Weissman. 2003. WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J. Biol. Chem. 278:43169-43177. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan, N. P., Y. E. Whang, J. L. Mohler, and H. S. Earp. 2005. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 65:10514-10523. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan, N. P., Y. Liu, S. Majumder, M. R. Warren, C. E. Parker, J. L. Mohler, H. S. Earp, and Y. E. Whang. 2007. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 104:8438-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmor, M. D., and Y. Yarden. 2004. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23:2057-2070. [DOI] [PubMed] [Google Scholar]
- 29.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 30.Satoh, T., J. Kato, K. Nishida, and Y. Kaziro. 1996. Tyrosine phosphorylation of ACK in response to temperature shift-down, hyperosmotic shock, and epidermal growth factor stimulation. FEBS Lett. 386:230-234. [DOI] [PubMed] [Google Scholar]
- 31.Sem, K. P., B. Zahedi, I. Tan, M. Deak, L. Lim, and N. Harden. 2002. ACK family tyrosine kinase activity is a component of Dcdc42 signaling during dorsal closure in Drosophila melanogaster. Mol. Cell. Biol. 22:3685-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, F., Q. Lin, Y. Gu, C. Childress, and W. Yang. 2007. Activated Cdc42-associated kinase 1 (ACK1) is a component of EGF receptor signaling complex and regulates EGF receptor degradation. Mol. Biol. Cell 18:732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder, P. M., J. C. Steines, and D. R. Olson. 2004. Relative contribution of Nedd4 and Nedd4-2 to ENaC regulation in epithelia determined by RNA interference. J. Biol. Chem. 279:5042-5046. [DOI] [PubMed] [Google Scholar]
- 34.Sorkina, T., M. Miranda, K. R. Dionne, B. R. Hoover, N. R. Zahniser, and A. Sorkin. 2006. RNA interference screen reveals an essential role of Nedd4-2 in dopamine transporter ubiquitination and endocytosis. J. Neurosci. 26:8195-8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371-2380. [PMC free article] [PubMed] [Google Scholar]
- 36.Sudol, M., and T. Hunter. 2000. NeW wrinkles for an old domain. Cell 103:1001-1004. [DOI] [PubMed] [Google Scholar]
- 37.Teo, M., L. Tan, L. Lim, and E. Manser. 2001. The tyrosine kinase ACK1 associates with clathrin-coated vesicles through a binding motif shared by arrestin and other adaptors. J. Biol. Chem. 276:18392-18398. [DOI] [PubMed] [Google Scholar]
- 38.Thanos, C. D., K. E. Goodwill, and J. U. Bowie. 1999. Oligomeric structure of the human EphB2 receptor SAM domain. Science 283:833-836. [DOI] [PubMed] [Google Scholar]
- 39.Tsubuki, S., Y. Saito, M. Tomioka, H. Ito, and S. Kawashima. 1996. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J. Biochem. 119:572-576. [DOI] [PubMed] [Google Scholar]
- 40.van Bemmelen, M. X., J. S. Rougier, B. Gavillet, F. Apotheloz, D. Daidie, M. Tateyama, I. Rivolta, M. A. Thomas, R. S. Kass, O. Staub, and H. Abriel. 2004. Cardiac voltage-gated sodium channel Nav1.5 is regulated by Nedd4-2 mediated ubiquitination. Circ. Res. 95:284-291. [DOI] [PubMed] [Google Scholar]
- 41.van der Horst, E. H., Y. Y. Degenhardt, A. Strelow, A. Slavin, L. Chinn, J. Orf, M. Rong, S. Li, L. H. See, K. Q. Nguyen, T. Hoey, H. Wesche, and S. Powers. 2005. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc. Natl. Acad. Sci. U. S. A. 102:15901-15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woelk, T., B. Oldrini, E. Maspero, S. Confalonieri, E. Cavallaro, P. P. Di Fiore, and S. Polo. 2006. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 8:1246-1254. [DOI] [PubMed] [Google Scholar]
- 43.Xu, D., A. Makkinje, and J. M. Kyriakis. 2005. Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J. Biol. Chem. 280:2924-2933. [DOI] [PubMed] [Google Scholar]
- 44.Yang, W., and R. A. Cerione. 1997. Cloning and characterization of a novel Cdc42-associated tyrosine kinase, ACK2, from bovine brain. J. Biol. Chem. 272:24819-24824. [DOI] [PubMed] [Google Scholar]
- 45.Yang, W., C. G. Lo, T. Despenza, and R. A. Cerione. 2001. ACK2 directly interacts with clathrin and inhibits AP-2 mediated receptor endocytosis. J. Biol. Chem. 276:17468-17473. [DOI] [PubMed] [Google Scholar]
- 46.Yang, W., J. M. Jansen, Q. Lin, S. Canova, R. A. Cerione, and C. Childress. 2004. Interaction of activated Cdc42-associated tyrosine kinase ACK2 with HSP90. Biochem. J. 382:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, W., Q. Lin, J. L. Guan, and R. A. Cerione. 1999. Activation of the Cdc42-associated tyrosine kinase-2 (ACK-2) by cell adhesion via integrin beta1. J. Biol. Chem. 274:8524-8530. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., K. A. Pickin, R. Bose, N. Jura, P. A. Cole, and J. Kuriyan. 2007. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, R., S. V. Patel, and P. M. Snyder. 2007. Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC. J. Biol. Chem. 282:20207-20212. [DOI] [PubMed] [Google Scholar]