Abstract
RNA uridylylation is critical for the expression of the mitochondrial genome in trypanosomes. Short U tails are added to guide RNAs and rRNAs, while long A/U heteropolymers mark 3′ ends of most mRNAs. Three divergent mitochondrial terminal uridylyl transferases (TUTases) are known: RET1 catalyzes guide RNA (gRNA) uridylylation, RET2 executes U insertion mRNA editing, and MEAT1 associates with the editosome-like complex. However, the activities responsible for 3′ uridylylation of rRNAs and mRNAs, and the roles of these modifications, are unclear. To dissect the functions of mitochondrial TUTases, we investigated the effects of their repression and overexpression on abundance, processing, 3′-end status, and in vivo stability of major mitochondrially encoded RNA classes. We show that RET1 adds U tails to gRNAs, rRNAs, and select mRNAs and contributes U's into A/U heteropolymers. Furthermore, RET1's TUTase activity is required for the nucleolytic processing of gRNA, rRNA, and mRNA precursors. The U tail's presence does not affect the stability of gRNAs and rRNAs, while transcript-specific uridylylation triggers 3′ to 5′ mRNA decay. We propose that the minicircle-encoded antisense transcripts, which are stabilized by RET1-catalyzed uridylylation, may direct a nucleolytic cleavage of multicistronic precursors.
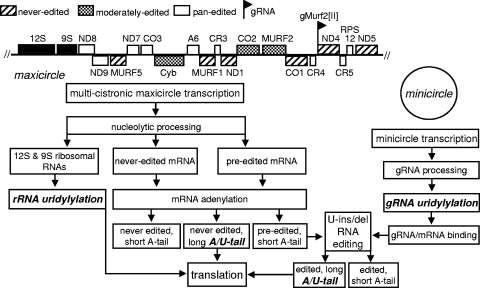
The mitochondrion of kinetoplastid protozoans contains a catenated DNA network composed of few maxicircles and thousands of minicircles. rRNA and protein-encoding genes are tightly packed within a conserved region of the maxicircle and are transcribed as a polycistronic precursor(s). Pre-rRNAs and pre-mRNAs are excised from the precursor through an unknown mechanism; intriguingly, mature 5′ and 3′ ends of the adjacent RNAs often overlap. The detection of correctly processed contiguous molecules that intersect in the precursor (23) not only indicates a precise cleavage mechanism, but also suggests a stochastic choice of cleavage sites. Six of the 18 annotated pre-mRNAs contain open reading frames and are referred to as never-edited mRNAs. The remaining 12 are designated preedited, as they must undergo U insertion/deletion editing to create translatable mRNAs. Additional protein diversity may be introduced by alternative editing of the “housekeeping” mRNAs (31, 32). Most mRNAs are also polyadenylated (9). The length of the A tail appears to correlate with the editing status; preedited and partially edited mRNAs possess only short A tails, while fully edited mRNAs have both short and long tails (17). Hence, the following mRNA types exist in trypanosome mitochondria: never-edited with short (∼20 nucleotides [nt]) and long (∼250 nt) tails, preedited with short tails, and edited with short and long tails (Fig. 1).
FIG. 1.
Major RNA classes in trypanosome mitochondria. Two transcript types are produced independently from maxicircle and minicircle DNA: multicistronic mRNA-rRNA precursors and gRNA precursors, respectively. RNAs above the thick line are transcribed from the same strand; RNAs shown below are encoded in the opposite DNA strand. The multicistronic precursor is processed by an unknown nuclease into rRNAs and never-edited and preedited pre-mRNAs. gRNAs maintain a triphosphate at the 5′ end but undergo 3′ processing and uridylylation. gRNA binding to preedited mRNAs initiates the U insertion/deletion editing process, which proceeds in the 3′ to 5′ direction, inserting or deleting U's to generate an open reading frame. Completion of the mRNA editing triggers the addition of an A/U heteropolymer to the existing short A tail. RNA 3′ uridylylation steps are shown in boldface type.
The uracil insertion/deletion RNA editing is directed by trans-acting guide RNAs (gRNAs). These ∼60-nt molecules are transcribed primarily from the minicircles, and are posttranscriptionally 3′-uridylylated (12). Uridylylation of gRNAs by RNA-editing TUTase 1 (RET1) was shown to be indispensable to the editing process (4, 5). However, it remains unknown whether inhibition of editing in RET1 RNA interference (RNAi) cells is caused by the loss of gRNA's U tail, which would imply a functional role of this structure in the editing reactions. Alternatively, the loss of the entire gRNA molecule would indicate the U tail's role in gRNA stability. A different class of RNA molecules, 9S and 12S rRNAs, also receive 10- to 15-nt-long U tails (1). However, the functions of these elements in rRNA biogenesis, decay, or translation processes and the identity of the U-adding enzyme are unclear.
We have previously identified the kinetoplast poly(A) polymerase 1 (KPAP1), an enzyme responsible for adenylation of mitochondrial mRNAs, and demonstrated that short (∼20 nt) A tails are required for the maintenance of some never-edited and most edited mRNAs (17). In contrast, the loss of A tails had no effect on steady-state levels of preedited transcripts. Short A tails are extended as random A/U (∼70/30% ratio) heteropolymers upon completion of the RPS12 mRNA editing. However, uncertainty about the function of A/U extensions and the identity of the U-contributing enzyme remains.
Recombinant RET1 is a highly processive enzyme, and it seems likely that uridylylation reactions in vivo are tightly controlled (7). RET1 also has a high affinity for single-stranded RNAs (7) and participates in multiple high-molecular-mass complexes (4). In addition to RET1, two other mitochondrial TUTases have been identified. The RET2 is a subunit of the RNA-editing core complex (RECC), also referred to as the 20S editosome or l-complex (2, 33). This TUTase is responsible for the U insertion mRNA-editing activity of the RECC (4, 16). The mitochondrial editosome-like complex-associated TUTase (MEAT1) is present in mitochondrial extracts in unassociated and complex-bound forms. MEAT1 RNAi inhibited cell growth, which suggests an essential function (8). To conclude, U additions are ubiquitous in mitochondrial RNAs, but their specific roles in RNA biogenesis and decay, and the nature of TUTases responsible for most 3′ modifications, remain ambiguous.
In this work, we investigated the effects of uridylylation on the steady-state abundance, processing, and in vivo stability of major RNA classes, which are encoded in two distinct mitochondrial genomes, minicircles and maxicircles (Fig. 1). Inducible dominant negative overexpression and RNAi cell lines for RET1, RET2, and MEAT1 have been analyzed. Parallel experiments were performed in KPAP1 RNAi cells. We show that RET1 adds continuous short U tails to gRNAs, rRNAs, and select mRNAs. RET1 repression led to a rapid decline in the steady-state abundance of all gRNAs tested and both rRNAs. Most unexpectedly, this was caused by blocked processing of gRNA and rRNA precursors and not by the accelerated decay of nonuridylylated RNAs. We found that select mRNAs (ND1, MURF1, and MURF5) do not possess short A tails, which are required for the maintenance of most mitochondrial mRNAs (17). Instead, the steady-state levels of these molecules are downregulated by 3′ uridylylation. Finally, loss of RET1 led to an accumulation of unprocessed precursors for some preedited and never-edited mRNAs. Our data provide evidence that RET1's uridylyl transferase activity shapes the mitochondrial transcriptome by acting in at least three modes: (i) by adding continuous U tails to gRNAs, rRNAs, and mRNAs; (ii) by incorporating U's into mRNA's long A/U tails; and (iii) indirectly, by affecting the processing of pre-gRNAs and maxicircle-encoded multicistronic precursors. RET2 functions appear to be confined to U insertion mRNA editing, and MEAT1 repression produced no discernible effects in assays used throughout this study.
MATERIALS AND METHODS
Trypanosome culture, RNAi, and RET1 overexpression.
The RET1 RNAi expression plasmid was generated by cloning a gene fragment (444 to 1,025 bp) into a p2T7-177 vector that allows for tetracycline-inducible expression (45). The construct was transfected into procyclic 29-13 Trypanosoma brucei strains (46), followed by clonal selection of phleomycin-resistant cell lines. RNAi was performed as described previously (44). The N-terminal mitochondrial importation signal of gBP21 (positions 1 to 20) (22) was added to the full-length TbRET1 gene, which was then cloned as a C-terminally TAP-tagged fusion into a pLew 79-based vector (46). Expression of RET1-TAP fusions and depletion of RET1 and KPAP1 proteins by RNAi was verified by Western blotting.
RNA stability assays.
RNAi clones were selected by limiting dilution and stabilized in SDM79 media with 10% serum, 50 μg/ml of hygromycin, 30 μg/ml of G418, and 2.5 μg/ml of phleomycin. RNAi was induced with 5 μg/ml of tetracycline in 100 ml culture (6 × 105 cells/ml), and cultivation was continued for 40 to 42 h, typically reaching a cell density of ∼5 × 106/ml. Cells were collected in 15-ml aliquots, quickly washed with ice-cold phosphate-buffered saline (PBS), and frozen in liquid nitrogen. Actinomycin D and ethidium bromide were added at 20 and 10 μg/ml, respectively. The change in relative abundance was calculated assuming that the gRNA/tRNA ratio at the time of actinomycin D addition was 100%.
RNA analysis and cRT-PCR.
Total RNA was isolated using a modified procedure (13). Detailed methods for quantitative reverse transcription-PCR (qRT-PCR), Northern blotting, and gRNA labeling and oligonucleotides used in this study are described in the supplemental material. Membranes and gels were exposed to phosphor storage screens, and volume quantitation was performed with the Quantity One software package (Bio-Rad). The change in relative abundance was calculated assuming that the ratio between analyzed transcripts and loading control in mock-induced cells was 100%. For circular RT-PCR (cRT-PCR), total RNA was purified from cells induced for 55 h. Real-time-PCR-grade RNA (5 μg) was circularized with 25 U of T4 RNA ligase in 25 μl manufacturer-supplied buffer (NEB) at 37°C for 45 min. The reaction mix was diluted to 100 μl and extracted with phenol and phenol-chloroform. Approximately 2.5 μg of circularized RNA was used for cDNA synthesis and amplification with the SuperScript III one-step RT-PCR system as recommended by the manufacturer (Invitrogen). PCR products were gel purified and cloned, and at least 96 clones were sequenced.
GRBC1/2 reconstitution, purification, UV cross-linking, and EMSA.
Details of GRBC1/2 coexpression and purification are described in the supplemental material. Templates for T7 transcription of gND7[506] with 15 U's and gND7[506] with no U tails were PCR amplified with two overlapping oligonucleotides. RNAs were transcribed with T7 RNA polymerase in the presence of [α-32P]ATP and purified on a 15% polyacrylamide-8 M urea gel. RNA (100,000 cpm plus 0 to 5 μg of fragmented yeast tRNA) was heated at 80°C for 2 min in 75 mM KCl, incubated on ice for 2 min, and mixed with 0.1 μg of protein in 10 μl of 20 mM Tris-HCl (pH 7.8), 75 mM KCl, and 7 mM magnesium acetate. Reaction mixtures were incubated at 27°C for 10 min and UV irradiated on ice for 5 min in HL-2000 HybriLinker (UVP). RNase A (5 μg) and RNase T1 (10 U) were added, and reaction mixtures were incubated at 50°C for 1 h. Samples were separated on an 8 to 16% SDS gel and transferred to a nitrocellulose membrane, which was stained with Sypro Ruby to detect proteins and exposed to a phosphor storage screen. For electrophoretic mobility shift assay (EMSA), binding reactions were set up with 0.1 pmol of RNA and 1 μg of fragmented yeast RNA and mixtures were incubated in 10 μl of cross-linking buffer with GRBC for 10 min at 27°C. RNA-protein complexes were separated by 8% PAGE in 50 mM Tris-HEPES buffer (pH 7.8).
RESULTS
Overexpression of active and inactive RET1 produces a growth inhibition phenotype.
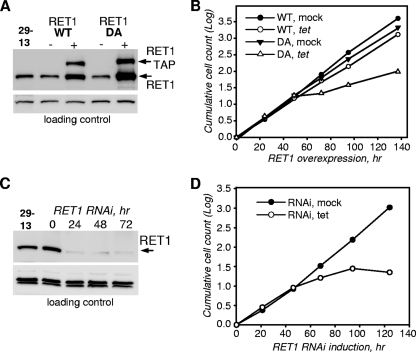
RET1 TUTase participates in heterogeneous high-molecular-mass (500 to 2,000 kDa) complexes, and its inhibition by RNAi arrests cell division and blocks RNA editing (4, 5). To distinguish the effects of protein ablation by RNAi, which may cause degradation of associated complexes and loss of RET1's RNA binding capacity, from those depending on its TUTase activity, we have developed inducible dominant negative cell lines of procyclic (insect)-form T. brucei. Mutation in the catalytic metal binding site (D471A, here referred to as DA) abolishes the catalysis but does not affect RET1's folding or RNA binding properties (7). For unknown reasons, the full-length untagged, TAP- or FLAG-tagged proteins were not imported into the mitochondria (not shown). To solve this problem, a mitochondrial import signal derived from the MRP1 (gBP21) protein (22) was added to the N terminus of the TAP-tagged RET1. Upon induction, the DA variant expressed at an ∼30% higher level than did the wild type (WT) (Fig. 2A), but both proteins were localized in the mitochondria, as verified by subcellular fractionation (not shown). The apparent increase in the level of endogenous protein in cells overexpressing tagged RET1 was most likely caused by a partial proteolysis of the RET1-TAP fusion proteins (not shown). Overexpression of the WT protein led to its incorporation into high-molecular-mass complexes (see Fig. S1A in the supplemental material) and produced a moderate inhibition of cell division, while cells expressing the mutant protein displayed a dramatically reduced growth rate (Fig. 2B). A dominant negative mutation in RET2, D97A, resulted in a strong growth inhibition, while overexpression of the active enzyme had no apparent effect on the cell division time (G. E. Ringpis and R. Aphasizhev, unpublished data). A growth phenotype was undetectable in cell lines expressing WT or D65A-mutated MEAT1 (not shown).
FIG. 2.
Overexpression of catalytically active or inactive TAP-tagged RET1 proteins produces a growth inhibition phenotype. (A) Immunoblot analysis of tet-inducible RET1 overexpression. Parasite cells (5 × 106) were boiled in SDS loading buffer, separated on an 8 to 16% SDS polyacrylamide gel, and subjected to immunoblotting with polyclonal affinity-purified anti-RET1 antibody. 29-13 is the parental strain of procyclic T. brucei used to derive inducible RNAi and overexpression cell lines (46). β-Tubulin was used as the loading control. (B) Growth kinetics of suspension cell cultures after mock induction (closed symbols) or addition of tetracycline (open symbols). Cells were induced at 106/ml and diluted every 24 h to the same density or left undiluted if less than one division occurred. (C) Immunoblot analysis of RET1 knockdown by inducible RNAi. Cells were analyzed as described for panel A. KPAP1 was used as loading control. (D) Cell growth kinetics after RET1 RNAi induction.
Inhibition of RET1 activity results in loss of gRNAs and accumulation of gRNA precursors.
gRNAs can be selectively labeled by vaccinia virus guanylyltransferase in the presence of [α-32P]GTP (12), which is characteristic of primary transcripts. We have reported that RET1 RNAi inhibits editing (5), which is preceded by the shortening of GTP-labeled products (4). Hence, it is possible that the loss of U tail rendered gRNAs inactive in editing, which would be consistent with the U tail's proposed role in gRNA-mRNA hybridization (11, 12). We have reexamined changes in length and relative abundance of the maxicircle-encoded gMurf2[II] and several minicircle-encoded gRNAs and in RET1 RNAi and DA cell lines.
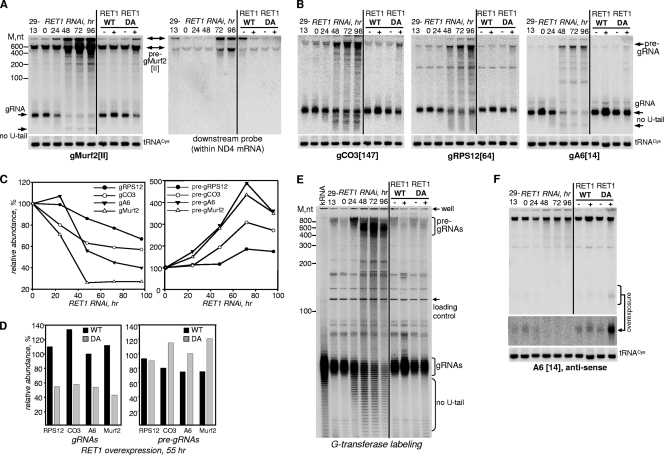
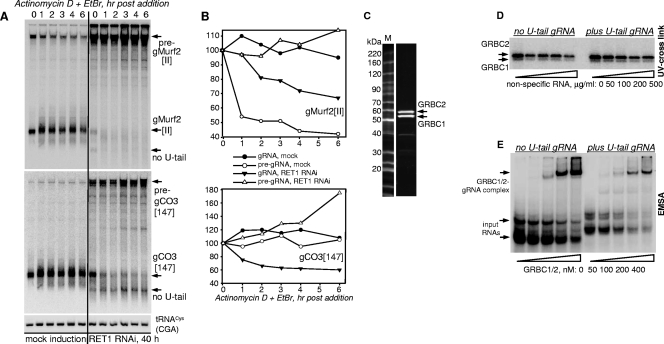
A robust knockdown of RET1 occurred at 24 h of RNAi (Fig. 2C) while changes in RNA levels became apparent after 48 h of induction. A complete arrest of cell division at ∼96 h was followed by massive cell death after 120 h of RNAi (Fig. 2D). High-resolution Northern blotting showed a 50 to 80% decline in abundance, depending on gRNA species, and the appearance of nonuridylylated gRNAs (Fig. 3A and B). Surprisingly, larger pre-gRNA molecules (600 to 800 nucleotides [nt]) accumulated with the progression of RNAi (Fig. 3C). The transcription of the gMurf2[II] gene was suggested to occur independently of mRNA precursors (14). Indeed, as seen in Fig. 3A, in which the same membrane was probed for gMurf2[II] (left panel) and then reprobed with oligonucleotide hybridizing 500 nt downstream (right panel), the maxicircle-encoded gRNA is also transcribed as a precursor, which accumulates upon RET1 repression.
FIG. 3.
Maxicircle- and minicircle-encoded gRNA precursors accumulate in RET1-depleted parasites. (A) Northern blotting of maxicircle-encoded gMurf2[II]. Total RNA was isolated from RNAi cells at the indicated induction time points. DA and WT RET1 proteins were expressed for 72 h. Probes were designed for the gRNA sequence (left panel) or toward a region located 500 nt downstream within the ND4 gene in the maxicircle DNA (right panel). Double-headed arrows show precursors. Positions of mature and nonuridylylated gRNAs are indicated by arrows. (B) Northern blotting of minicircle-encoded gRNAs. Quantitative changes in relative abundance of uridylylated gRNAs and pre-gRNAs during RNAi progression and in DA cells are shown in panels C and D, respectively. (E) Selective labeling of primary transcripts. The RNA samples analyzed in panels A and B were 5′ labeled with [α-32P]GTP in the presence of vaccinia virus guanylyltransferase and separated on 9% polyacrylamide-8 M urea gel. kRNA indicates RNA that was isolated from mitochondrial fraction purified by Renografin density gradient. Loading control (unidentified cytosolic RNA) and the gel wells are shown by arrows. (F) Northern blotting detection of antisense minicircle transcripts. A DNA copy of the gA6[14], minus the U tail, was 5′ labeled and hybridized with the membrane used for analysis of gRNA A6[14] (B, right panel). The overexposed region covers the migration area of gRNAs.
To determine whether pre-gRNAs accumulate on a global scale, the overall abundance and size distribution of primary transcripts were assayed via 5′ labeling with guanylyltransferase in the presence of [α-32P]GTP (Fig. 3E). The relative gRNA abundance decreased by ∼65% after 96 h of RNAi induction, concomitant with the appearance of nonuridylylated forms, and increased by ∼70% in cells expressing active RET1. In contrast, pre-gRNAs increased in RNAi cells by more than 2-fold as a heterogeneous 600- to 800-nt population and moderately declined in cells overexpressing WT RET1 (see Fig. S1B in the supplemental material). RNAi inhibition of RET2, MEAT1, or KPAP1 had no effects on gRNAs (data not shown).
Antisense transcription may determine the position of the gRNA precursor cleavage.
Our experiments demonstrated that, irrespective of their genomic location, gRNAs are transcribed as 600- to 800-nt-long precursors. The fact that the mature gRNAs maintain their 5′ triphosphates implies the removal of the long 3′ trailer. RET1's enzymatic activity is directly responsible for gRNA 3′ uridylylation, but is also involved, presumably indirectly, in the processing of gRNA precursors. We hypothesized that the inhibition of pre-gRNA processing in RET1 RNAi and DA cells was caused by the loss of RNA molecules which are transcribed from the opposite DNA strand in the minicircle. It is possible that these molecules might direct the nucleolytic cleavage of gRNA precursors and are stabilized by RET1-catalyzed uridylylation. We next tested whether such molecules are transcribed as antisense to gRNAs. The same samples analyzed for gRNA A6[14] as shown in Fig. 3B were hybridized with a complementary probe, a DNA copy of the gRNA (Fig. 3F). Most surprisingly, two types of transcripts were detected: abundant 600- to 800-nt-long RNAs and gRNA-sized molecules. Furthermore, the level of long antisense RNAs declined by ∼50% with progression of RET1 RNAi, as opposed to accumulation of the similarly sized pre-gA6[14] (Fig. 3B and C). Accordingly, overexpression of the active RET1 led to a moderate pre-gA6[14] reduction (Fig. 3D) and a 2-fold buildup of the long antisense transcript. The short antisense RNAs showed a RET1-dependent pattern similar to that of gRNA with a notable exception: a 3-fold accumulation in the dominant negative cell line.
Although the unequivocal functional link between the antisense transcripts and pre-gRNA processing remains to be firmly established, our results suggest that the double-stranded region formed by the overlapping 5′ ends of the opposing 600- to 800-nt transcription units may constitute a substrate for the processing nuclease (see Fig. 8). The opposite effects of RET1 repression on the pre-gRNA and long antisense transcript indicate that the RET1 activity is required to maintain the latter molecules, presumably by U tail addition. Collectively, these findings bring up an essential question of the U tail's function in vivo.
FIG. 8.
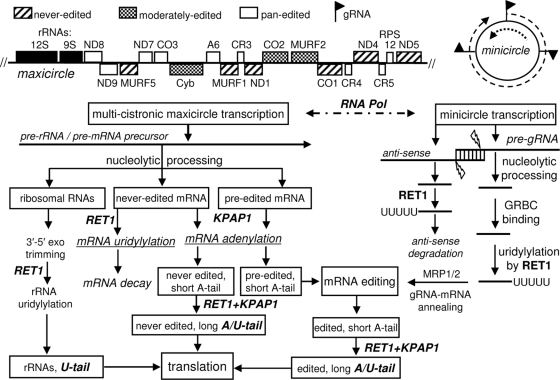
Model for RNA processing in trypanosome mitochondria. A schematic diagram of RET1-catalyzed reactions outlines their multiple roles in the formation of all known mitochondrial RNA classes. MRP1/2, a complex of mitochondrial RNA binding proteins 1 and 2 (3) which is thought to promote gRNA-mRNA annealing (28); GRBC, gRNA binding complex (44); KPAP1, mitochondrial poly(A) polymerase (17).
Loss of the U tail does not affect gRNA stability.
Mature gRNAs possess remarkably homogenous encoded 3′ ends (41). This suggests a precise gRNA processing mechanism, which requires RET1's activity (Fig. 3), and generates a substrate (nonuridylylated gRNA) for the subsequent addition of 15 to 20 U's by RET1. The decreased steady-state level in RET1 RNAi and DA cells may be attributed to inhibition of gRNA processing or accelerated decay of nonuridylylated gRNAs, or both. To distinguish these possibilities, we have compared the in vivo gRNA decay in control and RET1 RNAi cells. Mitochondrial transcription was inhibited in mock-induced cells and after 40 h of RET1 RNAi. Total RNA was analyzed for maxicircle- (Murf2[II]) and minicircle-encoded (gCO3[147]) gRNAs. Cysteine tRNA, which is imported into the mitochondria, served as a normalization control (44). Both uridylylated and nonuridylylated forms of gMurf2[II] and gCO3[147] were present in RET1 RNAi cells (Fig. 4A). The level of gCO3[147] precursor was at the limit of detection in mock-treated cells, but in RET1 RNAi cells its relative abundance increased by more than 5-fold and remained constant for the duration of the assay. The pre-gMurf2[II] underwent a similar increase and remained virtually unchanged for at least 4 h. In the mock-treated cells, ∼50% of pre-gMurf2[II] fraction was processed in 2 h after inhibition of transcription indicating nucleolytic processing was unaffected by transcription blockade (Fig. 4B). In summary, these data indicate an abrogation of the pre-gRNA processing by RET1 RNAi.
FIG. 4.
Loss of gRNAs in RET1 RNAi cells is caused by inhibited pre-gRNA processing and not by accelerated decay of gRNAs lacking 3′ U tails. (A) gRNA decay in T. brucei cells depleted of RET1. After 40 h of RNAi induction, actinomycin D and ethidium bromide were added to inhibit transcription. Cells were collected at indicated time intervals by quick centrifugation. Total RNA was separated on 10% polyacrylamide-8 M urea gel, transferred onto membrane, and sequentially probed for gRNAs and nuclearly encoded mitochondrial tRNACys(GCA) (34). (B) Quantitation of blotting signals from panel A. The change in relative abundance was calculated assuming that the gRNA/tRNA ratio in mock-induced cells at the time of actinomycin D addition was 100%. (C) Reconstitution of the GRBC1/2 complex. GRBC1 and His6-tagged GRBC2 were coexpressed in pDUET-1 vector in E. coli. The stoichiometric complex was purified by three sequential chromatographic steps as described in the supplemental material. Final Sepharose Q fraction was separated on an 8 to 16% SDS polyacrylamide gel and stained with Sypro Ruby. (D) gND7[506] with and without a U tail was uniformly labeled with [α-32P]ATP during T7 transcription. Labeled RNA was incubated with a purified GRBC1/2 complex in the presence of nonspecific RNA and irradiated by short UV light. After digestion with RNase A, label transfer was monitored by separating cross-linked products on an SDS gel and subsequently transferring onto a nitrocellulose membrane. (E) The same RNAs as in panel D were 5′ labeled, incubated with increasing concentrations of GRBC1/2 complex in the presence of 0.1 mg/ml of nonspecific RNA, and separated on an 8% polyacrylamide gel in Tris-HEPES buffer (pH 7.8). Gels were fixed and exposed to phosphor storage screen.
The time-resolved accumulation of nonuridylylated gRNAs, most obvious for gCO3 (∼220% at 3 h time point) (Fig. 4A), agrees with preservation of these molecules in the steady-state population (Fig. 3A and B). Because mitochondrial transcription, pre-gRNA processing, and U addition were blocked in RET1 RNAi cells treated with actinomycin D, we concluded that the U tail was rapidly removed while the gRNA itself remained unaffected. Therefore, the U tail does not significantly contribute to gRNA stability. These observations are consistent with in organello experiments reporting rapid turnover of the [α-32P]UTP-labeled total gRNA population (36) and the presence of the U-specific 3′-5′ exonuclease in mitochondrial extract (6). We also note that the nonuridylylated gRNAs were virtually undetectable in mock-induced cells, suggesting an efficient coupling of the 3′ nucleolytic processing and uridylylation (Fig. 4A, mock induction panel).
Nonuridylylated gRNAs are stabilized via binding to GRBC1/2 complex.
The RNAi repression of either GRBC1 or -2 subunits of the gRNA binding complex leads to an accelerated gRNA decay in vivo and, consequentially, to a uniform loss of gRNAs. However, neither accumulation of gRNA precursors nor appearance of gRNAs without U tails was detected (44). Because nonuridylylated gRNAs are apparently stable in vivo (Fig. 4A), we next investigated whether GRBC1 or -2, which does not have recognizable RNA binding motifs, indeed binds gRNAs, and how the U tail's presence affects binding. GRBC1 and -2 proteins were insoluble in heterologous expression systems and copurified from T. brucei with at least 20 other proteins (not shown). However, coexpression of both subunits in Escherichia coli produced a stable, stoichiometric particle, which was purified to homogeneity (Fig. 4C). The apparent native molecular mass of GRBC1/2 (∼180 kDa, estimated by size exclusion chromatography, and ∼220 kDa, based on electrophoretic gel mobility) suggests a heterotetrameric α2β2 organization. The short UV-induced cross-linking with uniformly labeled gRNAs demonstrated that GRBC2 is the RNA binding subunit of the complex but provided no indications of the U tail's positive contribution to the cross-linking efficiency (Fig. 4D). EMSA showed that GRBC1/2 forms a discrete complex with gND7[506]. The apparent dissociation constants for both uridylylated and tailless molecules were virtually identical (200 ± 20 nM, Fig. 4E), further suggesting that binding to the GRBC1/2 particle, and not U tail addition by RET1, is the most probable factor preventing rapid gRNA decay in the mitochondria.
RET1 activity is required for maxicircle transcript processing.
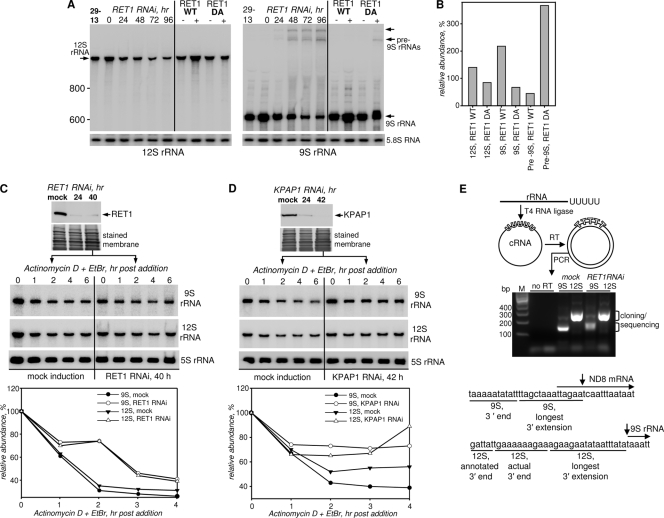
We began analyzing maxicircle transcripts with rRNAs. To test changes in their steady-state abundance, total RNA was separated on 5% PAGE-8 M urea and probed for 9S and 12S rRNAs (Fig. 5A). Quantitation of Northern blots at the 72-h RNAi time point demonstrated a decline of these RNAs by 90% and 70%, respectively. Expression of the dominant negative mutant for 72 h produced similar effects, indicating that the observed loss of rRNAs was caused by the inhibition of TUTase activity (Fig. 5B). Most surprisingly, hybridization with the 9S probe visualized an accumulation of molecules exceeding 2,000 nt in length (Fig. 5A, right panel). However, probing for the 12S rRNA's 3′ end (140 nt upstream of 9S rRNA) did not detect these precursors (Fig. 5A, left panel). Hence, the fragment which is removed from the pre-9S rRNA is located downstream from the mature rRNA sequence. The combined abundance of the putative precursor and 9S rRNA declined by ∼50%, suggesting that the loss of 9S rRNA may not be fully accounted for by the inhibition of 9S RNA release from the precursor. Therefore, we next asked if the rebuilding of a U tail by RET1 is required for the stability of processed rRNAs.
FIG. 5.
RET1 activity is required for rRNA processing. (A) Northern blotting analysis of rRNAs in RET1 RNAi and overexpression cell lines. 9S rRNA precursors are indicated by arrows. (B) Quantitation of blotting signals for 9S and 12S rRNA in parasites overexpressing WT and DA RET1 proteins. The mitochondrial rRNA ratio to 5.8S cytoplasmic rRNA in mock-induced cells was assumed to be 100%. A combined abundance of both bands shown by arrows was calculated for 9S pre-rRNA. (C and D) rRNA decay in T. brucei cells depleted of RET1 and KPAP1, respectively. At the time of transcription block, protein levels were reduced by more than 90%, as determined by quantitative immunoblotting. Cells were collected at the indicated time intervals by quick centrifugation. Total RNA was separated on 5% polyacrylamide-8 M urea gel, transferred onto membrane, and sequentially probed for mitochondrial rRNAs and cytoplasmic 5S rRNA. (E) Mapping of 3′ processing intermediates by cRT-PCR. RET1 RNAi was induced for 55 h. PCR products obtained in the diagrammed experiment were separated on a 1.2% agarose gel. DNA fragments indicated by brackets were cloned and sequenced. Arrows show positions of the initial endonucleolytic cleavages at 12S-9S rRNA and 9S rRNA-ND8 mRNA junctions.
The in vivo decay assay was adopted for rRNAs, as follows. RNAi was induced for 40 h to achieve a >90% depletion of RET1, and mitochondrial transcription was then inhibited with ethidium bromide and actinomycin D (Fig. 5C). In the control, both rRNAs declined linearly for 2 h, reaching ∼30% of the initial levels. The residual population remained stable for the remaining 2 h of the assay. A more complex decay pattern was observed in RET1-depleted cells; after an initial drop to ∼70% over a period of 1 h, rRNAs remained stable for another hour. In any event, at the terminal point of the assay, the two RNAs reached similar levels whether or not RET1 was removed by RNAi.
Because rRNAs receive U tails and because mRNAs, presumably excised from the same precursor, are adenylated, we next investigated whether repression of mitochondrial poly(A) polymerase KPAP1 would affect rRNA decay. Adenylation is required for maintenance of some never-edited and edited mRNAs but is dispensable for preedited molecules (17) and even stimulates their decay in vitro (21). Surprisingly, inhibition of KPAP1 led to a moderate stabilization of mature 9S and 12S rRNAs (Fig. 5D). To conclude, it is unlikely that the decline of 9S and 12S rRNAs in RET1-depleted cells was caused by the accelerated decay of nonuridylylated molecules. Our data suggest that the degradation of stalled intermediates, whose processing depends on RET1 activity indirectly, is a primary reason for the loss of rRNAs upon RET1 repression. rRNAs were unaffected by RET2 and MEAT1 RNAi (not shown).
Mature rRNA 3′ ends are produced by endonucleolytic, exonucleolytic, and RET1 activities.
Close spacing of rRNA genes in the maxicircle (Fig. 1) and the size of 9S rRNA 3′ precursors (Fig. 5A) suggest that the two rRNAs are either transcribed as independent units or precisely split in the common precursor. To analyze the consequences of RET1 repression on 5′- and 3′-end processing, total RNA was circularized with T4 RNA ligase and subjected to reverse transcription and PCR amplification in a procedure known as cRT-PCR (Fig. 5E). In the resultant product, any extensions beyond mature termini are flanked with 5′ and 3′ encoded regions. Unexpectedly, RET1 RNAi led to minor lengthening of cRT-PCR products. At the sequencing level, 5′ ends of both rRNAs were homogenous in all PCR products (not shown). The encoded 3′ end of the 12S RNA observed in our experiments was 11 nucleotides longer than previously annotated (40), but still uniform, terminating with 6 to 20 U's (see Fig. S2A [mock panels] and Table S1 in the supplemental material). In contrast, RET1 repression caused an accumulation of molecules terminating with an additional 18 or fewer encoded nucleotides, some of which were uridylylated. The longest unprocessed 3′ end observed in multiple pre-12S rRNA clones matched precisely the 5′ end of the 9S rRNA (Fig. 5E); shortened versions of this linker region were also observed (see Fig. S2A in the supplemental material). At the 9S rRNA-ND8 mRNA junction, the longest, 15-nt-long intermediate extended into the previously mapped ND8 mRNA 5′ untranslated region (UTR) by 6 nucleotides (Fig. 5E). Thus, sequencing analysis indicated a position of the initial endonucleolytic cleavage in the precursor and suggested that the mature 3′ end is created by subsequent exonucleolytic trimming. The lack of a hybridization signal corresponding to 12S rRNA precursor is likely due to its size and/or faster decay and not the 9S rRNA-containing species.
We have not identified a single clone bearing 5′ to 3′ rRNA truncation or extensive 3′ to 5′ degradation intermediates in RET1 RNAi cells. This is consistent with an earlier conclusion that the loss of rRNAs is not elicited by the decreased stability of processed nonuridylylated molecules. Apparently, the mature 5′ ends of both rRNA molecules are generated by a precise endonucleolytic cleavage, whereas the 3′ ends are produced by sequential cleavage, 3′-5′ trimming, and 3′ uridylylation. The fact that U tails are still present in some clones obtained from RET1 RNAi most likely reflects stability of the rRNAs present at the time of RNAi induction. Parallel analysis performed on MEAT1-depleted parasites (see Fig. S2B in the supplemental material) or RET2 RNAi cells (not shown) produced no evidence of rRNA processing or uridylylation defects and, therefore, no indications that MEAT1 or RET2 TUTases may target rRNAs.
It seems plausible that the precise cleavage of the maxicircle precursor between 12S and 9S rRNAs may be also directed by trans-acting antisense RNAs, which require RET1-catalyzed uridylylation for stability. Searches of the KISS database (30) with a wublastn-modified matrix allowing G-U base pairing indeed identified several short patches of complementarity with minicircle-derived sequences. An example of an imperfect duplex that may hybridize in the vicinity of a putative 12S-9S cleavage site is shown in Fig. S2C in the supplemental material.
RET1 functions in mRNA 3′ uridylylation and processing.
RET1 repression ablates gRNAs, thereby inhibiting mRNA editing, but it has been assumed that never-edited mRNAs are not subjected to RET1-dependent alterations. Indeed, the overall abundance of CO1 mRNA is not significantly affected by RET1 RNAi, as determined by poisoned primer extension analysis (5). These experiments, however, do not explain the inhibition of CO1 mRNA translation in RET1 RNAi cells (29). To uncover possible roles of RET1 in mRNA processing and decay, we examined the consequences of RET1 repression on these processes.
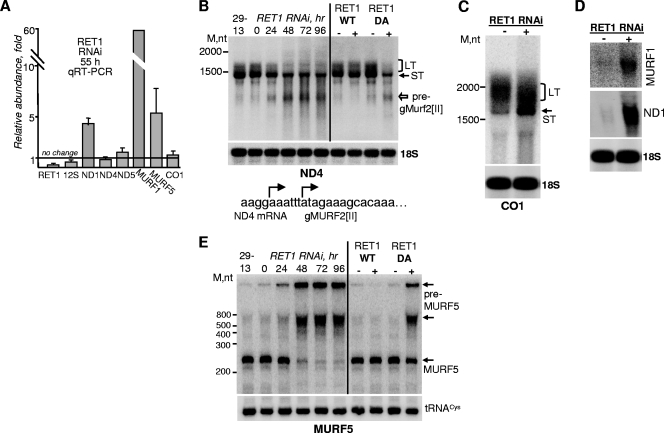
The qRT-PCR survey of all annotated never-edited mRNAs demonstrated that the relative abundance of some transcripts (CO1, ND4, and ND5) remained unchanged or slightly increased, while other mRNAs (ND1, MURF1, and MURF5) were upregulated by ∼5- to 50-fold upon RET1 knockdown (Fig. 6A). Poisoned primer extension and qRT-PCR assays are limited to relatively short regions of targeted RNAs, which prompted us to analyze never-edited transcripts by Northern blotting. Remarkably, hybridization with the ND4 mRNA-specific probe (positions 566 to 766), which corresponds to the downstream probe shown in Fig. 3A, detected a decline of the long-tailed mRNA form and the accumulation of ∼800-nt-long fragments (Fig. 6B). The 5′ end of the ND4 gene is unusual because it contains an intragenic gene for Murf2[II] gRNA. Therefore, the appearance of additional fragments most likely signals the accumulation of a nonprocessed gMurf2[II] precursor. The collapse of the mRNA's long tail, on the other hand, reflects RET1's involvement in the synthesis of this structure. To verify this hypothesis on another transcript, we analyzed CO1 mRNA. As seen from Fig. 6C, a compact 250- to 300-nt-long tail partially collapsed upon RET1 knockdown. These data indicate that at least some never-edited mRNAs possess long tails composed of A's and U's, like those in fully edited RPS12 mRNA (17).
FIG. 6.
RET1 activity is involved in mRNA processing and 3′ uridylylation. (A) qRT-PCR analysis of never-edited mRNAs, 12S rRNA, and RNAi-targeted RET1 transcript after 55 h of RNAi induction. RNA levels were normalized to β-tubulin mRNA. Error bars represent the standard deviation from at least three replicates. The thick line at “1” stands for no change in mRNA's relative abundance, while bars above or below it represent an increase or decrease, respectively. (B) Total RNA from RET1 RNAi and DA cells was separated on a 1.6% formaldehyde agarose gel and probed for ND4 mRNA. mRNAs with long tails (LT) are indicated by brackets; short-tailed (ST) forms are shown by arrows. gRNA and mRNA transcription start sites identified previously (14) are diagrammed below. The white arrow points to a putative gMurf2[II] precursor. (C and D) Total RNA isolated after RET1 RNAi induction for 55 h was analyzed as in panel B. (E) Northern blotting of MURF5 mRNA in RET1 RNAi and DA cells. Total RNA was separated on a 5% polyacrylamide-8 M urea gel.
Similar analysis applied to never-edited mRNAs, whose steady-state abundance increased in the RET1 RNAi cells, produced strikingly different results. In agreement with qRT-PCR data, Northern blotting detected significant upregulation of ND1 and MURF1 mRNAs, but no discernible differences in their gel mobilities (Fig. 6D). Because the KPAP1-added short A tails are required for the maintenance of some never-edited and edited molecules, typified by CO1 and RPS12 mRNAs, respectively, we next determined whether RET1-regulated mRNAs possess short A tails. A cRT-PCR amplification and sequencing of ND1 mRNA's termini provided no evidence for the presence of short A tails (120 clones analyzed). Instead, in mock-induced cells, we observed mostly 3′ degradation intermediates lacking predicted stop codons, while ∼50% of nondegraded UTRs terminated with nonencoded U tails (see Fig. S3 in the supplemental material). In RET1 RNAi cells, more than 80% of all clones contained heterogeneous 3′ UTRs extending beyond the stop codon, but less than 3% were uridylylated. It is possible that ND1 and MURF1 mRNAs possess only long tails, which were undetected by cRT-PCR. To conclude, ND1 and most likely MURF1 represent the first examples of mitochondrial mRNAs whose abundance is negatively affected by 3′ uridylylation. It remains to be established whether this is a developmentally regulated phenomenon; MURF1 mRNA, for example, is significantly upregulated in the bloodstream parasites (10).
We have shown previously that preedited RPS12 mRNA accumulates in RET1-depleted cells. In the same experiment, we observed the appearance of at least two longer molecules that contained preedited RPS12 sequences (17). Blotting analysis of the MURF5 RNA provided another example of disrupted pre-mRNA processing (Fig. 6E). Two longer fragments (∼800 and >2,000 nt) accumulated in RET1 RNAi and the DA cells, while the molecule corresponding to the annotated MURF5's length (234 nt) constituted a minor fraction. The increase in the combined mRNA and pre-mRNA abundance is consistent with qRT-PCR assessment. Finally, KPAP1 RNAi knockdown or RNase H treatment in the presence of oligo(dT) had no effect on MURF5 mRNA gel mobility, suggesting that this RNA, similar to ND1 and MURF1, does not have a short A tail (not shown). In summary, we found that RET1 activity is involved in two distinct modes of mRNA uridylylation. Extension of KPAP1-added short A tails into long A/U tail heteropolymers, typified by CO1 and ND4 mRNA, has no apparent detrimental effects on mRNA abundance, while addition of a continuous U tail downregulates targeted transcripts. In addition, RET1 activity is indirectly involved in the processing of select mRNA precursors.
3′ uridylylation stimulates mRNA decay.
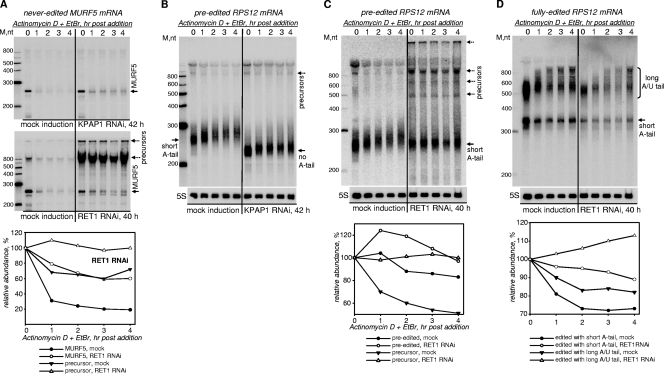
KPAP1 RNAi leads to the decline of most mitochondrial mRNAs (17). To determine whether RET1 activity influences in vivo decay of mRNA precursors, mRNAs lacking short A tail, and adenylated mRNAs, we have adopted the arrested transcription assay to assess mRNA stability in RET1 RNAi cells. MURF5 was chosen as the representative never-edited, nonadenylated transcript whose processing is inhibited by RET1 RNAi. Preedited RPS12 mRNA was selected as an example of a molecule which has only a short A tail, and the fully edited RPS12 represented mRNA populations with both short A tails and long A/U tails.
The processing, length, and stability of the MURF5 transcript were unaffected by KPAP1 RNAi (Fig. 7A, upper panel), which further confirms a lack of 3′ adenylation for this molecule. Upon transcription arrest, MURF5 precursors were efficiently processed in mock-induced cells (>80% decline in 2 h) but remained virtually unchanged in RET1-depleted parasites (Fig. 7A, lower panel). Surprisingly, the decay of processed MURF5 RNA—although this molecule is less abundant in RET1 RNAi cells because of blocked processing—was decelerated by RET1 RNAi. In fact, the RET1-induced degradation may have been underestimated in this assay because of the concurrent precursor processing, which was replenishing the mature molecules in mock-induced cells. We conclude that the steady-state levels of ND1, MURF1, and MURF5 transcripts, unlike most other mitochondrial mRNAs, are downregulated via uridylylation-stimulated 3′-5′ mRNA degradation.
FIG. 7.
RET1-catalyzed 3′ uridylylation promotes mRNA decay. (A) MURF5 mRNA decay in T. brucei cells depleted of KPAP1 (upper panel) and RET1 (lower panel). After RET1 RNAi or KPAP1 RNAi induction, actinomycin D and ethidium bromide were added to inhibit transcription. Total RNA was isolated from cells collected at the indicated time points and separated on a denaturing 5% polyacrylamide gel. Quantitation was performed in reference to cytoplasmic 5S rRNA (normalization panels are shown in panels B and C). The graph below Northern blotting panels represents relative abundances at each time point, which were calculated assuming the mRNA/5S rRNA ratio in mock-induced cells at the time of actinomycin D addition to be 100%. (B and C) Preedited RPS12 mRNA decay in KPAP1 and RET1 RNAi cells, respectively. Total RNA was isolated from KPAP1- and RET1-depleted cells at the indicated time points after the transcriptional arrest and hybridized with a probe against preedited RPS12 mRNA. Positions of pre-RPS12 mRNA are indicated by arrows. (D) Fully edited RPS12 mRNA decay in RET1 RNAi cells. The same membrane as those in panels B and C was hybridized with a probe specific for a fully edited RPS1 mRNA.
KPAP1 RNAi led to an accumulation of nonadenylated preedited RPS12 RNA (Fig. 7B, time zero). Under the conditions of transcriptional arrest, the editing apparently proceeded throughout the assay, causing multiple internal U additions, as reflected by the lengthening of the molecule and a decreasing hybridization signal. In agreement with our previous model, the KPAP1-added short A tail is required to maintain RNAs in which editing has advanced beyond the first editing block (17). Indeed, nonadenylated mRNAs edited by insertion of only a few U's were no longer detectable in the KPAP1 RNAi cells (Fig. 7B, right panel). Although the concomitant editing and rapid degradation of edited nonadenylated mRNAs prevented decay assessment, these results further establish that both the editing and adenylation states of the mRNA contribute to its stability in the mitochondria.
As expected, RET1 RNAi inhibited precursor processing as well as the editing process. Consequently, upon transcriptional blockage, the preedited mRNA was neither produced nor converted into edited form (Fig. 7C). Based on minor changes in its relative abundance, we conclude that although one or two U's are typically found scattered within short A tails (15, 17), the RET1-catalyzed uridylylation does not significantly affect the stability of preedited mRNAs.
Although the editing process is blocked in RET1 RNAi cells, the residual fully edited mRNAs with short A tails and long A/U tails declined more slowly than did the corresponding populations in mock-induced cells (Fig. 7D). The electrophoretic mobilities of short-A-tailed molecules were identical in the two panels, while long-tailed molecules were marginally shorter in RET1 RNAi cells at time zero. Collectively, these data demonstrate that RET1 is responsible for U incorporation into A/U tails found on fully edited mRNAs. The destabilizing effects of random U addition, however, are less dramatic than short-U-tail-induced downregulation of never-edited ND1, MURF1, and MURF5 transcripts.
DISCUSSION
In this study, we report that RNA uridylylation reactions catalyzed by the founding member of the TUTase family, RNA editing TUTase 1, are directly responsible for adding short U tails to gRNAs, rRNAs, and select mRNAs. RET1 also incorporates uracil residues into long A/U tails, which are found on most mitochondrial mRNAs. Furthermore, we show that RET1's TUTase activity is indirectly involved in the nucleolytic processing of precursor molecules for all major structural and functional RNA classes encoded by the mitochondrial genome of T. brucei.
3′ uridylylation and nucleolytic processing of kinetoplast DNA (kDNA) transcripts.
It is well accepted that a single-subunit RNA polymerase transcribes both strands of the maxicircle DNA from undefined promoters and produces multicistronic transcripts (Fig. 8). The mature forms of adjacent transcripts often overlap within a precursor, indicating that alternative cleavage produces the 3′ end of the upstream RNA at the expense of the 5′ region in the downstream unit and vice versa. The nature of the processing nuclease is unknown, yet we were surprised to observe accumulation of distinct precursors containing 9S rRNA, some preedited and never-edited mRNAs, and gRNAs in RET1-depleted parasites. Persistent attempts to detect RET1's nuclease activity were unsuccessful (I. Aphasizheva, unpublished data), but the possibility of such intrinsic activity remained. Here, we have used a single-amino-acid dominant negative mutation to demonstrate that nucleolytic processing depends on RET1's TUTase (poly[U] polymerase) activity.
As expected, the growth inhibition phenotype induced by RET1 DA overexpression was less severe than that of RNAi. This is consistent with a more-modest manifestation of the dominant negative effects at the RNA level but does not exclude a possibility that RET1 targets RNA species which have not been analyzed here. Nonetheless, it appears that the processing defects triggered by RET1 repression are indirect and are caused by inhibited uridylylation of RNAs which are required for the nucleolytic cleavage.
We propose that the nucleolytic processing of the multicistronic RNA precursors in kinetoplast mitochondria is directed (guided) by short, gRNA-like antisense molecules. This would explain the alternative cleavage of precursors by stochastic binding of different antisense RNAs. It is plausible that these RNAs are stabilized by RET1-catalyzed 3′ uridylylation and are rapidly degraded in RET1-depleted cells. The identity of the processing nuclease and its possible association with editing complexes (18) await further investigation.
gRNA genes may act as bidirectional promoters.
Although the mechanistic link between the antisense transcripts and pre-gRNA processing remains to be firmly established, our results suggest that the double-stranded region formed by the overlapping 5′ ends of the opposing 600- to 800-nt transcription units may constitute a substrate for the processing nuclease (Fig. 8). The opposite effects of RET1 repression on the pre-gRNA A6[14] and long antisense transcript indicate that the RET1 activity is required to maintain the latter molecules. The hybridization properties of DNA probes for both gRNA- and antisense-containing sequences are virtually identical; thus, it appears that the gRNA-sized antisense RNAs are present at a much lower level. In fact, asymmetric degradation of antisense molecules would be required to liberate functional gRNA. The increase of short antisense RNAs in RET1 DA, but not in RNAi, cells (Fig. 3F) led us to speculate that these molecules, which are normally rapidly turned over by the U tail addition, are protected from degradation because of binding to the inactive RET1. Therefore, our data suggest that both gRNA and gRNA-sized antisense transcript are uridylylated following the cleavage reaction but then enter different processing pathways—gRNAs are channeled to the editing complex, and antisense molecules are targeted for degradation.
The source of antisense RNAs for maxicircle transcript processing remains speculative since they can be produced by transcription in the vicinity of cleavage sites or encoded elsewhere in the genome. Relatively few events would be required to split maxicircle-encoded multicistronic precursors into two rRNAs and ∼18 pre-mRNAs. Indeed, searches of the existing databases identified short matches between minicircle DNA sequences and the cleavage sites in maxicircle transcripts (see Fig. S2C in the supplemental material). However, this mechanism is less realistic for hundreds, if not thousands, of minicircle-encoded pre-gRNAs which accumulate on a global scale in RET1-depleted cells (Fig. 3E). It is possible that the gRNA gene may be transcribed in both directions such that antisense transcripts overlap with their 5′ ends. In this scenario, the 5′ end of an antisense molecule would define a cleavage site in pre-gRNA and vice versa. Considering the length of gRNA precursors (600 to 800 nt), these transcription units traverse almost the entire minicircle. Because T. brucei minicircles encode up to five gRNAs, the primary transcript must contain more than one gRNA sequence. It remains to be established whether the 5′ triphosphate is necessary for gRNA's function and/or stability. If this is indeed the case, then only the most 5′-end gRNA can be processed into a functional molecule. In any event, the metabolic fate of the 3′ trailer remains to be elucidated.
Functions of the U tail.
The majority of gRNAs were identified by comparing edited mRNAs and the corresponding pseudogenes. On the other hand, direct sequencing of short RNAs from mitochondria of Leishmania tarentolae (24) and T. brucei (25, 30) showed that gRNAs specifying known editing sites constitute only a fraction of the total short RNA population and that most short RNAs are uridylylated. The two established components of gRNA biogenesis, RET1 and the GRBC1/2 complex, interact in an RNA-dependent manner, and their knockdowns ablate mature gRNAs (4, 44). Here, we demonstrated that RET1 is involved in gRNA maturation indirectly at the nucleolytic processing step and directly through U tail addition. The disruption of the former process accounts for the gRNA loss in RET1 RNAi, while inhibition of the latter reaction does not affect gRNA stability. The RNA binding properties of the reconstituted purified GRBC1/2 complex, combined with the results of our previous studies (44), indicate that this RNA binding particle is not involved in the precursor processing. Instead, binding to GRBC1/2 is likely to be required to prevent gRNA's rapid degradation regardless of its 3′ uridylylation status.
Interplay of mRNA adenylation and uridylylation.
The first indications that uridylylation may promote mRNA decay came from reports of the UTP-stimulated degradation of adenylated RNAs in organello (26, 37). This phenomenon was no longer observed in the mitochondrial fraction isolated from RET1-depleted parasites (38). Identification of mitochondrial poly(A) polymerase demonstrated that short A tails are required and sufficient for maintenance of most mRNAs. Cloning of long A/U heteropolymers, however, indicated that both poly(A) polymerase and TUTase activities are involved in synthesis of these structures (17). Here, we present evidence that RET1 participates in two distinct modes of mRNA 3′ uridylylation: formation of long A/U tails and transcript-specific addition of continuous short U tails.
Because RET1 RNAi not only ablates editing but also inhibits processing, thereby affecting abundance of preedited, never-edited, and edited mRNAs regardless of their 3′-end status, we resorted to in vivo stability assays to demonstrate that uridylylation generally accelerates mRNA decay. The U depletion from long A/U tails caused their partial collapse, and a moderate stabilization of respective mRNA species. RET1 RNAi triggered the significant increase in the steady-state levels of ND1 and MURF1 transcripts and caused decelerated decay of the MURF5 mRNA. This designates a transcript-specific addition of continuous short U tails as a mechanism of selective posttranscriptional gene repression. Hence, stimulating RET1 activity in organello by adding UTP may have led to a nonspecific mRNA 3′ uridylylation, followed by rapid degradation. It is not clear whether UTP-stimulated mRNA decay represents a biologically relevant regulatory circuit (26, 38), but it is evident that the nature of RET1 activators and inhibitors needs further investigation.
Conclusion.
The understanding of RNA uridylylation as a ubiquitous mechanism of eukaryotic gene regulation is still in its early stages. Several TUTase activities have been characterized to different extents in human cells (reviewed in reference 19). The best-studied example is U6 snRNA uridylylation, which presumably generates a binding site for SM-like proteins (42, 43). Report of the uridylylation-triggered degradation of histone mRNAs in the cytoplasm revealed how the abundance of these nonadenylated transcripts correlates with DNA synthesis (27). The alternative pathway of pre-let-7 microRNA (miRNA) maturation, which blocks processing by Dicer and stimulates precursor degradation, has been linked to the addition of 10 to 15 U's (20). In plants, U addition precedes degradation of the mRNA's fragment produced by the miRNA-directed cleavage (39). Finally, Cid1-catalyzed transcript-specific uridylylation of polyadenylated mRNAs stimulates decapping by recruiting the Lsm1-7 complex in Schizosaccharomyces pombe (35). The pervasive influence of uridylylation on the mitochondrial transcriptome in one of the most ancient eukaryotic organisms, T. brucei, may reflect the interplay of poly(A) and poly(U) polymerases at the RNA's 3′ end as a primordial means of regulating gene expression.
Supplementary Material
Acknowledgments
We thank members of our laboratory and Yongsheng Shi for stimulating discussions and reading the manuscript.
This work was supported by the NIH grant AI064653 to R.A.
Footnotes
Published ahead of print on 19 January 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adler, B. K., M. E. Harris, K. I. Bertrand, and S. L. Hajduk. 1991. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 11:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, G. Gao, A. M. Simpson, X. Kang, A. M. Falick, S. Sbicego, and L. Simpson. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, and L. Simpson. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9:62-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aphasizhev, R., I. Aphasizheva, and L. Simpson. 2003. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 100:10617-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. [DOI] [PubMed] [Google Scholar]
- 6.Aphasizhev, R., and L. Simpson. 2001. Isolation and characterization of a U-specific 3′-5′ exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276:21280-21284. [DOI] [PubMed] [Google Scholar]
- 7.Aphasizheva, I., R. Aphasizhev, and L. Simpson. 2004. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J. Biol. Chem. 279:24123-24130. [DOI] [PubMed] [Google Scholar]
- 8.Aphasizheva, I., G. E. Ringpis, J. Weng, P. D. Gershon, R. H. Lathrop, and R. Aphasizhev. 2009. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA 15:1322-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat, G. J., D. J. Koslowsky, J. E. Feagin, B. L. Smiley, and K. Stuart. 1990. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61:885-894. [DOI] [PubMed] [Google Scholar]
- 10.Bhat, G. J., A. E. Souza, J. E. Feagin, and K. Stuart. 1992. Transcript-specific developmental regulation of polyadenylation in Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 52:231-240. [DOI] [PubMed] [Google Scholar]
- 11.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 12.Blum, B., and L. Simpson. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell 62:391-397. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Clement, S. L., M. K. Mingler, and D. J. Koslowsky. 2004. An intragenic guide RNA location suggests a complex mechanism for mitochondrial gene expression in Trypanosoma brucei. Eukaryot. Cell 3:862-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker, C. J., and B. Sollner-Webb. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61:1001-1011. [DOI] [PubMed] [Google Scholar]
- 16.Ernst, N. L., B. Panicucci, R. P. Igo, Jr., A. K. Panigrahi, R. Salavati, and K. Stuart. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell 11:1525-1536. [DOI] [PubMed] [Google Scholar]
- 17.Etheridge, R. D., I. Aphasizheva, P. D. Gershon, and R. Aphasizhev. 2008. 3′ Adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 27:1596-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grams, J., M. T. McManus, and S. L. Hajduk. 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 19:5525-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guschina, E., and B. J. Benecke. 2008. Specific and non-specific mammalian RNA terminal uridylyl transferases. Biochim. Biophys. Acta 1779:281-285. [DOI] [PubMed] [Google Scholar]
- 20.Heo, I., C. Joo, J. Cho, M. Ha, J. Han, and V. N. Kim. 2008. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32:276-284. [DOI] [PubMed] [Google Scholar]
- 21.Kao, C. Y., and L. K. Read. 2005. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell. Biol. 25:1634-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koller, J., U. F. Muller, B. Schmid, A. Missel, V. Kruft, K. Stuart, and H. U. Goringer. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749-3757. [DOI] [PubMed] [Google Scholar]
- 23.Koslowsky, D. J., and G. Yahampath. 1997. Mitochondrial mRNA 3′ cleavage polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 90:81-94. [DOI] [PubMed] [Google Scholar]
- 24.Madej, M. J., J. D. Alfonzo, and A. Huttenhofer. 2007. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 35:1544-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madej, M. J., M. Niemann, A. Huttenhofer, and H. U. Goringer. 2008. Identification of novel guide RNAs from the mitochondria of Trypanosoma brucei. RNA Biol. 5:84-91. [DOI] [PubMed] [Google Scholar]
- 26.Militello, K. T., and L. K. Read. 2000. UTP-dependent and -independent pathways of mRNA turnover in Trypanosoma brucei mitochondria. Mol. Cell. Biol. 20:2308-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen, T. E., and W. F. Marzluff. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22:50-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller, U. F., L. Lambert, and H. U. Goringer. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 20:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nebohácová, M., D. A. Maslov, A. M. Falick, and L. Simpson. 2004. The effect of RNA interference down-regulation of RNA editing 3′-terminal uridylyl transferase (TUTase) 1 on mitochondrial de novo protein synthesis and stability of respiratory complexes in Trypanosoma brucei. J. Biol. Chem. 279:7819-7825. [DOI] [PubMed] [Google Scholar]
- 30.Ochsenreiter, T., M. Cipriano, and S. L. Hajduk. 2007. KISS: the kinetoplastid RNA editing sequence search tool. RNA 13:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochsenreiter, T., M. Cipriano, and S. L. Hajduk. 2008. Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PLoS ONE 3:e1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsenreiter, T., and S. L. Hajduk. 2006. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 7:1128-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panigrahi, A. K., A. Schnaufer, N. L. Ernst, B. Wang, N. Carmean, R. Salavati, and K. Stuart. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paris, Z., M. A. Rubio, J. Lukes, and J. D. Alfonzo. 2009. Mitochondrial tRNA import in Trypanosoma brucei is independent of thiolation and the Rieske protein. RNA 15:1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rissland, O. S., and C. J. Norbury. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16:616-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan, C. M., C. Y. Kao, D. A. Sleve, and L. K. Read. 2006. Biphasic decay of guide RNAs in Trypanosoma brucei. Mol. Biochem. Parasitol. 146:68-77. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, C. M., K. T. Militello, and L. K. Read. 2003. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 278:32753-32762. [DOI] [PubMed] [Google Scholar]
- 38.Ryan, C. M., and L. K. Read. 2005. UTP-dependent turnover of Trypanosoma brucei mitochondrial mRNA requires UTP polymerization and involves the RET1 TUTase. RNA 11:763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, B., and H. M. Goodman. 2004. Uridine addition after microRNA-directed cleavage. Science 306:997. [DOI] [PubMed] [Google Scholar]
- 40.Sloof, P., J. Van den Burg, A. Voogd, R. Benne, M. Agostinelli, P. Borst, R. Gutell, and H. Noller. 1985. Further characterization of the extremely small mitochondrial ribosomal RNAs from trypanosomes: a detailed comparison of the 9S and 12S RNAs from Crithidia fasciculata and Trypanosoma brucei with rRNAs from other organisms. Nucleic Acids Res. 13:4171-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiemann, O. H., and L. Simpson. 1996. Analysis of the 3′ uridylylation sites of guide RNAs from Leishmania tarentolae. Mol. Biochem. Parasitol. 79:229-234. [DOI] [PubMed] [Google Scholar]
- 42.Trippe, R., E. Guschina, M. Hossbach, H. Urlaub, R. Luhrmann, and B. J. Benecke. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12:1494-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trippe, R., B. Sandrock, and B. J. Benecke. 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 26:3119-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng, J., I. Aphasizheva, R. D. Etheridge, L. Huang, X. Wang, A. M. Falick, and R. Aphasizhev. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell 32:198-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickstead, B., K. Ersfeld, and K. Gull. 2002. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 125:211-216. [DOI] [PubMed] [Google Scholar]
- 46.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.