Abstract
MicroRNAs (miRNAs) repress gene expression posttranscriptionally by inhibiting translation and by expediting deadenylation so as to trigger rapid mRNA decay. Their regulatory influence is mediated by the protein components of the RNA-induced silencing complex (RISC), which deliver miRNAs and siRNAs to their mRNA targets. Here, we present evidence that CCR4-NOT is the deadenylase that removes poly(A) from messages destabilized by miRNAs in human cells. Overproducing a mutationally inactivated form of either of the catalytic subunits of this deadenylase (CCR4 or CAF1/POP2) significantly impedes the deadenylation and decay of mRNA targeted by a partially complementary miRNA. The same deadenylase initiates the degradation of “off-target” mRNAs that are bound by an imperfectly complementary siRNA introduced by transfection. The greater inhibitory effect of inactive CAF1 or POP2 (versus inactive CCR4) suggests a predominant role for this catalytic subunit of CCR4-NOT in miRNA- or small interfering RNA (siRNA)-mediated deadenylation. These effects of mi/siRNAs and CCR4-NOT can be fully reproduced by directly tethering RISC to mRNA without the guidance of a small RNA, indicating that the ability of RISC to accelerate deadenylation is independent of RNA base pairing. Despite its importance for mi/siRNA-mediated deadenylation, CCR4-NOT appears not to associate significantly with RISC, as judged by the failure of CAF1 and POP2 to coimmunoprecipitate detectably with either the Ago or TNRC6 subunit of RISC, a finding at odds with deadenylase recruitment as the mechanism by which RISC accelerates poly(A) removal.
It is increasingly clear that microRNAs (miRNAs) play a crucial role in eukaryotic gene regulation. In metazoans, these small (∼22-nucleotide) untranslated RNAs base pair with mRNAs to which they are partially complementary, downregulating their expression by a combination of two posttranscriptional mechanisms: translational repression and poly(A) tail removal leading to rapid mRNA degradation (1, 11, 16, 17, 32-34). In the rare instances in which a miRNA is fully complementary to its target, it can also destabilize the mRNA by directing endonucleolytic cleavage at the site where it binds (38, 41).
miRNAs influence gene expression not through an intrinsic activity but rather through the activity of the protein complex that delivers them to the mRNAs they target (8, 25). This RNA-induced silencing complex (RISC) contains multiple components, including at a minimum a specificity-determining miRNAm or small interfering RNA (RNA), an Ago protein to which the small RNA binds, and a TNRC6 protein. In humans, there are four Ago paralogs (Ago1 to Ago4), one of which (Ago2) has an endonuclease activity capable of cleaving the mRNA strand of mRNA-miRNA or mRNA-siRNA duplexes that are fully complementary (18, 22). In addition, there are three TNRC6 paralogs (TNRC6A to TNRC6C; TNRC6A is also known as GW182). When tethered to a reporter mRNA via a heterologous RNA-binding domain, each of the four human Ago proteins can downregulate reporter gene expression in a miRNA-independent manner by repressing the translation and reducing the cellular concentration of the reporter mRNA (27, 35). These effects appear to be mediated by the TNRC6 proteins with which the Ago proteins interact, since tethering TNRC6 protein fragments that are unable to bind Ago has similar consequences (4, 15, 42).
The mechanisms by which RISC represses translation and expedites deadenylation and decay are not clear. For example, whereas some findings suggest that RISC inhibits translation initiation by disrupting cap recognition (13, 21, 28, 30, 39), others indicate that it acts at a subsequent step in translation (19, 23, 24, 26, 29, 31). Similarly, it is not known how RISC accelerates poly(A) removal, which is the first and rate-limiting step of miRNA-mediated mRNA degradation, although the affinity of TNRC6 for poly(A)-binding protein (PABP) appears to be important for that event (9, 40).
Human cells contain three deadenylases (CCR4-NOT, PAN2-PAN3, and PARN). CCR4-NOT is a multimeric complex comprising nine subunits, two of which are catalytically active: CCR4a (CNOT6) or its paralog CCR4b (CNOT6L) and CAF1 (CNOT7) or its paralog POP2 (CNOT8/CALIF) (14). In contrast, PAN2-PAN3 is a heteromultimer containing a single catalytically active subunit (PAN2), and PARN is a homodimer (20, 36, 37). A role for CCR4-NOT in miRNA-mediated deadenylation in Drosophila was deduced from the inhibitory effect of depleting the NOT1 subunit of that deadenylase (2), but the deadenylase stimulated by miRNAs in human cells has not been identified. Nor is it known whether RISC stimulates poly(A) removal by recruiting a deadenylase or by making the autonomous interaction of that deadenylase with poly(A) more productive.
We present evidence here that CCR4-NOT is the deadenylase whose degradative action is stimulated when mRNA base pairs with a partially complementary miRNA or siRNA in human cells. We further show that tethering RISC subunits to mRNA likewise accelerates mRNA decay by promoting poly(A) removal by CCR4-NOT. Despite the ability of RISC to stimulate deadenylation by CCR4-NOT, these two protein complexes appear to have little if any affinity for one another.
MATERIALS AND METHODS
Plasmids.
Plasmids pBG, pBG+L7, pNHA-Ago1, pNHA-Ago2, pNHA-Ago3, pNHA-Ago4, pNHA-Ago2-D597A, pHA-Ago2, and pSVα1-GAPDH have been described previously (6, 34, 35). The reporter plasmids pBG+boxB and pBG+GM were constructed by inserting a DNA fragment encoding either a single bacteriophage lambda boxB element (GCTAGCAACTACTAAACTGGGCCCTGAAGAAGGGCCCATATCGAGGATATCTAGA) or an element (GM) partially complementary to siEGFP (GCTAGCTCTACGGCAAGCTATACCTGAAGTTCAAATTCTAGA), respectively, between the NheI and XbaI sites of plasmid pBG. Plasmid pNHA-TNRC6A was constructed from plasmid pCI-λNV5 (10) by inserting tandem DNA fragments encoding a hemagglutinin (HA) epitope tag (YPYDVPDYA), a hexaglycine linker, and human TNRC6A (GW182) downstream of a sequence encoding a high-affinity variant of the boxB-binding domain of the lambda N protein (MNARTRRRERRAEKQAQWKAAN). Plasmids pV5-CCR4a, pV5-CCR4b, pV5-CAF1, pV5-POP2, pV5-PAN2, pV5-PARN, pV5-CCR4a-mut, pV5-CAF1-mut, pV5-POP2-mut, pV5-PAN2-mut, and pV5-PARN-mut were constructed from plasmid pCI-λNV5 by inserting DNA encoding a V5 epitope tag (GKPIPNPLLGLDST), a tetraglycine linker, and human CCR4a, CCR4b, CAF1, POP2, PAN2, PARN, CCR4a-E240A, CAF1-D40A/E42A, POP2-D40A/E42A, PAN2-D977A, or PARN-D28A/D382A, respectively (37), between a cytomegalovirus promoter and a simian virus 40 polyadenylation signal. The protein product of plasmid pCAF1-V5 was identical to that of pV5-CAF1 except that the tetraglycine linker and V5 epitope tag were fused to the C terminus of CAF1 instead of the N terminus. Plasmids pSR-HA-CCR4a and pSR-HA-CCR4b, which encoded N-terminally HA-tagged CCR4a and CCR4b, respectively, have been described previously (37).
Testing deadenylase mutants for a dominant-negative phenotype.
HeLa cells cultured in 35-mm wells were transiently transfected with DNA mixtures (1.5 μg) as described previously (34). The DNA mixtures contained a β-globin reporter plasmid (pBG or pBG+L7; 1.0 μg), a plasmid encoding a wild-type deadenylase or its functionally inactive mutant counterpart (pV5-CCR4a, pV5-CCR4a-mut, pV5-CAF1, pV5-CAF1-mut, pV5-POP2, pV5-POP2-mut, pV5-PAN2, pV5-PAN2-mut, pV5-PARN, or pV5-PARN-mut; 0.1 to 0.4 μg), and a plasmid encoding a constitutively transcribed α-globin-GAPDH (AG-GAPDH) mRNA chimera (pSVα1-GAPDH; 0.1 μg), which served as an internal standard. Transcription of the reporter genes from a c-fos promoter was transiently induced by serum stimulation, and the deadenylation and decay of the resulting BG or BG+L7 mRNAs were monitored by oligonucleotide-directed RNase H cleavage and Northern blot analysis as previously described (34).
Depleting HeLa cells of CAF1 and POP2 by RNA interference.
HeLa cells growing in 60-mm wells were transiently transfected with an siRNA targeting a specific deadenylase subunit (siCAF1, siPOP2, or siCAF1+siPOP2; 150 pmol) or with a nonspecific control siRNA (siNC) by using Lipofectamine 2000 (Invitrogen) according to manufacturer's protocol. The following siRNAs were used siCAF1, 5′-AACAAGUCUAC-3′ and 5′-GGUGUAAUGUAGACUUGUUAA-3′; siPOP2, 5′-UAUAACUGUAACUGAGCACGA-3′ and 5′-GUGCUCAGUUACAGUUAUAUU-3′; and siNC, 5′-ACGUGACACGUUCGGAGAAUU-3′ and 5′-UUCUCCGAACGUGUCACGUUU-3′. At 24 h after transfection, the cells were treated with trypsin and transferred to 35-mm wells, and the deadenylation and decay of BG+L7 mRNA were examined as described above.
Real-time RT-PCR analysis of knockdown efficiency.
The knockdown efficiency of siRNAs was measured by using a StepOnePlus instrument (ABI) to perform real-time reverse transcription-PCR (RT-PCR) on RNA samples as described previously (35). The PCR mixtures were heated to 95°C for 30 s and then subjected to 40 amplification cycles (15 s at 95°C, 1 min at 60°C), and the data were analyzed with StepOne software v2.1 (ABI). Endogenous GAPDH mRNA served as an internal standard. The following primers were utilized: hCAF1 primers CTCTGTGAAGGGGTCAAATGGT and TAACTGTTCTGCCACCTCCTGTAA, hPOP2 primers CTGAACCTTTTCTTCCCATCCA and CTGTGCCTAAGCCATAGAGCC, hCCR4a primers TCTCGCAACCTCAAATCCAG and CATTGGTGGTTCCATTCTTCC, hCCR4b primers CTGCCGTCAGCATCATTCAC and TCCATCATATCCACGCTCCTT, and hGAPDH primers CGCTCTCTGCTCCTCCTGTT and CCATGGTGTCTGAGC GATGT.
Deadenylation resulting from off-target RNA interference.
To test for deadenylation caused by off-target RNA interference, HeLa cells growing in 35-mm wells were transiently transfected with DNA mixtures containing pBG+GM (0.75 μg), pSVα1-GAPDH (0.05 μg), and siEGFP (80 pmol; 5′-GAACUUCAGGGUCAGCUUGCCG-3′ and 5′-GCAAGCUGACCCUGAAGUUCAU-3′). CAF1 and POP2 were depleted as described above. The deadenylation and decay of BG+GM mRNA after transient induction of transcription were monitored as previously described (34).
Deadenylation mediated by tethered Ago.
To test for deadenylation mediated by Ago tethering, HeLa cells growing in 35-mm wells were transiently transfected with DNA mixtures (1.5 μg) containing a β-globin reporter plasmid (pBG or pBG+boxB; 0.75 μg), a plasmid encoding Ago1, Ago2, Ago3, Ago4, or Ago2mut (Ago2-D597A) fused at the amino terminus to a λ N peptide and an HA epitope tag (pNHA-Ago1, pNHA-Ago2, pNHA-Ago3, pNHA-Ago4, or pNHA-Ago2-D597A; 0.2 to 0.7 μg), and a plasmid encoding a constitutively transcribed α-globin-GAPDH (AG-GAPDH) mRNA chimera (pSVα1-GAPDH; 0.05 μg), which served as an internal standard. To test deadenylase mutants for a dominant-negative effect in the context of Ago tethering, HeLa cells were transiently transfected with DNA mixtures containing pBG+boxB (0.75 μg), pNHA-Ago2 (0.5 μg), and pSVα1-GAPDH (0.05 μg), with or without pV5-CAF1-mut or pV5-POP2-mut (0.05 to 0.20 μg). The deadenylation and decay of BG and BG+boxB mRNA after transient induction of transcription were monitored as previously described (34).
Coimmunoprecipitation.
HEK 293 cells were transiently transfected with plasmids that encoded an N-terminally V5-tagged deadenylase protein and either FLAG-tagged Ago2 or HA-tagged GW182. Alternatively, the cells were transfected with plasmids that encoded CAF1 bearing a V5 tag at either the N or C terminus and N-terminally HA-tagged Ago2, GW182, TNRC6B, CCR4a, CCR4b, or β-galactosidase. At 36 h after transfection, the cells were washed with phosphate-buffered saline (PBS) and harvested in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5% NP-40, 0.2 μg/ml of RNase A [Roche], and EDTA-free protease inhibitor cocktail [one tablet per 10 ml of cell extract; Roche]). After rotation at room temperature for 15 min, each lysate was clarified by centrifugation at 14,000 × g for 15 min at 4°C, mixed with EZview Red Anti-FLAG or Anti-HA Affinity Gel (Sigma), and incubated for 3 h with gentle rotation at 4°C. The resin was washed with Tris-buffered saline, and the immunocomplexes were eluted in 60 to 150 μl of Laemmli buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% [vol/vol] glycerol, 0.002% bromophenol blue) and analyzed by electrophoresis and immunoblotting.
Immunoblotting.
Cells were harvested and lysed, and the lysates were separated on 8% polyacrylamide-SDS gels and transferred by electroblotting, as previously described (33). The blots were probed for 1 h at room temperature in PBS-T with mouse monoclonal anti-HA antibodies (Sigma) diluted 1:10,000, mouse monoclonal anti-V5 antibodies (Santa Cruz Biotechnology) diluted 1:1,000, or rabbit polyclonal antiactin antibodies (CoWin Biotech) diluted 1:2,000; incubated with a secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories) diluted 1:15,000; and developed with an Immun-Star HRP chemiluminescence kit (Bio-Rad).
RESULTS
Identification of the deadenylase responsible for miRNA-directed poly(A) removal in human cells.
Our previous studies have shown that a β-globin reporter bearing a 3′ untranslated region (3′ UTR) element (L7) partially complementary to let-7 miRNA (BG+L7 mRNA) undergoes rapid deadenylation and decay in HeLa cells due to its interaction with endogenous let-7 (34). To determine which of the three human deadenylases is responsible for removing the BG+L7 poly(A) tail, we tested inactive forms of the catalytic subunit(s) of each deadenylase for a dominant-negative effect when overproduced in vivo. Specifically, HeLa cells were cotransfected with genes encoding BG+L7 mRNA, AG-GAPDH mRNA (an internal standard), and either wild-type or catalytically inactive human CCR4a, CAF1, POP2, PAN2, or PARN. Transcription of the reporter from its c-fos promoter was transiently induced, and let-7-mediated degradation of BG+L7 mRNA was monitored as a function of time by Northern blot analysis (Fig. 1A).
FIG. 1.
Dominant-negative effect of catalytically inactive CCR4-NOT subunits. The deadenylation and decay of BG+L7 mRNA were monitored after transiently inducing its synthesis in HeLa cells transfected with a gene encoding a wild-type or mutationally inactivated (mut) V5-tagged deadenylase subunit (CAF1, CAF1-D40A/E42A; POP2, POP2-D40A/E42A; CCR4a, CCR4a-E240A; PAN2, PAN2-D977A; or PARN, PARN-D28A/D382A) and an AG-GAPDH gene. (A) Decay of BG+L7 mRNA. Equal amounts of cytoplasmic RNA isolated at time intervals after transient induction of transcription were analyzed by Northern blotting to detect BG+L7 mRNA. For comparison, the slow decay of BG mRNA lacking a let-7-responsive L7 element was also examined in the absence of deadenylase subunit overproduction (BG, top left). (B) Graphs of the concentration of BG+L7 mRNA as a function of time. All values were normalized to AG-GAPDH mRNA, a constitutively transcribed internal standard. (C) Deadenylation of BG+L7 mRNA. The cytoplasmic RNA samples analyzed in panel A were subjected to site-specific cleavage by RNase H in the presence of an oligodeoxynucleotide complementary to codons 74 to 81 within the coding region. The 3′ BG+L7 RNA fragments thereby produced were analyzed by electrophoresis and blotting, using markers (M) that corresponded in size to reporter mRNA 3′ fragments bearing no poly(A) or a 160-nucleotide poly(A) tail. (D) Immunoblot analysis of ectopically expressed V5-tagged deadenylase subunits. β-Actin served as an internal standard.
Overproducing inactive forms of either of the catalytic subunits of CCR4-NOT markedly slowed the decay rate of BG+L7 mRNA, with greater effects observed for CAF1-mut and POP2-mut than for CCR4a-mut (Fig. 1B and D). In contrast, overproducing catalytically inactive PAN2-mut or PARN-mut had no inhibitory effect. None of the wild-type deadenylase subunits altered the decay rate of BG+L7 mRNA when overproduced.
To show that the impediment to decay was a consequence of impaired deadenylation, the Northern blot analyses were repeated after subjecting the RNA samples to oligonucleotide-directed RNase H cleavage near the middle of BG+L7 mRNA so as to improve the electrophoretic resolution of messages bearing poly(A) tails of different lengths (Fig. 1C). Consistent with their effect on mRNA degradation, CAF1-mut and POP2-mut each brought BG+L7 deadenylation almost to a halt, and CCR4a-mut significantly slowed poly(A) removal, whereas PAN2-mut and PARN-mut had no effect. We conclude that CCR4-NOT is the deadenylase whose action is stimulated by miRNAs in human cells.
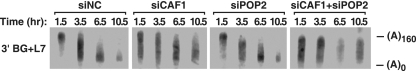
The dominant-negative effect of both CAF1-mut and POP2-mut showed that overproducing an inactive form of either protein can disrupt deadenylase function but did not distinguish which of the corresponding wild-type paralogs normally makes a greater contribution to miRNA-mediated deadenylation in HeLa cells. To make this distinction, we examined the effect of reducing the biosynthesis of CAF1 and/or POP2 by RNA interference. Transfection of HeLa cells with either siCAF1 or siPOP2 had the intended effect, diminishing the concentration of the message that encoded the targeted deadenylase subunit, as judged by real-time RT-PCR analysis. Importantly, knocking down the expression of CAF1 significantly slowed let-7-mediated poly(A) removal from BG+L7 mRNA, whereas knocking down POP2 to a similar extent did not (Fig. 2), a finding consistent with the possibility that CAF1 may normally predominate over POP2 as a catalytic subunit of the CCR4-NOT deadenylase in HeLa cells.
FIG. 2.
Effect of CAF1 or POP2 depletion on miRNA-mediated deadenylation. The deadenylation of BG+L7 mRNA was monitored after transiently inducing its synthesis in HeLa cells transfected with an siRNA complementary to CAF1 or POP2 mRNA or with a noncomplementary control siRNA (siNC). The CAF1- or POP2-specific siRNAs reduced the concentration of CAF1 mRNA by 70% and the concentration of POP2 mRNA by 67%, as determined by real-time RT-PCR.
Deadenylation as an off-target effect of RNA interference.
Synthetic siRNAs can reduce the cellular concentration not only of their intended mRNA targets to which they are fully complementary but also of “off-target” mRNAs to which they are only partially complementary (3, 12). To identify the mechanism of this off-target effect, we transfected HeLa cells with a double-stranded siRNA (siEGFP) and determined its effect on the decay of a BG mRNA derivative (BG+GM) bearing a single copy of a partially complementary element in the 3′ UTR (Fig. 3A). siEGFP accelerated the deadenylation of BG+GM mRNA, leading to its rapid degradation once the shortest poly(A) tails had reached a length of 20 to 30 nucleotides or less (Fig. 3B and C). As observed for let-7 and BG+L7 mRNA, off-target deadenylation of BG+GM mRNA by siEGFP was slowed by using RNA interference to deplete CAF1 (Fig. 3D). These results indicate that transfected siRNAs destabilize off-target mRNAs by inducing their deadenylation by CCR4-NOT.
FIG. 3.
Off-target deadenylation mediated by an siRNA. The deadenylation and decay of BG+GM mRNA were monitored after transiently inducing its synthesis in HeLa cells transfected with a partially complementary (siEGFP) or noncomplementary (siNC) siRNA. (A) RNA duplex expected for siEGFP base paired with element GM. (B) Decay of BG+GM mRNA. Equal amounts of cytoplasmic RNA isolated at time intervals after transient induction of transcription were analyzed by Northern blotting. AG-GAPDH mRNA served as an internal standard. (C) Deadenylation of BG+GM mRNA. The cytoplasmic RNA samples analyzed in panel B were subjected to site-specific cleavage by RNase H and Northern blot analysis. (D) Effect of CAF1 or POP2 depletion on siEGFP-mediated deadenylation of BG+GM mRNA.
Deadenylation and decay mediated by tethered RISC.
The multiplicity of functionally redundant Ago and TNRC6 subunits in RISC has complicated efforts to investigate their function in mammalian cells by RNAi depletion, leading researchers to use other analytical methods for this purpose. For example, there is general agreement that translational repression by RISC can be achieved not only by binding a RISC-associated miRNA to a message that is partially complementary but also by tethering a protein subunit of RISC (Ago or TNRC6) directly to mRNA via a heterologous RNA-binding domain to which that subunit has been fused (2, 4, 15, 27, 35, 42). In contrast, there have been conflicting reports as to whether tethered RISC can also reproduce the destabilizing influence of miRNAs (27, 35). Because all of those conclusions about mRNA destabilization were merely inferences deduced from changes in steady-state mRNA concentration, we decided to address this question more definitively by directly monitoring the time course of mRNA decay.
Each of the four human Ago paralogs was tethered to BG mRNA by fusing the RNA-binding domain of the lambda N protein and an epitope tag to Ago (N-HA-Ago1-4) and inserting a single boxB binding site for that domain into the BG 3′ UTR. The degradation of the resulting BG+boxB messages was monitored in HeLa cells as a function of time after transient induction of transcription. Tethering Ago1, Ago2, Ago3, Ago4, or a catalytically inactive form of Ago2 (Ago2mut) markedly destabilized BG+boxB mRNA (Fig. 4A and D). In each case, the rate of decay was quantitatively similar, irrespective of the endonucleolytic activity of the tethered Ago paralog. No destabilization was caused by an Ago2 chimera that lacked the boxB-binding domain (HA-Ago2) and therefore was unable to bind the reporter. As observed for miRNA-mediated mRNA decay, degradation was triggered by deadenylation, which was greatly accelerated by tethering Ago and its associated proteins (Fig. 4B). Moreover, deadenylation mediated by tethered RISC could be impeded by overproducing a catalytically inactive CAF1 or POP2 mutant (Fig. 4C). These findings show that tethered RISC expedites mRNA degradation and that it does so by the same mechanism as RISC that has been recruited via partial miRNA-mRNA complementarity.
FIG. 4.
Deadenylation mediated by tethered RISC. The deadenylation and decay of BG+boxB mRNA were monitored after transiently inducing its synthesis in HeLa cells transfected with a gene encoding HA-tagged human Ago1, Ago2, Ago3, Ago4, or catalytically inactive Ago2mut (Ago2-D597A) fused (N-HA-Ago) or not fused (HA-Ago) to the boxB-binding domain of the bacteriophage lambda N protein. (A) Decay of BG+boxB mRNA. Equal amounts of cytoplasmic RNA isolated at time intervals after transient induction of transcription were analyzed by Northern blotting. AG-GAPDH mRNA served as an internal standard. (B) Deadenylation of BG+boxB mRNA. The cytoplasmic RNA samples analyzed in panel A were subjected to site-specific cleavage by RNase H and Northern blot analysis. (C) Dominant negative effect of catalytically inactive CAF1 or POP2 on deadenylation mediated by tethered N-HA-Ago2. (D) Immunoblot analysis of ectopically expressed HA-tagged Ago proteins. β-Actin served as an internal standard.
Meager affinity of CCR4-NOT for RISC.
In principle, RISC could hasten the deadenylation of mRNAs with which it associates either by recruiting CCR4-NOT to those messages or by making the autonomous encounters of that deadenylase with mRNA more productive. The former mechanism would require physical association of the deadenylase with RISC, whereas the latter would not. To address this point, we performed coimmunoprecipitation experiments with epitope-tagged Ago2, GW182, and TNRC6B to determine the degree to which they associate with each of the three mammalian deadenylases. HeLa cells were cotransfected with a gene encoding FLAG-tagged Ago2 or HA-tagged GW182 and a gene encoding V5-tagged TNRC6B or Ago2 (positive control), CCR4a, CCR4b (a CCR4a paralog), CAF1, POP2, PAN2, PARN, or firefly luciferase (negative control). Alternatively, the cells were cotransfected with a gene encoding HA-tagged Ago2, GW182, TNRC6B, CCR4a, CCR4b, or β-galactosidase (negative control) and a gene encoding CAF1 that was V5-tagged at its amino or carboxyl terminus. Cell extracts were treated first with RNase A to digest RNA and then with monoclonal anti-FLAG or anti-HA antibodies. In each case, the immunoprecipitate was analyzed by electrophoresis and immunoblotting with monoclonal anti-V5 antibodies to identify proteins that associate with Ago2, GW182, or TNRC6B in an RNA-independent manner (Fig. 5). As expected, Ago2 was observed to copurify with the RISC subunits GW182 and TNRC6B but not with PAN2 or PARN, which also did not copurify detectably with GW182. Importantly, no coimmunoprecipitation of CAF1 or POP2 with either Ago2 or GW182 could be detected, even after a long exposure (Fig. 5A and B), whereas coimmunoprecipitation of CAF1 with CCR4a and CCR4b was readily detectable (Fig. 5C and D). CAF1 failed to coimmunoprecipitate with GW182, TNRC6B, or Ago2 irrespective of the location of the epitope tag on CAF1 (Fig. 5C and D) or whether RNase A treatment was omitted (data not shown). Although a long exposure revealed faint bands suggesting some copurification of CCR4a and CCR4b with Ago2, their level of association was <1% of that observed for TNRC6B, and neither coimmunoprecipitated detectably with GW182. The similar results obtained by epitope tagging multiple subunits of both RISC and CCR4-NOT and by epitope tagging CAF1 at two different locations make it unlikely that the tags prevented interaction of the two complexes. We conclude that, despite its role in miRNA-dependent poly(A) shortening, the CCR4-NOT deadenylase associates little if at all with RISC.
FIG. 5.
Failure of deadenylase subunits to coimmunoprecipitate with RISC. HEK 293T cells were cotransfected with genes encoding a FLAG- or HA-tagged RISC subunit and either a V5-tagged deadenylase subunit (POP2, CAF1, CCR4a, CCR4b, PAN2, or PARN), another V5-tagged RISC subunit (positive control), or V5-tagged firefly luciferase (negative control). The FLAG- or HA-tagged RISC subunit was immunoprecipitated, and copurifying V5-tagged proteins were detected by immunoblotting. Alternatively, HEK 293T cells were cotransfected with a gene encoding CAF1 with a V5 epitope tag at either the N terminus (V5-CAF1) or C terminus (CAF1-V5) and a second gene encoding an HA-tagged RISC subunit (Ago2, GW182, or TNRC6B), CCR4-NOT subunit (CCR4a or CCR4b), or β-galactosidase (negative control). The HA-tagged protein was immunoprecipitated, and immunoblotting was performed to detect any copurifying V5-tagged CAF1. (A) V5-tagged proteins that copurify with FLAG-Ago2. (B) V5-tagged proteins that copurify with N-HA-GW182 (TNRC6A). (C) HA-tagged proteins with which N-terminally tagged V5-CAF1 copurifies. (D) HA-tagged proteins with which C-terminally tagged CAF1-V5 copurifies. HA- or V5-tagged proteins were also detected in cell extracts to verify their production. The ubiquitous band marked with an asterisk is the cross-reacting monoclonal anti-FLAG or anti-HA antibody used for immunoprecipitation.
DISCUSSION
Our data indicate that CCR4-NOT is the deadenylase that removes poly(A) from messages targeted by miRNAs in human cells, thereby triggering rapid mRNA degradation. Overproducing a mutationally inactivated form of either of the two catalytic subunits of this deadenylase (ccry or CAF1/POP2) significantly impedes the deadenylation of RISC-associated mRNAs, whereas overproducing catalytically inactive forms of the other two human deadenylases, PAN2-PAN3 and PARN, has no effect. These findings in human cells are consistent with the effect of depleting CAF1 or NOT1 in Drosophila cells (2, 7) and with recent independent reports based on studies in mouse fibroblasts and mouse ascites cell extracts (5, 9).
Human CCR4-NOT comprises nine subunits, two of which have intrinsic deadenylase activity (14). Each of those catalytically active subunits exists in two paralogous forms in humans: CCR4a/CCR4b and CAF1/POP2. Overproducing inactive CAF1 or POP2 causes a markedly greater impediment to poly(A) removal than overproducing CCR4a, whereas excess production of the active forms of these proteins has no effect. These findings indicate that, of the two catalytic subunits in the CCR4-NOT complex, CAF1/POP2 makes a greater contribution to miRNA-mediated deadenylation than does CCR4. Furthermore, using RNA interference to reduce the cellular concentration of CAF1 significantly slows deadenylation, whereas knocking down POP2 synthesis to a similar extent does not, results that suggest a greater role for CAF1 than POP2 in poly(A) removal directed by miRNAs in HeLa cells. This difference may simply reflect a possible molar excess of CAF1 over POP2 in this cell type, since the failure of deadenylation to be impeded by wild-type POP2 overproduction argues against an intrinsic disparity in the ability of these two paralogs to degrade poly(A).
The regulatory effects of miRNAs are mediated by the protein components of RISC, which deliver miRNAs to the messages they target (8, 25). In principle, RISC could expedite poly(A) removal either by recruiting CCR4-NOT to mRNA, thereby increasing its concentration in the vicinity of the poly(A) tail, or by altering the poly(A) tail and its associated proteins in a way that increases the efficacy of deadenylation by CCR4-NOT without raising its local concentration. The former mechanism implies a physical association of RISC with CCR4-NOT, either direct or mediated by one or more other proteins specifically recruited by RISC. However, our coimmunoprecipitation data suggest that, notwithstanding a recent report to the contrary (9), RISC does not associate significantly with CCR4-NOT or with either of the other two human deadenylases. Thus, despite the importance of the CAF1/POP2 subunit of CCR4-NOT for miRNA-mediated deadenylation, neither of these proteins was found to interact detectably with either Ago or TNRC6. Moreover, only trace amounts of CCR4a and CCR4b were observed to coimmunoprecipitate with Ago (corresponding to <1% of the level at which TNRC6 immunoprecipitated), and neither copurified detectably with TNRC6, the apparent effector subunit of RISC. It is possible that the weak association of CAF1 with Ago2 that was recently reported (9) was an indirect consequence of bridging by the ubiquitous mRNP component PABP (40). We conclude that RISC likely facilitates deadenylation by making the autonomous encounters of CCR4-NOT with poly(A) more productive. For example, the reported ability of TNRC6 to displace eIF4G from PABP (40) may render poly(A) more susceptible to deadenylase attack. Such disruption of the eIF4G-mediated interaction of the cap-binding complex with PABP might also help to account for the inhibitory effect of miRNAs on translation, although it would not explain their ability to repress both cap-independent translation and the translation of mRNAs that lack a poly(A) tail as effectively as they repress the translation of capped, polyadenylated mRNAs (7, 19, 26, 34).
A complication of using RNA interference as a genetic tool is the propensity of transfected siRNAs to downregulate not only their intended targets but also many other mRNAs with which they can base pair with imperfect complementarity (3, 12). Our results indicate that these off-target effects are, at least in part, a consequence of mRNA destabilization resulting from poly(A) tail removal by CCR4-NOT. These findings support the view that synthetic siRNAs downregulate off-targets by the same mechanisms used by endogenous miRNAs to repress mRNAs to which they are partially complementary.
The multiplicity of functionally redundant Ago and TNRC subunits of RISC in mammalian cells has made tethering a valuable strategy for examining the respective contributions of those proteins and their component domains to gene silencing by miRNAs. Previously, we have shown that tethering any of the four human Ago proteins to mRNA, presumably as a complex with TNRC6, reduces both the concentration and translation efficiency (protein synthesis per message) of that mRNA (35). Our present data show that this effect on mRNA concentration is a consequence of expedited mRNA decay triggered by rapid deadenylation. Moreover, deadenylation mediated by tethered Ago involves CCR4-NOT, the same deadenylase whose action is stimulated by miRNAs. Thus, in every respect, the effects of recruiting RISC to mRNA by tethering its Ago subunit mimic those of recruiting RISC by miRNA base pairing to an imperfectly complementary mRNA element. These findings confirm the validity of RISC tethering as a surrogate methodology for investigating miRNA-mediated silencing.
Acknowledgments
We thank Ann-Bin Shyu for providing cDNA clones of wild-type and mutant deadenylase subunits.
This research was supported by a grant to J.G.B. from the National Institutes of Health (GM79477) and by grants to L.W. from the National Natural Science Foundation of China (30970618) and the Chinese Academy of Sciences (2008OHTP01).
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553-563. [DOI] [PubMed] [Google Scholar]
- 2.Behm-Ansmant, I., J. Rehwinkel, T. Doerks, A. Stark, P. Bork, and E. Izaurralde. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birmingham, A., E. M. Anderson, A. Reynolds, D. Ilsley-Tyree, D. Leake, Y. Fedorov, S. Baskerville, E. Maksimova, K. Robinson, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. 3′UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3:199-204. [DOI] [PubMed] [Google Scholar]
- 4.Chekulaeva, M., W. Filipowicz, and R. Parker. 2009. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA 15:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. Y., D. Zheng, Z. Xia, and A. B. Shyu. 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16:1160-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. Y. A., and A. B. Shyu. 1994. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol. Cell. Biol. 14:8471-8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eulalio, A., E. Huntzinger, T. Nishihara, J. Rehwinkel, M. Fauser, and E. Izaurralde. 2009. Deadenylation is a widespread effect of miRNA regulation. RNA 15:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulalio, A., F. Tritschler, and E. Izaurralde. 2009. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 15:1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian, M. R., G. Mathonnet, T. Sundermeier, H. Mathys, J. T. Zipprich, Y. V. Svitkin, F. Rivas, M. Jinek, J. Wohlschlegel, J. A. Doudna, C. Y. Chen, A. B. Shyu, J. R. Yates III, G. J. Hannon, W. Filipowicz, T. F. Duchaine, and N. Sonenberg. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 35:868-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehring, N. H., G. Neu-Yilik, T. Schell, M. W. Hentze, and A. E. Kulozik. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell 11:939-949. [DOI] [PubMed] [Google Scholar]
- 11.Giraldez, A. J., Y. Mishima, J. Rihel, R. J. Grocock, S. Van Dongen, K. Inoue, A. J. Enright, and A. F. Schier. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312:75-79. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21:635-637. [DOI] [PubMed] [Google Scholar]
- 13.Kiriakidou, M., G. S. Tan, S. Lamprinaki, M. De Planell-Saguer, P. T. Nelson, and Z. Mourelatos. 2007. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 129:1141-1151. [DOI] [PubMed] [Google Scholar]
- 14.Lau, N. C., A. Kolkman, F. M. van Schaik, K. W. Mulder, W. W. Pijnappel, A. J. Heck, and H. T. Timmers. 2009. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 422:443-453. [DOI] [PubMed] [Google Scholar]
- 15.Lazzaretti, D., I. Tournier, and E. Izaurralde. 2009. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, R. C., R. L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843-854. [DOI] [PubMed] [Google Scholar]
- 17.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769-773. [DOI] [PubMed] [Google Scholar]
- 18.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437-1441. [DOI] [PubMed] [Google Scholar]
- 19.Lytle, J. R., T. A. Yario, and J. A. Steitz. 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′UTR as in the 3′UTR. Proc. Natl. Acad. Sci. U. S. A. 104:9667-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangus, D. A., M. C. Evans, N. S. Agrin, M. Smith, P. Gongidi, and A. Jacobson. 2004. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 24:5521-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathonnet, G., M. R. Fabian, Y. V. Svitkin, A. Parsyan, L. Huck, T. Murata, S. Biffo, W. C. Merrick, E. Darzynkiewicz, R. S. Pillai, W. Filipowicz, T. F. Duchaine, and N. Sonenberg. 2007. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317:1764-1767. [DOI] [PubMed] [Google Scholar]
- 22.Meister, G., M. Landthaler, A. Patkaniowska, Y. Dorsett, G. Teng, and T. Tuschl. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15:185-197. [DOI] [PubMed] [Google Scholar]
- 23.Nottrott, S., M. J. Simard, and J. D. Richter. 2006. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 13:1108-1114. [DOI] [PubMed] [Google Scholar]
- 24.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 25.Peters, L., and G. Meister. 2007. Argonaute proteins: mediators of RNA silencing. Mol. Cell 26:611-623. [DOI] [PubMed] [Google Scholar]
- 26.Petersen, C. P., M. E. Bordeleau, J. Pelletier, and P. A. Sharp. 2006. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21:533-542. [DOI] [PubMed] [Google Scholar]
- 27.Pillai, R. S., C. G. Artus, and W. Filipowicz. 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10:1518-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309:1573-1576. [DOI] [PubMed] [Google Scholar]
- 29.Seggerson, K., L. Tang, and E. G. Moss. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243:215-225. [DOI] [PubMed] [Google Scholar]
- 30.Thermann, R., and M. W. Hentze. 2007. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature 447:875-878. [DOI] [PubMed] [Google Scholar]
- 31.Wang, B., A. Yanez, and C. D. Novina. 2008. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl. Acad. Sci. U. S. A. 105:5343-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wightman, B., I. Ha, and G. Ruvkun. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855-862. [DOI] [PubMed] [Google Scholar]
- 33.Wu, L., and J. G. Belasco. 2005. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 25:9198-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, L., J. Fan, and J. G. Belasco. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U. S. A. 103:4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, L., J. Fan, and J. G. Belasco. 2008. Importance of translation and nonnucleolytic Ago proteins for on-target RNA interference. Curr. Biol. 18:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, M., M. Reuter, H. Lilie, Y. Liu, E. Wahle, and H. Song. 2005. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 24:4082-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita, A., T. C. Chang, Y. Yamashita, W. Zhu, Z. Zhong, C. Y. Chen, and A. B. Shyu. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054-1063. [DOI] [PubMed] [Google Scholar]
- 38.Yekta, S., I. H. Shih, and D. P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304:594-596. [DOI] [PubMed] [Google Scholar]
- 39.Zdanowicz, A., R. Thermann, J. Kowalska, J. Jemielity, K. Duncan, T. Preiss, E. Darzynkiewicz, and M. W. Hentze. 2009. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol. Cell 35:881-888. [DOI] [PubMed] [Google Scholar]
- 40.Zekri, L., E. Huntzinger, S. Heimstadt, and E. Izaurralde. 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29:6220-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng, Y., R. Yi, and B. R. Cullen. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. U. S. A. 100:9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zipprich, J. T., S. Bhattacharyya, H. Mathys, and W. Filipowicz. 2009. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 15:781-793. [DOI] [PMC free article] [PubMed] [Google Scholar]