Abstract
Both cis-diamminedichloroplatinum(II) (cisplatin or cis-DDP) and trans-diamminedichloroplatinum(II) form covalent adducts with DNA. However, only the cis isomer is a potent anticancer agent. It has been postulated that the selective action of cis-DDP occurs through specific binding of nuclear proteins to cis-DDP-damaged DNA sites and that binding blocks DNA repair. We find that a very abundant nuclear protein, the linker histone H1, binds much more strongly to cis-platinated DNA than to trans-platinated or unmodified DNA. In competition experiments, H1 is shown to bind much more strongly than HMG1, which had been previously considered a major candidate for such binding in vivo.
Keywords: anticancer drugs, DNA adducts, HMG1, linker histones
cis-Diamminedichloroplatinum(II) (cisplatin or cis-DDP) is a potent chemotherapeutic agent widely used in the clinical practice to treat several types of human malignancies. The therapeutic effect is believed to arise as a consequence of cis-DDP binding to DNA (1), but it cannot be solely explained on the basis of DNA binding because a number of closely related compounds, among which is included the geometric isomer trans-DDP, are not effective agents although they also damage DNA. The differential biological effect of different Pt compounds may lie in the differential processing of different Pt-DNA adducts by the cell. The realization that different adducts may be processed differently suggested that certain proteins might enhance or block DNA repair by specifically interacting with cis-DDP-modified DNA; this has focused research toward identifying such proteins.
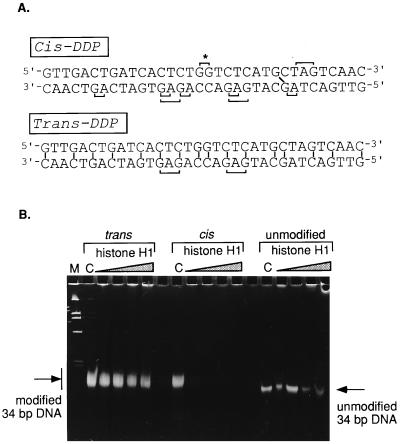
cis-DDP causes the formation of two major intrastrand DNA adducts, 1,2-d(GpG) and d(ApG) cross-links in which the two chloride ions of cis-DDP are replaced by the N7 atoms of guanine and adenine. An intrastrand cross-link may also be formed, at a much lower frequency, at d(GpXpG), where X is any base. Other minor adducts may also arise, including interstrand cross-links involving guanine residues on opposite strands. Biochemical and structural analyses of both intra- and interstrand cis-DDP adducts (2, 3) reveal major distortions of the DNA double helix, including bending and unwinding (for review, see ref. 4). The therapeutically inactive trans-DDP is incapable of forming the 1,2-(GpG) and d(ApG) adducts for stereochemical reasons. It does form 1,3-intrastrand links and also cross-links opposite strands, but although cis-DDP reacts with guanine residues in d(GpC), the trans-isomer preferentially cross-links complementary guanine and cytosine residues (5). The different kinds of cross-links created by the cis- and trans-DDP are illustrated schematically for one specific DNA fragment used in this work (see Fig. 2A).
Figure 2.
(A) Nucleotide sequence of the double-stranded 34-bp fragment used as a substrate for modification. The possible sites of intra- and interstrand cross-linking by either cis-DDP or trans-DDP are marked on the sequence. (B) Electrophoretic analysis of the binding of increasing amounts of histone H1 to trans-modified, cis-modified, or unmodified 34-bp fragments. Lanes C contain the respective DNA controls without H1; the successive lanes contain increasing amounts of histone H1 (expressed as one molecule of H1 per number of bp) as follows: 1/64, 1/32, 1/16, and 1/10. Lane M contains pUC19/HinfI marker.
Recent years have witnessed the discovery of a number of cellular proteins, mostly with still unidentified in vivo functions, that recognize and bind selectively to cis-DDP-modified DNA. These include the relatively abundant chromatin nonhistone proteins HMG1 and HMG2 (6, 7), the human structure-specific recognition protein 1 SSRP1 (8, 9), the yeast intrastrand cross-link recognition protein Ixr1 (10), and human UBF (11). All these proteins belong to the HMG1-box protein family in that they contain one or multiple copies of a specific DNA-binding motif, HMG1 box, first described in the HMG1 and HMG2 proteins (for review, see ref. 12). The HMG1 box is known to bind to distorted DNA structures and bend and unwind DNA upon binding (13–16).
Herein we present evidence that an even more abundant chromatin protein, the linker histone H1, also binds preferentially to cis-Pt-modified DNA. Linker histones, so named because of their binding to linker DNA between successive nucleosomes in the chromatin fiber, share with HMG1/2 many DNA binding properties (for a recent review, see ref. 17). Thus, for example, they unwind DNA (18) and bind preferentially to four-way junction DNA (19, 20). The linker histones compete with HMG1 for four-way junction DNA, exhibiting higher affinity of binding (20, 21). These similarities in DNA binding properties and the observation that HMG1 and 2 preferentially bind to cis-DDP-modified DNA prompted us to examine histone H1 with respect to its binding to cis- and trans-DDP-modified DNA.
MATERIALS AND METHODS
Chemical Reagents.
Platinum compounds (cis- and trans-DDP) were purchased from Sigma. Restriction endonuclease AvaI was obtained from New England BioLabs. The 123-bp DNA ladder was purchased from GIBCO/BRL Life Technologies.
DNA Samples and Chemical Modifications.
The 123-bp fragment was purified from 1.5% agarose gels by using the fragment isolation kit from Qiagen (Chatsworth, CA), after AvaI digestion of the 123-bp DNA ladder. Single-strand DNA fragments 34 nucleotides long were custom-synthesized with an Applied Biosystems 380B DNA synthesizer using the phosphoramidite technique and annealed to form double-stranded DNA fragments. DNA concentration was determined spectrophotometrically using an extinction coefficient of 20 (ml per cm per mg, at 260 nm). The purity of the oligomer was checked by either 15% polyacrylamide gel or 3.3% agarose gel electrophoresis (22). Modification of DNA samples was performed in 10 mM Tris⋅HCl, pH 7.5/5 mM NaCl containing DNA (0.1 μg/μl) and Pt compounds (stock solutions at 10−4 M, kept at 0–4°C for 2–3 weeks) added at an input molar ratio of 0.025 of DDP per nucleotide. This is a commonly used ratio (e.g., ref. 6), and has, in preliminary experiments, resulted in a saturation level of modification by both the cis and trans isomers. The saturation level was defined as the concentration of DDP at which no further retardation and broadening of the modified DNA bands was observed upon electrophoresis in polyacrylamide gels (e.g., Fig. 1). Incubation was for 18–24 h in the dark at 37°C. The modified samples were used within 1 week after modification.
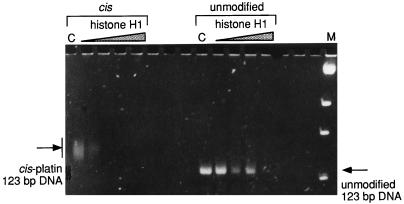
Figure 1.
Binding of histone H1 to isolated 123-bp fragment, either cis-modified or unmodified. Lanes C contain the respective DNA control; the successive lanes contain increasing amounts of histone H1 (expressed as one molecule of H1 per number of bp) as follows: 1/160, 1/64, 1/32, and 1/16. Lane M contains pUC19/BstNI marker.
Purification of Proteins.
Histone H1 was isolated from mouse liver nuclei under nondenaturing conditions on CM Sephadex C25 columns (23). HMG1 from chicken erythrocytes was obtained from a 0.5 M NaCl extract of crude nuclei by chromatography on a Polybuffer-exchange column (PBE94, Pharmacia) according to ref. 24. Protein concentration was determined spectrophotometrically by using the following extinction coefficients: 1.85 (ml per cm per mg, at 230 nm) for H1 (25) and 1.0 (ml per cm per mg, at 280 nm) for HMG1 (26). The purity of protein preparations was verified by SDS/PAGE (27).
Protein–DNA Binding Assay.
The binding reactions were performed in 15-μl mixtures containing 0.2 μg of DNA, 10 mM Tris⋅HCl (pH 7.8), 20 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, and increasing amounts of protein. Incubation was for 30 min at room temperature. Samples were analyzed on nondenaturing polyacrylamide gels in TAE buffer [40 mM Tris⋅HCl, pH 8.3/25 mM acetate/1 mM EDTA (22)]; current was kept below 15 V/cm to avoid overheating. DNA was visualized by staining with either ethidium bromide or silver. Gels were photographed by using either black/white no. 667 positive or type 55 positive/negative Polaroid films (Imaging Products, Simi Valley, CA).
Competition Experiments.
For oligonucleotide competition, 10 ng of radioactively labeled cis-modified 34-bp fragment was incubated with 10 ng of histone H1 in the absence or presence of increasing amounts of unlabeled competitor 34-bp fragment, as specified in Fig. 3. The binding of H1 to the radioactive fragment was monitored by gel electrophoresis in 3% agarose gels and autoradiography. For competition experiments between H1 and HMG1, 1 μg of cis-DDP-modified 34-bp fragment was incubated with 2 μg of HMG1 in 5 mM Tris⋅HCl, pH 7.8/10 mM NaCl/0.05 mM phenylmethylsulfonyl fluoride for 60 min at room temperature. The mixtures were then incubated with increasing amounts of H1 for 60 min and analyzed by 15% nondenaturing polyacrylamide gels in 20 mM Tris⋅HCl, pH 8.3/12.5 mM acetate/0.5 mM EDTA.
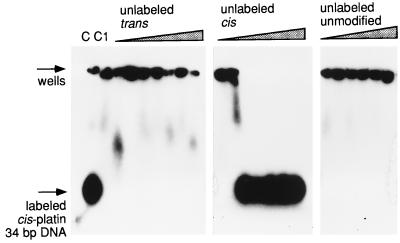
Figure 3.
Competition between unlabeled trans-DDP-modified, cis-DDP-modified, and unmodified 34-bp fragments for binding to histone H1 bound to labeled cis-DDP-modified 34-bp fragment. Lane C contains the labeled cis-DDP-modified 34-bp fragment only; lane C1 contains the same fragment bound to histone H1, in the absence of competitor DNA. The amount (fold molar excess) of cis- and trans-competitor DNA in consecutive lanes is as follows: 1, 5, 10, 20, 50, 100, and 200; the unmodified competitor was added at 5-, 10-, 20-, 100-, and 200-fold excess.
We have defined the apparent dissociation constants of H1 or HMG1 binding to cis-modified DNA as the protein concentration at which half of the DNA remains in the unbound state. At least in the case of the H1-DNA reaction, this cannot represent a true dissociation constant because the reaction is much more complex than a simple 1:1 association. Nevertheless, the apparent values should indicate the relative affinities of the two proteins for the cis-damaged DNA.
RESULTS AND DISCUSSION
Histone H1 Binds Preferentially to cis-DDP-Modified DNA.
On the basis of a number of existing similarities in the binding of HMG1,2 and linker histones (H1, H5, and their like) to DNA, we hypothesized that these histones may also preferentially bind to cis-platinated DNA. To that end, we studied the interaction of histone H1 with a number of globally platinated DNA fragments by deoxyribonucleoprotein gel electrophoresis.
The first fragment of choice was the commercially available 123-bp DNA ladder that has been used as a substrate for platination in earlier work (6, 28). The globally cis-platinated fragment is modified at multiple sites, as evidenced by the retardation and significant broadening of the band of the modified fragment in polyacrylamide gels (Fig. 1, compare lanes 1 and 9). When the globally cis-modified and unmodified 123-bp fragments were titrated with increasing amounts of histone H1, a very strong preference for the histone to bind to the platinum-modified fragment was observed. This preference was usually recognized as a disappearance of the fragment from the gel, with ethidium bromide-stained material trapped in the wells. The formation of insoluble H1–DNA complexes is a frequently observed phenomenon (refs. 29 and 30 and earlier work reviewed in ref. 31) and is probably related to the existence of two DNA binding sites in the globular domain of the protein (32) that facilitates the formation of insoluble network aggregates between the protein and the DNA.
The titration experiments were repeated with a synthetic 34-bp fragment that contains the sequence used by Lippard and colleagues (2) in their crystallographic study of the structure of a defined cis-platinated dodecamer modified at one specific site, d(GpG), at the center of the fragment. The dodecamer was elongated on both the 5′ and 3′ ends with randomly selected sequences to introduce additional sites of platination on both strands and sites of interstrand modifications (see Fig. 2A for possible sites of modification). This was done to enhance the sensitivity of detection by providing for multiple sites for both cis-DDP and trans-DDP binding.
When the 34-bp synthetic fragment was modified with either cis- or trans-Pt and then titrated with increasing amounts of H1, we saw a clear preference for H1 binding to the cis-platinated fragment over the trans-platinated one: the former disappeared from the gel very early during the titration, but the latter did not seem to change much in intensity even at the highest protein/DNA ratio tested (Fig. 2B). That the trans-platination reaction had been successful was evident from the fact that the electrophoretic mobility of the trans-DDP-treated fragment was reduced and the band was significantly broadened as compared with the unmodified DNA band, as would be expected on the basis of the presence of multiple modified sites. The band corresponding to the unmodified fragment diminished slightly in intensity even at relatively high histone/DNA ratios, indicating much weaker histone binding than that of the cisplatinated fragment.
Competition Between Modified and Unmodified DNA Fragments for H1 Binding.
The preference detected in incubation mixtures that contained only one type of DNA (unmodified or cis- or trans-platinated) was confirmed in direct competition experiments. In these experiments, a histone H1–DNA complex was formed between radioactively labeled cis-platinated 34-bp fragment in the absence or presence of increasing amounts of the same unlabeled fragment that had been (i) cis-platinated, (ii) trans-platinated, or (iii) unmodified. The results (Fig. 3) show that in the absence of competitor DNA, the radioactively labeled cis-modified 34-bp fragment was shifted to the well by addition of H1. This shift could not be reversed by addition of up to a 200-fold excess of unlabeled unmodified or trans-modified fragments. In contrast, the radioactivity in the wells moved back to the position of the free oligonucleotide if the competition was performed with even low ratios of unlabeled cis-platinated fragment. Almost 50% of the bound cis-modified 34-bp fragment was competed out by an approximately 3-fold excess of the same unlabeled fragment; at 5-fold excess, the competition was complete. We do not believe that the absolute concentrations necessary for each competition can be simply interpreted in such an assay, because one is attempting to compete out DNA fragments forming complex insoluble protein–DNA networks with soluble fragments. Nevertheless, the relative values obtained with the differently modified DNAs clearly signify a much higher affinity of H1 for the cis-DDP-modified DNA than for either the trans-DDP-modified or unmodified DNA of the same sequence.
Competition Between H1 and HMG1 for Binding to cis-Platinated DNA Fragments.
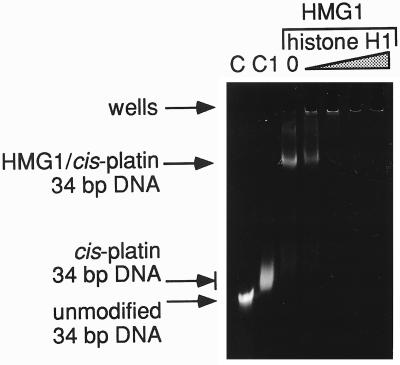
How does the affinity of binding of H1 to cis-platinated DNA compare with that of HMG1? To answer this question, we performed direct competition experiments between the two proteins. We made use of the observation that under the conditions of our assay, HMG1 can form soluble complexes that can be readily observed as a band shift (Fig. 4, lane 3). On the other hand, H1 forms highly aggregated complexes that do not enter such gels. Thus, a competition between the two proteins can be monitored as a transfer of DNA between the soluble HMG1-mediated complex and the H1-mediated complex in the well. A complex was formed between HMG1 and cis-platinated 34-bp fragment; this was further incubated with increasing amounts of H1. As seen in Fig. 4, H1 competed very effectively against HMG1 for the modified fragment. Even at a 1:20 ratio of H1 to HMG1, a substantial portion of the DNA is displaced from the HMG1-shifted band into larger aggregation (Fig. 4, lane 4). At a 1:4 ratio, competition is complete (Fig. 4, lane 5). This result corroborates the independent estimates for the apparent dissociation constants obtained for both HMG1 and for H1 binding from the types of experiments shown in Figs. 1 and 2 (2 × 10−6 and 1 × 10−7 M, respectively).
Figure 4.
Competition between HMG1 and H1 for cis-DDP-modified 34-bp fragment. Lanes C and C1 contain the unmodified 34-bp DNA and the cis-DDP-modified 34-bp DNA, respectively. The successive lanes contain the HMG1/cis-DDP-modified 34-bp complex with a molar ratio of H1 to HMG1 of 0, 0.06, 0.3, 0.6, and 1.2.
The higher affinity of cis-DDP-modified DNA for linker histone as compared with the HMG1/2 class, together with the much higher concentration of this histone in the nucleus (33) leads to the conclusion that linker histones, rather than HMG1/2 proteins will be the most likely occupants of sites of cis-DDP-DNA lesions in the cell.
What is the possible physiological significance of such a preference of linker histone for cis-DDP-modified DNA? It has been argued that some of the nuclear proteins (see Introduction) that have been shown to preferentially bind to cis-DDP-modified DNA might act to block repair of the damaged DNA. This might explain the specific lethality of the cis isomer. Of all the proteins that have now been shown to have this preference, linker histones are by far the most abundant in the nucleus. Furthermore, there is suggestive evidence that H1 levels in the nucleus are under dynamic regulation (for review, see ref. 34) and that the relatively large cytoplasmic pool of this histone combined with its fast transmembrane transport (35, 36) may serve as a readily available source of the protein even in the absence of its synthesis. It may be of relevance to note that the only other abundant chromatin protein stored in the cytoplasm is HMG1/2 (37, 38), and a role for this protein in the biological activity of cis-Pt has been suggested by several groups.
Although these experiments do not distinguish possible differences in H1 affinity between particular types of cis-Pt adducts, they clearly establish the point that H1 is very strongly bound to some of these. The fact that this binding is stronger than that of the HMG1/2, hitherto the prime candidate for in vivo protection of cis-DDP adducts from repair, should probably refocus investigations into the mode of action of cis-Pt as an anticancer agent.
Acknowledgments
This research was supported by National Institutes of Health Grant GM50276 and by Fogarty International Research Collaboration Award (FIRCA TW00568-02) to K.v.H. and J.Z.
ABBREVIATION
- cis-DDP
cis-diamminedichloroplatinum(II)
References
- 1.Sherman S E, Lippard S J. Chem Rev. 1987;87:1153–1181. [Google Scholar]
- 2.Takahara P M, Rosenzweig A C, Frederick C A, Lippard S J. Nature (London) 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 3.Yang D, van Boom S S G E, Reedijk J, van Boom J H, Wang A H-J. Biochemistry. 1995;34:12912–12920. doi: 10.1021/bi00039a054. [DOI] [PubMed] [Google Scholar]
- 4.Lilley D M J. J Biol Inorg Chem. 1996;1:189–191. [Google Scholar]
- 5.Kaspárková J, Brabec V. Biochemistry. 1995;34:12379–12387. doi: 10.1021/bi00038a035. [DOI] [PubMed] [Google Scholar]
- 6.Pil P M, Lippard S J. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 7.Hughes E N, Engelsberg B N, Billings P C. J Biol Chem. 1992;267:13520–13527. [PubMed] [Google Scholar]
- 8.Toney J H, Donahue B A, Kellett P J, Bruhn S L, Essigmann J M, Lippard S J. Proc Natl Acad Sci USA. 1989;86:8328–8332. doi: 10.1073/pnas.86.21.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruhn S L, Pil P M, Essigmann J M, Housman D E, Lippard S J. Proc Natl Acad Sci USA. 1992;89:2307–2311. doi: 10.1073/pnas.89.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown S J, Kellett P J, Lippard S J. Science. 1993;261:603–605. doi: 10.1126/science.8342024. [DOI] [PubMed] [Google Scholar]
- 11.Treiber D K, Zhai X, Jantzen H M, Essigmann J M. Proc Natl Acad Sci USA. 1994;91:5672–5676. doi: 10.1073/pnas.91.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin M, Reeves R. Prog Nucleic Acids Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 13.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 14.Grosschedl R. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 15.Sheflin L G, Spaulding S W. Biochemistry. 1989;28:5658–5664. doi: 10.1021/bi00439a048. [DOI] [PubMed] [Google Scholar]
- 16.Sheflin L G, Fucile N W, Spaulding S W. Biochemistry. 1993;32:3238–3248. doi: 10.1021/bi00064a005. [DOI] [PubMed] [Google Scholar]
- 17.Zlatanova J, van Holde K. Prog Nucleic Acids Res Mol Biol. 1996;52:217–259. doi: 10.1016/s0079-6603(08)60968-x. [DOI] [PubMed] [Google Scholar]
- 18.Ivanchenko M, Hassan A, van Holde K, Zlatanova J. J Biol Chem. 1996;271:32580–32585. doi: 10.1074/jbc.271.51.32580. [DOI] [PubMed] [Google Scholar]
- 19.Varga-Weisz P, van Holde K, Zlatanova J. J Biol Chem. 1993;268:20699–20700. [PubMed] [Google Scholar]
- 20.Hill D A, Reeves R. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga-Weisz P, van Holde K, Zlatanova J. Biochem Biophys Res Commun. 1993;203:1904–1911. doi: 10.1006/bbrc.1994.2410. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Banchev T, Srebreva L, Zlatanova J. Biochim Biophys Acta. 1991;1073:230–232. doi: 10.1016/0304-4165(91)90208-x. [DOI] [PubMed] [Google Scholar]
- 24.Adachi Y, Mizuno S, Yoshida M. J Chromatogr. 1990;530:39–46. doi: 10.1016/s0378-4347(00)82300-2. [DOI] [PubMed] [Google Scholar]
- 25.Camerini-Otero R D, Sollner-Webb B, Felsenfeld G. Cell. 1976;8:333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- 26.Kohlstaedt L A, Cole R D. Biochemistry. 1994;33:570–575. doi: 10.1021/bi00168a023. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Kane S A, Lippard S J. Biochemistry. 1996;35:2180–2188. doi: 10.1021/bi952240a. [DOI] [PubMed] [Google Scholar]
- 29.Krylov D, Leuba S, van Holde K, Zlatanova J. Proc Natl Acad Sci USA. 1993;90:5052–5056. doi: 10.1073/pnas.90.11.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaneva J, Schroth G P, van Holde K, Zlatanova J. Proc Natl Acad Sci USA. 1995;92:7060–7064. doi: 10.1073/pnas.92.15.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zlatanova J, Yaneva J. DNA Cell Biol. 1991;10:239–248. doi: 10.1089/dna.1991.10.239. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 33.van Holde K E. Chromatin. New York: Springer; 1988. [Google Scholar]
- 34.Zlatanova J, van Holde K. J Cell Sci. 1992;103:889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]
- 35.Zlatanova J, Srebreva L, Banchev T, Tasheva B, Tsanev R. J Cell Sci. 1990;96:461–468. doi: 10.1242/jcs.96.3.461. [DOI] [PubMed] [Google Scholar]
- 36.Breeuwer M, Goldfarb D S. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- 37.Bustin M, Neihart N K. Cell. 1979;16:181–189. doi: 10.1016/0092-8674(79)90199-5. [DOI] [PubMed] [Google Scholar]
- 38.Einck L, Soares N, Bustin M. Exp Cell Res. 1984;152:287–301. doi: 10.1016/0014-4827(84)90631-1. [DOI] [PubMed] [Google Scholar]