Abstract
14-3-3 proteins regulate many cellular functions, including proliferation. However, the detailed mechanisms by which they control the cell cycle remain to be fully elucidated. We report that one of the 14-3-3 isoforms, 14-3-3τ, is required for the G1/S transition through its role in ubiquitin-independent proteasomal degradation of p21. 14-3-3τ binds to p21, MDM2, and the C8 subunit of the 20S proteasome in G1 phase and facilitates proteasomal targeting of p21. This function of 14-3-3τ may be deregulated in cancer. The overexpression of 14-3-3τ is frequently found in primary human breast cancer and correlates with lower levels of p21 and shorter patient survival. Tenascin-C, an extracellular matrix protein involved in tumor initiation and progression and a known 14-3-3τ inducer, decreases p21 and abrogates adriamycin-induced G1/S arrest. It has been known that p21 is required for a proper tamoxifen response in breast cancer. We show that the overexpression of 14-3-3τ inhibits tamoxifen-induced p21 induction and growth arrest in MCF7 cells. Together, the findings of our studies strongly suggest a novel oncogenic role of 14-3-3τ by downregulating p21 in breast cancer. Therefore, 14-3-3τ may be a potential therapeutic target in breast cancer.
14-3-3 proteins are a family of about 30-kDa dimeric well-conserved α-helical phosphoserine/threonine binding proteins. They contain seven mammalian isoforms (β, ɛ, γ, η, σ, τ, ζ) and are able to bind to multiple protein ligands. The 14-3-3 binding proteins are very diverse; therefore, 14-3-3 is involved in many different cellular processes, including mitogenesis, DNA damage checkpoint, cell cycle control, and apoptosis (12). Most 14-3-3 ligands require phosphorylation to bind to 14-3-3; and their consensus motifs are R(S/Ar)XpSXP (mode 1), RX(Ar/S)XpSXP (mode 2) (12, 46), and (pS/pT)X1-2-COOH (mode 3) (13). However, this consensus is not absolutely required, since a few 14-3-3 binding ligands have sequences significantly different from the sequences of these motifs or do not even require phosphorylation for binding (12, 46).
In general, 14-3-3 proteins play a role in promoting survival and repressing apoptosis (33). However, each isoform may have unique functions in certain physiological contexts. For example, 14-3-3τ binds to ATM-phosphorylated E2F1 during DNA damage and promotes E2F1 stability, leading to the induction of E2F1 proapoptotic target genes such as p73, Apaf1, and caspases (44). Like other 14-3-3 isoforms, however, there appears to be a role for 14-3-3τ in cell survival as well. The deletion of 14-3-3τ in mice leads to embryonic lethality, probably due to developmental arrest (25). Examination of 14-3-3τ+/− mice reveals a role for 14-3-3τ in cardiomyocyte survival (25). This is probably due to its activity that antagonizes ASK1 and sequesters BAD and FOXO family members. However, whether and how 14-3-3τ is involved in cell cycle progression remain poorly understood.
In the study described here, we investigated the role of 14-3-3τ in cell cycle control and uncovered its involvement in the regulation of the cyclin-dependent kinase inhibitor p21(Waf1/Cip1). p21 is a p53 target gene and a major regulator that mediates p53-dependent G1 arrest and senescence. The turnover of the p21 protein is under very tight control. p21 can be degraded through both ubiquitin-dependent and ubiquitin-independent mechanisms. In the ubiquitin-dependent pathway, Skp2 and CRL4(Cdt2) are responsible for p21 degradation in S phase (1, 5, 23, 30, 48), whereas APC/CCdc20 controls the degradation of p21 in prometaphase (3). There are also data demonstrating that Skp2 is not required for basal p21 ubiquitylation and degradation (8). p21 can also be directly targeted to the proteasome for degradation without ubiquitylation (38). This process is mediated by an interaction between p21 and the C8 α subunit of the 20S proteasome (40) and can be promoted by MDM2 and MDMX (20, 21, 49). The MDM2/MDMX-regulated degradation of p21 occurs at the G1 and early S phases (21). The stability of the p21 protein is also regulated by heat shock proteins. An Hsp90 binding protein, WISp39, recruits Hsp90 to p21 and protects p21 from degradation during DNA damage (19). Therefore, it appears that several different regulators control the stability of the p21 protein, probably depending on the phases of the cell cycle and cellular contexts.
Given the evidence of both ubiquitin-dependent and -independent degradation of p21, Pagano and colleagues proposed that both mechanisms of degradation of p21 in different protein complexes may occur in cells (3). It has been shown that free p21, but not cyclin E/cdk2-bound p21, can be degraded by the proteasome in vitro without ubiquitylation (4, 26, 27). Thus, when p21 is complexed with cdk2, it may be degraded by the ubiquitin-dependent pathway, while the ubiquitin-independent mechanism may target free p21 for degradation. It has been shown that this process involves the REGγ proteasome complex (7, 26), which forms the 11S lid and activates the 20S catalytic core proteasome. However, the regulatory mechanisms for the ubiquitin-independent degradation of p21 remain unclear.
In the present study, we identify 14-3-3τ as the protein that regulates the ubiquitin-independent proteasomal degradation of p21 in G1 phase. We demonstrate a direct role for 14-3-3τ in the 20S-mediated p21 degradation via facilitation of an interaction between p21, MDM2, and C8 in vitro. This new role of 14-3-3τ might have an important clinical implication. The extracellular matrix tenascin-C induces 14-3-3τ and degrades p21 through the induction of 14-3-3τ and ameliorates adriamycin-induced cell cycle arrest. 14-3-3τ is often overexpressed in breast cancer, and its overexpression is associated with the downregulation of p21 and shorter patient survival. Through the downregulation of p21, 14-3-3τ overexpression also leads to tamoxifen resistance in MCF7 breast cancer cells.
MATERIALS AND METHODS
Cell culture and transfection.
MDA-MB231 cells, HEK293 cells, MCF7 cells, primary human foreskin fibroblasts (HFFs), Skp2−/− MEF cells, and p53−/− MDM2−/− MEF cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (50 IU/ml), and streptomycin (50 μg/ml) in a humidified incubator with 5% CO2 at 37°C. The mouse ts85 mouse mammary carcinoma cell line was maintained in DMEM containing 10% FBS. The ts85 cell line expresses a temperature-sensitive E1 ubiquitin-activating enzyme and was thus maintained at 33°C. Isogenic colon carcinoma cell line HCT116 with either a p21−/−, p53−/−, or p21+/+ p53+/+ status and human osteosarcoma cell line U2OS were maintained in McCoy's 5A cells. The cells were transfected by the calcium phosphate method, with Lipofectamine 2000 (Invitrogen), or with a Gene Pulser Xcell electroporation system (Bio-Rad).
Recombinant plasmids.
The construction of pCMV-SPORT6-14-3-3τ and pSUPER-si14-3-3τ has been described previously (44). The 19-nucleotide (nt) target sequence for small interfering 14-3-3τ (si14-3-3τ) is 5′-GGACTATCGGGAGAAAGTG-3′, that for si14-3-3τ#2 is 5′-CGAGGAGCGCAACCTGCTC-3′, that for si14-3-3τ#3 is 5′-GAACGTGGTCGGGGGCCGC-3′, and that for small interfering green fluorescent protein (siGFP) is 5′-GACCCGCGCCGAGGTGAAG-3′; for a random small interfering scrambled sequence (siScr), the target sequence is 5′-GCGCGCTTTGTAGGATTCG-3′. The target sequences were constructed with either the pSUPER or the pSUPERIOR.puro vector (OligoEngine). pCMV-SPORT6-14-3-3τ was used to generate a silent mutation resistant to 14-3-3τ small interfering RNA (siRNA) by changing 4 nucleotides (5′-GCTGATTAAGGACTACAGAGAAAAAGTGGAGTCCGAG-3′, in which the 4 changed nucleotides are underscored) by using a QuikChange site-directed mutagenesis kit (Stratagene). The mutation was verified by sequencing. To construct pGEX6p1-human p21, p21 cDNA was amplified by PCR with primers 5′-CTCTCCGGATCCATGTCAGAACCGGCTGGG-3′ and 5′-CTCTCTAGACTCGAGTTAGGGCTTCCCTTGGAG-3′. The PCR product was digested with BamHI/XhoI, cloned into pGEX6p1, and verified by sequencing. EYFP1 (or EYFP2)-p21 was directly obtained by cloning the p21 PCR product after BspE1/XbaI digestion into the pcDNA 3.1 yellow fluorescent protein 1 (EYFP1; containing EYFP amino acids [aa] 1 to 158) vector or the pcDNA3.1 yellow fluorescent protein 2 (EYFP2; containing EYFP aa 159 to 239) vector (36). EYFP1 (or EYFP2)-14-3-3τ was obtained by moving 14-3-3τ from pCMVTag2C-14-3-3τ by EcoRI/ApaI digestion into pEGFPC1. The BspE1 fragment from pEGFPC1-14-3-3τ containing 14-3-3τ cDNA was then cloned into pcDNA3.1EYFP1 (or pcDNA3.1EYFP2) zipper vectors. pGEX4T-MDM2 was obtained from Mien-Chie Hung through Addgene.
Recombinant adenovirus construction.
The adenovirus type 1 4-3-3τ construct (Ad14-3-3τ) was constructed by moving the siRNA-resistant 14-3-3τ cDNA from pCMV-SPORT6-14-3-3τ by XhoI/NotI digetsion to pShuttle-CMV, followed by recombination with the AdEasy vector. To construct Ad-si14-3-3τ and Ad-siScr (siRNA against a scrambled sequence), the NotI/SalI fragments of the pSUPER constructs containing the H1 RNA gene promoter and si14-3-3τ or siScr were cloned into pShuttle, followed by recombination with the AdEasy vector. The viruses were purified by CsCl banding.
Establishment of stable cell lines with inducible expression of si14-3-3τ.
For establishment of stable cell lines with the inducible expression of si14-3-3τ and siRNA against a control sequence from GFP, U2OS cells were transfected with the Tet repressor (TetR) expression vector (pcDNA6/TR) and were subjected to selection with 5 μg/ml blasticidin 48 h later to obtain U2OS-TREX cells. The resistant cells were maintained in McCoy's 5A medium containing 5 μg/ml blasticidin. U2OS-TREX cells were transfected with pSUPERIOR-puro si14-3-3τ and pSUPERIOR-puro siGFP. Stable clones were isolated by use of a selection marker (puromycin) and were maintained in medium containing puromycin. As these cells have TetR, the expression of siRNA is repressed in the absence of tetracycline. The short hairpin RNAs (shRNAs) were induced by treating these cells with 1 μM doxycycline (Dox). The cells were then split and counted with a Beckman Coulter counter every 3 days. Each arm contained three independent samples, and each sample was counted two to four times.
Immunoprecipitation, Western blot analysis, and immunocytochemical staining.
The transfected cells were harvested in TNN buffer (50 mM Tris, 0.25 M NaCl, 5 mM EDTA, 0.5% NP-40), and immunoprecipitation was carried out as described previously (27). The specific signals were detected with appropriate antibodies. The antibodies specific to 14-3-3τ (antibody C-17), 14-3-3σ (antibody N-14), 14-3-3η (antibody E-12), glutathione S-transferase (GST; antibody B-14), p27 (antibody M-197), MDM2 (antibody 2A10 or SMP14), and Skp2 (antibody N-19 or H-435) were purchased from Santa Cruz. The monoclonal antibody for 14-3-3τ (antibody 3B9) and the β-actin antibody were purchased from Sigma. To detect the purified ΔN14-3-3τ and 14-3-3τ proteins on the same membrane, an antibody raised against the full length of 14-3-3 (antibody FL-246; Santa Cruz) was used. The p21 antibody (antibody EA10) was purchased from EMD Biosciences, Inc. Anti-GAPDH was purchased from Alexis. The C8 antibody was purchased from BioMol. The 20S proteasome antibody was purchased from Zymed. For immunocytochemical staining, the cells were stained with a mouse anti-p21 antibody (antibody EA10; EMD Biosciences, Inc.), a rabbit 14-3-3τ antibody (antibody C17; Santa Cruz), and fluorescein isothiocyanate- or Texas Red X-conjugated secondary antibody.
GST pulldown assay.
GST-14-3-3τ, GST-ΔN14-3-3τ (lacking the N-terminal 126 aa) (44), GST-C8, GST-p21, GST-MDM2, and GST were induced and purified from Escherichia coli. The GST portion of GST-14-3-3τ and GST-p21 were excised by the use of PreScission protease, as described previously (44). For the pulldown assay, the reaction mixtures were incubated in TNN buffer at 4°C overnight. The GST proteins were pulled down with glutathione-Sepharose, and the bound proteins were analyzed by Western blotting with the appropriate antibodies.
In vitro p21 degradation assay.
One microgram of p21 was incubated with or without 1 μg of 14-3-3τ in 5 nM purified 20S proteasome (human) (PW8720; BioMol) at 37°C in a buffer containing 20 mM Tris-HCl, pH 7.1, 200 mM NaCl, 10 mM MgCl2, 0.25 mM ATP, and 1 mM dithiothreitol. The degradation time course was monitored by immunoblotting with a monoclonal antibody against p21. Purified CDK4-cyclin D1 complex was purchased from Cell Signaling.
Semiquantitative and real-time RT-PCR.
RNA was extracted with the TRIzol reagent (Invitrogen). Reverse transcription-PCR (RT-PCR) was performed with the following primer pairs: for 14-3-3τ, primers 5′-GGACTATCGGGAGAAAGTGG-3′ and 5′-TCCTGCACTGTCTGATGTCC-3′ (annealing temperature, 62°C; 20 cycles; PCR product, 463 bp); for p21, primers 5′-CTCAGAGGAGGCGCCATG-3′ and 5′-GGGCGGATTAGGGCTTCC-3′ (annealing temperature, 60°C; 25 cycles; PCR product, 527 bp), and for GAPDH, primers 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-AAATGAGCCCCAGCCTTCTCCATG-3′ (annealing temperature, 55°C; 20 cycles; PCR product, 325 bp).
We ensured linear amplification in all cases. The RT-PCR primer sequences and the condition for 18S have been described previously (47). Quantitative real-time PCR was performed in triplicate on an MX3005p thermal cycler (Stratagene) by the SYBR green dye method to track the progress of the reactions, with carboxy-X-rhodamine (ROX) dye being added as a reference. GAPDH was run in parallel with the test genes. The results were analyzed with MxPro (version 4.0) QPCR software (Stratagene).
BrdU incorporation assay.
To assay bromodeoxyuridine (BrdU) incorporation and evaluate the G1 arrest induced by adriamycin, the cells were incubated with BrdU (33 μM) for 5 h and were then probed with anti-BrdU antibody, according to the manufacturer's instructions (Becton Dickinson).
Frozen breast cancer tissues.
Frozen breast tumor tissues or healthy breast tissues were collected with informed consent through the University of Alabama at Birmingham (UAB) Tissue Procurement Core Facility under an approved institutional review board protocol. Samples 1 to 20, 1T, and 7T were collected between 1990 and 2003. The rest of the samples were collected between 2004 and 2007. All patients who provided the tissue samples were either diagnosed with breast cancer or their diagnosis was confirmed at the UAB Comprehensive Cancer Center. All patients underwent surgery at UAB. The majority of patients were monitored at UAB for further treatment, but some patients underwent follow-up with oncologists in their own geographical area. The frozen tissues were minced and lysed in 1% SDS-60 mM Tris HCl and boiled for 5 min. The lysates were briefly sonicated and clarified by centrifugation to remove insoluble tissues, followed by Western blotting.
Statistical analysis.
Descriptive statistics were used to summarize the patients' demographic and clinical characteristics by 14-3-3τ status (positive or negative). The mean, standard deviation, and range were applied for continuous variables; and the frequency and percentage were calculated for categorical variables. The differences in these characteristics between the 14-3-3τ-positive and -negative groups were compared by using the t test, the chi-square test, or Fisher's exact test. We analyzed the survival data using the Kaplan-Meier method. The difference in survival times by 14-3-3τ status was examined by the log-rank test. P values less than 0.05 were considered statistically significant. Statistical analyses were performed by using SAS (version 9.0) software (SAS Institute).
RESULTS
A critical role for 14-3-3τ in cell cycle progression and regulation of p21.
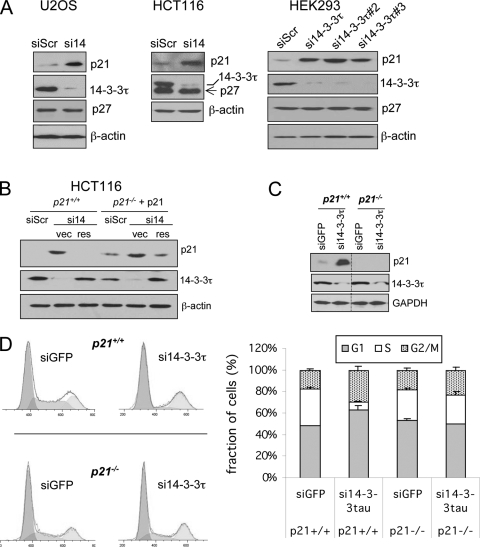
To test whether 14-3-3τ has a physiological role in the control of cell proliferation, we depleted 14-3-3τ in a breast cancer cell line (MDA-MB231 cells). The depletion of 14-3-3τ significantly impaired cell proliferation in this cell line (Fig. 1A). We also observed the induction of p21 in 14-3-3τ-depleted cells, which corresponded to the inhibition of cell growth (Fig. 1B). These results were confirmed in an osteosarcoma cell line (U2OS cells) with Dox-inducible siRNA. We established several U2OS cell lines that stably express inducible siRNA against 14-3-3τ or a control GFP sequence under the control of the tetracycline operon in pSUPERIOR.puro vector. Upon Dox treatment, 14-3-3τ was very effectively depleted in two independent clones, and the depletion of 14-3-3τ correlated temporarily very well with the induction of p21 over the course of Dox treatment (Fig. 1C). Like MDA-MB231, the depletion of 14-3-3τ also significantly inhibited cell proliferation (see Fig. S1 in the supplemental material). The defect in proliferation caused by 14-3-3τ depletion recovered once Dox was withdrawn (see Fig. S1B in the supplemental material), which corresponded to the recovery of both 14-3-3τ and p21 to their baseline levels (Fig. 1C). Consistent with the induction of p21, cell cycle analysis with these cell lines showed that 14-3-3τ-depleted cells were arrested at G1 phase (Fig. 1D). There was also no accumulation of apoptotic cells by 14-3-3τ depletion in these U2OS cells, in HEK293 cells (44), or in HCT116 cells (see the data presented in Fig. 2D).
FIG. 1.
Depletion of 14-3-3τ induces the p21 protein and G1 arrest. (A and B) MDA-MB231 cells were infected with AdsiGFP or Adsi14-3-3τ at a multiplicity of infection of 100 on days 0, 3, and 5. The cells were then counted for the preparation of growth curves (A) or were lysed for Western blot analysis (B). The data represent the mean cell number ± standard deviation for from 6 to 12 readings for each data point. The difference in the slope between the growth curves for AdsiGFP and Adsi14-3-3τ was significant (P < 0.001), according to the linear regression model. (C) U2OS inducible stable cell lines expressing siGFP and si14-3-3τ either were left untreated (without Dox [w/o Dox]) or were treated with 1 μg/ml of doxycycline (w Dox). #9 and #16 represent two independent clones. Doxycycline was withdrawn (w/d) from some of clone 16 on day 13. The cells were harvested for Western blot analysis on the indicated days. (D) 14-3-3τ depletion induces G1 arrest. The cell cycle profiles of the 14-3-3τ siRNA (clone 9) or control siGFP U2OS inducible stable cell lines used for the experiment whose results are presented in panel C were analyzed at day 14 by propidium iodide staining-flow cytometry. Right panels, fractions of each cell cycle phase (mean ± standard deviation) from three experiments.
FIG. 2.
Depletion of 14-3-3τ induces p21-dependent cell cycle arrest. (A) 14-3-3τ knockdown induces p21 but not p27 in U2OS, HCT116, and HEK293 cells. U2OS and HCT116 cells were transiently transfected with si14-3-3τ (si14) or siRNA against a scrambled sequence (siScr). HEK293 cells were transiently transfected with three different 14-3-3τ siRNAs. The cell lysates were harvested 48 h later and were then analyzed by Western blot analysis. In HCT116 cells, the membrane was sequentially probed with anti-p27 and anti-14-3-3τ antibodies. (B) p21+/+ HCT116 cells were transiently transfected with siScr or si14-3-3τ, along with an empty vector (vec) or a vector expressing siRNA-resistant 14-3-3τ (res). For p21−/− HCT116 cells, a p21-expressing vector (pCEP-p21) was cotransfected with siRNA or a rescue construct. The cell lysates were harvested 48 h later and were subjected to Western blot analysis. (C and D) 14-3-3τ depletion induces p21-mediated G1 arrest. p21+/+ and p21−/− HCT116 cells were infected by AdsiGFP or Adsi14-3-3τ for 2 days at a multiplicity of infection of 100. Protein was harvested for Western blot analysis (C). The lysates from both p21+/+ and p21−/− cells were run on the same blot with the same exposure time. The space lanes were excised for clarity of presentation. The cell cycle profile was analyzed by propidium iodide staining-flow cytometry (D). The bar graphs represent the fractions of each cell cycle phase (mean ± standard deviation) from three independent experiments. The P value for the difference of the S-phase fraction between siGFP and si14-3-3τ groups in p21+/+ cells was less than 0.001 (t test), whereas there was no difference in p21−/− cells (P = 0.57).
In addition to MDA-MB231 and U2OS cells, the depletion of 14-3-3τ also induced p21 in a colon cancer cell line (HCT116 cells), HEK293 cells (Fig. 2A), as well as many other cell types, including primary mouse embryonic fibroblasts (MEFs), primary human fibroblasts, and a breast cancer cell line (MCF7 cells) (data presented below), suggesting that the regulation of p21 by 14-3-3τ is general rather than cell line specific. The induction of p21 is truly due to the depletion of 14-3-3τ, since three different 14-3-3τ siRNAs all induced p21 (Fig. 2A, right panels), and p21 is restored to its baseline level when 14-3-3τ depletion was rescued with an RNA interference (RNAi)-resistant 14-3-3τ construct (Fig. 2B). Together, these results indicate a physiological role for 14-3-3τ in the regulation of p21 and cell cycle progression.
Depletion of 14-3-3τ induces p21-dependent G1 arrest.
Given the role of both p21 and p27 in the control of G1 progression, we also examined the expression of p27. As shown in Fig. 2A, while we observed the induction of p21 in 14-3-3τ-depleted cells, the p27 protein was not induced. To test whether the G1 arrest caused by 14-3-3τ depletion is a result of p21 induction, we depleted 14-3-3τ in a pair of isogenic p21+/+ and p21−/− HCT116 cells in which the p21 genes were disrupted by homologous recombination (43). The cell cycle effect of 14-3-3τ depletion was alleviated in the absence of p21 (Fig. 2C and D), indicating that the induction of p21 by 14-3-3τ depletion is largely responsible for G1 arrest.
14-3-3τ regulates p21 protein stability.
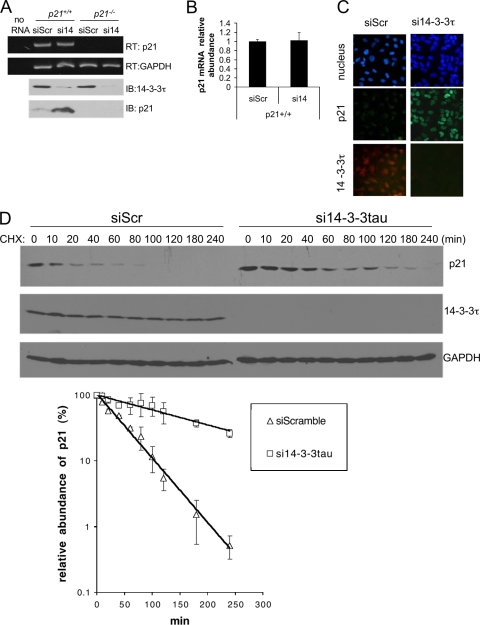
The effect of 14-3-3τ depletion was seen in a transfected cytomegalovirus (CMV) promoter-driven p21 in p21−/− HCT116 cells (Fig. 2B, right three lanes), indicating that the effect is not due to the activation of an endogenous p21 promoter. This was confirmed by measuring the p21 RNA levels. 14-3-3τ siRNA induced the p21 protein but not its RNA in both HCT116 cells (Fig. 3A and B) and HEK293 cells (see Fig. S2 in the supplemental material). Thus, the regulation is via a posttranscriptional mechanism. The depletion of 14-3-3τ also enhanced p21 induction upon treatment with the chemotherapeutic agent adriamycin (see Fig. S2 in the supplemental material), suggesting that 14-3-3τ dampens the induction of p21 by DNA damage. Unlike p27, which is degraded in the cytoplasm, the turnover of p21 occurs in the nucleus (38, 40). To rule out the possibility that 14-3-3τ depletion might lead to the retention of p21 in the cytosol and therefore the stabilization of p21, we performed immunocytochemical staining to investigate the cellular localization of p21 in 14-3-3τ-depleted cells. As shown in Fig. 3C, the p21 induced by 14-3-3τ depletion predominantly remained in the nucleus. This result also indicates that 14-3-3τ is not required for the nuclear localization of p21. Finally, we directly measured the half-life of p21 by cycloheximide treatment. Indeed, the depletion of 14-3-3τ stabilized the p21 protein (Fig. 3D). Taken together, these data establish a role for 14-3-3τ in the regulation of p21 protein stability.
FIG. 3.
Depletion of 14-3-3τ stabilizes the p21 protein. (A and B) RT-PCR and quantitative PCR analyses of p21 gene expression after 14-3-3τ knockdown. p21+/+ and p21−/− HCT116 cells were transfected with siScr or si14-3-3τ (si14). The cells were then harvested for RT-PCR (RT) or immunoblotting (IB) analysis. The “no RNA” lane represents a control RT-PCR without input RNA. (B) p21 transcript levels from the same samples described for panel A were measured by quantitative real-time PCR assay. (C) The p21 protein induced by 14-3-3τ depletion is located in the nucleus. The U2OS cell line was infected by Adsi14-3-3τ or control AdsiScr at a multiplicity of infection of 300. Two days later, cells were stained for p21 and 14-3-3τ. Nuclei were stained with Hoechst 33258. We counted at least 300 14-3-3τ-depleted cells, and p21 was located in the nucleus in all these cells. (D) 14-3-3τ regulates p21 stability. HCT116 cells were transfected with siScr or si14-3-3τ. Twenty-four hours later, the cells were treated with cycloheximide (CHX) at 20 μg/ml for the indicated time periods. The endogenous p21 and 14-3-3τ proteins as well as loading control GAPDH proteins were detected by Western blot analysis. (Lower panel) The p21 levels in three repeated experiments were quantified by densitometry, and the relative abundance of p21 following cycloheximide treatment compared with that seen without cycloheximide treatment was plotted.
14-3-3τ regulates p21 degradation through an ubiquitin-independent pathway.
The stability of p21 is regulated by both ubiquitin-dependent and ubiquitin-independent pathways. The ubiquitin-dependent proteolysis of p21 is mediated by three E3 ligases: Skp2 (5, 48), CRL4(Cdt2) (1, 23, 30), and APC/CCdc20 (3). The overexpression of Skp2 and APC/CCdc20 was reported to cause the degradation of p21 in cultured cells (3). To investigate whether the regulation of p21 by 14-3-3τ involves Skp2 or APC/CCdc20, we performed these degradation assays with 14-3-3τ-depleted cells. Consistent with the findings presented in the literature, the overexpression of Skp2 or APC/CCdc20 led to a decrease in p21 protein levels (Fig. 4A and B). However, the Skp2- or APC/CCdc20-mediated degradation of p21 was not affected by 14-3-3τ depletion. The degradation of p27 and cyclin A serves as a positive control for the E3 ligase activities of Skp2 and APC/CCdc20, respectively. It was also noted that the depletion of 14-3-3τ induced p21 but not p27 (Fig. 4A), consistent with the results shown in Fig. 2A. We also used a pair of primary Skp2+/+ and Skp2−/− MEFs to knock down and rescue the expression of 14-3-3τ. Consistent with published data (5, 8), the p21 level was higher in Skp2−/− MEFs, but it could still be induced by 14-3-3τ depletion in Skp2−/− MEFs (Fig. 4C). Thus, the regulation of p21 by 14-3-3τ does not involve Skp2 or APC/CCdc20.
FIG. 4.
14-3-3τ regulates p21 degradation through an ubiquitin-independent pathway. (A and B) p21+/+ HCT116 cells were transfected with siScr or si14-3-3τ, along with increasing amounts of vectors expressing Skp2 (A) or APC/CCdc20 (B). Twenty-four hours later, the proteins were harvested for immunoblotting. The p21 protein signals were quantified by densitometry. The relative abundance of the p21 protein compared with that in the 0-μg transfection control of the same group was plotted (right panels). The degradation of p27 and cyclin A served as a control for the activities of Skp2 and APC/CCdc20, respectively. (C) Skp2+/+ and Skp2−/− MEF cells were infected by AdsiGFP (siG), Adsi14-3-3τ (si14), or Adsi14-3-3τ, along with an siRNA-resistant 14-3-3τ cDNA (res), for 2 days. The cells were then harvested for Western blot analysis. (D) The ts85 ubiquitin-activating enzyme temperature-sensitive mutant cells were infected by AdsiGFP (siG) or Adsi14-3-3τ (si14) (for both, multiplicity of infection, 300) or by Adsi14-3-3τ along with Ad14-3-3τ (multiplicity of infection, 50) expressing an siRNA-resistant 14-3-3τ cDNA (res), for 2 days at 33°C. Some cells were then incubated at 39°C for 5 h or overnight. The cells were then harvested for Western blot analysis. (Left panels) Equal amounts of lysates were loaded in each lane; (right panels) smaller amounts of lysates from cells incubated at 39°C were loaded to evaluate the induction of p21 by 14-3-3τ siRNA.
To further investigate whether the 14-3-3τ-dependent degradation of p21 involves a functional ubiquitin-activating enzyme, we tested the effect of 14-3-3τ depletion in the ts85 cell line, which harbors a thermolabile ubiquitin-activating enzyme that abolishes the transfer of ubiquitin to target proteins at a nonpermissive temperature (39°C), thus disabling the ubiquitin-dependent protein degradation pathway (11). As shown in Fig. 4D, p21 was stabilized when ts85 cells were moved from the permissive temperature (33°C) to the nonpermissive temperature (39°C), under which condition the ubiquitin-activating enzyme has been inactivated, as shown by blocking of the effect of Skp2 siRNA on the p21 protein (see Fig. S3 in the supplemental material); nevertheless, the depletion of 14-3-3τ still induced p21. Since the incubation at 39°C greatly increased the p21 levels and some p21 signals might be outside the linear range of immunoblotting, we loaded smaller amounts of protein lysates from cells at 39°C in the SDS-polyacrylamide gel (Fig. 4D, right panels). The results showed a similar effect on p21 by 14-3-3τ, even after an overnight incubation at 39°C. Taken together, these data indicate that 14-3-3τ regulates p21 degradation through an ubiquitin-independent pathway.
The 14-3-3τ-dependent degradation of p21 involves MDM2.
The ubiquitin-independent degradation of p21 is mediated by an interaction between its C terminus and the C8 α subunit of the 20S proteasome (38, 40) and can be promoted by MDM2 (20, 49). To test the relationship of 14-3-3τ and MDM2 in p21 degradation, we made use of MDM2−/− p53−/− MEFs. Since the p21 level is very low in MDM2−/− p53−/− MEFs (probably because p21 is a p53 target), we transfected p21 into these cells along with 14-3-3τ siRNA. As shown in Fig. 5A, the reconstitution of MDM2 in MDM2−/− p53−/− cells inhibited the expression of exogenously transfected CMV promoter-driven p21. This is consistent with the increased turnover of p21 protein by MDM2 rather than the downregulation of the p21 promoter. Importantly, the effect of MDM2 was blocked when 14-3-3τ was depleted, but the effect was recovered once 14-3-3τ was rescued. Of note, 14-3-3τ siRNA failed to induce p21 in MDM2−/− p53−/− MEFs, suggesting that either p53 or MDM2 is required for the effect of 14-3-3τ siRNA. The regulation of p21 by 14-3-3τ is independent of the action of p53, since 14-3-3τ depletion induced p21 in p53−/− cells (Fig. 5B). Therefore, 14-3-3τ-dependent p21 degradation requires MDM2. We also performed MDM2-mediated degradation assays with 14-3-3τ-depleted cells. Consistent the findings presented in the literature (20, 49), the overexpression of MDM2 led to a decrease in p21 protein levels (Fig. 5C). The degradation of p53 serves as a positive control for the E3 ligase activities of MDM2. The depletion of 14-3-3τ significantly blocked the effect of MDM2 toward p21 but not p53, indicating that the MDM2-mediated degradation of p21 but not p53 requires 14-3-3τ. Taken together, these data strongly suggest that 14-3-3τ and MDM2 are involved in the same pathway for the regulation of p21 protein stability.
FIG. 5.
14-3-3τ and MDM2 are in the same pathway for the degradation of p21. (A) p53−/− MDM2−/− MEFs were transfected by siScr, si14-3-3τ (si14), or si14-3-3τ, along with siRNA-resistant 14-3-3τ cDNA (res). A p21 cDNA construct (pCEP-p21) and an MDM2 cDNA construct were cotransfected to the cells, as indicated. Cell lysates were then harvested for immunoblotting. (B) 14-3-3τ regulates p21 in a p53-independent manner. p53+/+ and p53−/− HCT116 cells were transfected with siScr or si14-3-3τ (si14). The 14-3-3τ in the si14-3-3τ-transfected cells was rescued (res) with an siRNA-resistant 14-3-3τ cDNA. In p53−/− cells, an empty vector (pCEP) or p21-expressing plasmid pCEP-p21 was cotransfected, as indicated. (C) p21+/+ HCT116 cells were transfected by siScr or si14-3-3τ, along with increasing amounts of MDM2-expressing vectors. Twenty-four hours later, the proteins were harvested for immunoblotting. The endogenous p21 protein was detected by Western blot analysis, and the signals were quantified by densitometry. The relative abundance of the p21 protein compared with that in the 0-μg transfection control of the same group was plotted (right panel). The degradation of p53 served as a control for the activity of MDM2.
14-3-3τ binds to p21, MDM2, and C8 to facilitate complex formation between p21, MDM2, and C8.
It was reported that MDM2 promotes p21-C8 binding (49) and mediates p21 turnover at G1 and early S phase (21). We tested whether 14-3-3τ associates with p21, MDM2, and C8 during G1 phase. Indeed, a coimmunoprecipitation experiment demonstrated the complex formation between 14-3-3τ, p21, C8, and MDM2 in G1 phase (Fig. 6A and B). Also noticed was the surge of this complex formation in the sample obtained 6 h after serum stimulation, coinciding with the decline in p21 levels in this experiment (Fig. 6B). To investigate the role of 14-3-3τ in this complex formation, we tested whether the interaction between p21 and C8 or MDM2 depends on 14-3-3τ. This is indeed the case, since the depletion of 14-3-3τ inhibited the complex formation between p21 and C8 and between p21 and MDM2, and rescue of the 14-3-3τ levels reconstituted their interactions (Fig. 6C). These results demonstrate a physiological role for 14-3-3τ in facilitating the interaction between p21, C8, and MDM2.
FIG. 6.
Physiological role for 14-3-3τ in p21, MDM2, and C8 complex formation. (A and B) 14-3-3τ binds to p21 in G1 phase. The HFFs were brought to quiescence by serum starvation and were then induced to the cell cycle by addition of 20% FBS for the indicated periods of time. The cells were fixed and stained with propidium iodide and were then analyzed by flow cytometry (A). The cell lysates from synchronized HFFs were then immunoprecipitated with normal mouse IgG or mouse anti-p21 antibody and were immunoblotted with the indicated antibodies (B). The input in the left panels represents 1/10 of the lysates for immunoprecipitation. The input, IgG, and anti-p21 lanes were blotted in parallel and exposed on the same film. (C) 14-3-3τ is required for complex formation between p21, C8, and MDM2. HCT116 cells were transfected with siScr or si14-3-3τ (si14). The 14-3-3τ in the si14-3-3τ-transfected cells was rescued with siRNA-resistant 14-3-3τ cDNA. The lysates were then immunoprecipitated with normal mouse IgG or mouse anti-p21 antibody and immunoblotted with the indicated antibodies. The input in the right panels represents 1/20 of the lysates used for immunoprecipitation. (D) The interaction between 14-3-3τ and p21 is inhibited during genotoxic stress. HCT116 cells were left untreated or were treated with adriamycin (5 μM) for 18 h. The cell lysates were then immunoprecipitated with rabbit anti-14-3-3τ, mouse anti-p21, or corresponding normal rabbit or mouse IgG and immunoblotted with the indicated antibodies. The relative intensities measured by densitometry are shown below each panel. (E) HCT116 cell lysates were immunoprecipitated with normal mouse IgG or mouse anti-14-3-3τ and immunoblotted with the indicated antibodies. (F) HCT116 cells were transfected with an empty vector or an MDM2 overexpression vector. The lysates were then immunoprecipitated with normal mouse IgG or mouse anti-14-3-3τ antibody and immunoblotted with the indicated antibodies.
The p21 protein is induced under stress conditions, such as those resulting from DNA damage due to the increase in its transcript level and protein stability. We therefore tested whether the interaction between p21 and 14-3-3τ is downregulated during DNA damage to allow the stabilization of p21. We performed reciprocal coimmunoprecipitation of endogenous 14-3-3τ and p21 in HCT116 cells. Upon adriamycin treatment, the amount of 14-3-3τ-associated p21 is reduced, despite the huge induction of total p21 protein (Fig. 6D, left panel). In the p21 immunoprecipitates, the p21 level in the adriamycin-treated sample is 6-fold higher but has only a marginal increase (1.2-fold) in the associated 14-3-3τ level (Fig. 6D, right panel), indicating a decrease in their levels of binding per p21 molecule. Thus, we conclude that their interaction is inhibited by adriamycin treatment.
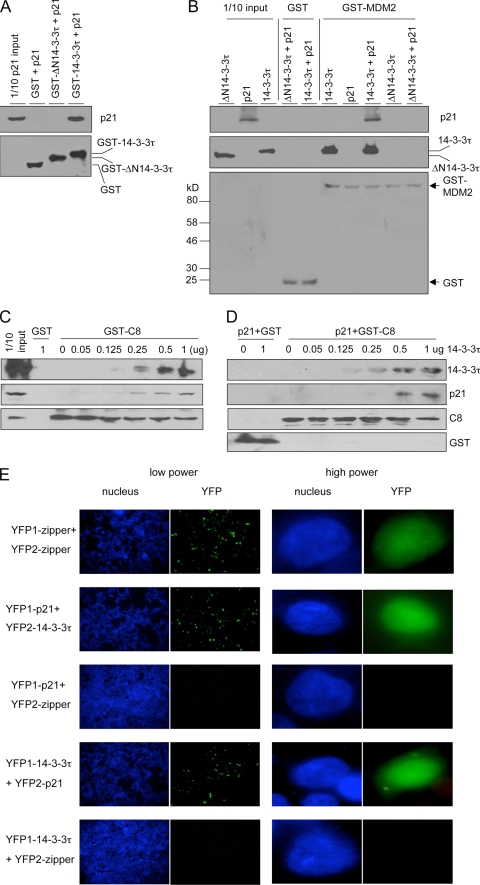
We postulated that 14-3-3τ facilitated the complex formation via its ability to interact with each protein individually. The interaction between endogenous 14-3-3τ and p21 was detected by p21 immunoprecipitation (Fig. 6B and C). In the same experiment, 14-3-3σ or 14-3-3η could not be detected in p21 immunoprecipitates (Fig. 6C). This result suggests a specific role for 14-3-3τ in the regulation of p21; however, future experiments are needed to clarify the role of other 14-3-3 family members in the regulation of p21. The interaction between 14-3-3τ and p21 and between 14-3-3τ and C8/20S was further supported by the results of reciprocal immunoprecipitation experiments (Fig. 6D and E; see also Fig. S4 in the supplemental material). 14-3-3τ also interacted with endogenous MDM2 as well as with overexpressed MDM2 (Fig. 6F). Using GST pulldown assays, we also showed direct interactions between purified 14-3-3τ and purified p21 (Fig. 7A), MDM2 (Fig. 7B), and C8 (Fig. 7C and D). A 14-3-3τ mutant (ΔN14-3-3τ) lacking the N terminus failed to interact with p21 (Fig. 7A) or MDM2 (Fig. 7B), indicating a requirement of the N terminus for these interactions. It is important to note that while GST-MDM2 could pull down 14-3-3τ, it did not pull down p21. However, when 14-3-3τ (but not ΔN14-3-3τ) was added to the reaction mixture, GST-MDM2 interacted with p21. Thus, 14-3-3τ interacts with both p21 and MDM2 and bridges their interaction. The data also provide a molecular mechanism for the requirement of 14-3-3τ in MDM2-induced p21 degradation, as observed in Fig. 5C.
FIG. 7.
14-3-3τ interacts with p21, MDM2, and C8 and mediates their complex formation. (A) Interaction of p21 and 14-3-3τ in vitro. Purified p21 protein was incubated with purified GST, GST-ΔN14-3-3τ, or GST-14-3-3τ; and the GST pulldown assay was performed. p21 and GST fusion proteins were detected by immunoblotting. (B) 14-3-3τ directly binds to MDM2 and bridges the interaction between MDM2 and p21. Purified 14-3-3τ, ΔN14-3-3τ, p21, p21 and 14-3-3τ, or p21 and ΔN14-3-3τ was incubated with GST or GST-MDM2. The GST pull-down assay was then performed, and the proteins were analyzed by immunoblotting. (C) Binding of p21 to GST-C8 is facilitated by 14-3-3τ in a dose-dependent manner. HCT116 cellular lysates (100 μg) were incubated with 2 μg of purified GST or GST-C8 and increasing amounts of purified 14-3-3τ protein overnight at 4°C. The GST-C8 pulldown assay was then performed, and the proteins associated with GST-C8 were probed with the indicated antibodies. The input represents 1/10 of the reaction mixture before it was loaded onto the beads. (D) The GST-C8 pulldown assay was performed as described for panel C, except that purified p21 protein (1 μg) was incubated with GST or GST-C8 in the absence or presence of increasing amounts of purified 14-3-3τ. (E) Bimolecular fluorescence complementation assay. HEK293T cells were transiently transfected with split YFP expression plasmids. The nuclei were stained with Hoechst 33258. The reconstituted YFP signals due to the protein-protein interaction between the YFP1 and YFP2 fusion proteins were captured on a Zeiss fluorescent microscope. Because of the limitation from the transfection efficiency, not every cell shows a YFP signal. YFP1-zipper and YFP2-zipper containing a zipper domain derived from GCN4 were used as positive controls for self-interaction and negative controls for other interactions.
14-3-3τ also plays a similar role in bridging p21 and C8. A GST-C8 pulldown assay demonstrated a direct interaction between purified 14-3-3τ and GST-C8 (Fig. 7C and D). To test whether the interaction between p21 and C8 could be facilitated by 14-3-3τ, we incubated p21 (either endogenous protein from HCT116 cells [Fig. 7C] or purified protein [Fig. 7D]) with GST-C8 in the presence of increasing amounts of 14-3-3τ and then performed a GST-C8 pulldown assay. Purified p21 interacted with GST-C8 poorly, if at all, but their interaction was enhanced by 14-3-3τ in a dose-dependent manner (Fig. 7C and D). It is worth noting that although p21 was shown to bind to MDM2 (49) and C8 (40), the binding reactions in those experiments contained reticulocyte lysates (for the in vitro translation of the p21 protein), which is known to contain 14-3-3 proteins (37). When we used purified p21 protein to interact with purified GST-MDM2 or GST-C8, they interacted poorly, but their interactions were enhanced by 14-3-3τ (Fig. 7B to D).
To detect the interaction between 14-3-3τ and p21 within cultured cells with minimal perturbation of the normal cellular environment, we performed a bimolecular fluorescence complementation assay (BiFC) to examine the interaction of 14-3-3τ and p21 (22). YFP1-p21 (containing EYFP aa 1 to 158 fused with p21) interacted with YFP2-14-3-3τ (EYFP aa 159 to 239 fused with 14-3-3τ) but not with a control zipper domain fusion protein (Fig. 7E; see also Fig. S5 in the supplemental material). A reciprocal combination of the BiFC assay, i.e., YFP1-14-3-3τ with YFP2-p21, demonstrated an interaction between 14-3-3τ and p21 as well. Consistent with a direct interaction between 14-3-3τ and p21, their interaction could also be seen in MDM2−/− p53−/− MEFs (see Fig. S5C in the supplemental material). These data also show that the interaction between p21 and 14-3-3τ occurred mainly in the nucleus. Taken together, these findings indicate that 14-3-3τ facilitates p21 binding to MDM2 and C8 via its ability to interact with each individual protein.
14-3-3τ directly promotes p21 degradation by the 20S proteasome.
Since 14-3-3τ facilitates the interaction between p21 and the C8 subunit of the 20S proteasome, it might directly promote the proteasome-mediated degradation of p21. To test for that, we performed an in vitro degradation assay in which purified p21 protein was incubated with purified 20S proteasome in the presence or absence of 14-3-3τ. As shown in Fig. 8A, 14-3-3τ indeed enhanced the 20S-mediated degradation of p21. The effect of 14-3-3τ is not because the 14-3-3τ-p21 complex is a better substrate for the 20S proteasome, since the 14-3-3τ protein itself was not degraded. The effect of 14-3-3τ was lost in mutant ΔN14-3-3τ (Fig. 8B), which did not bind to p21, supporting the fact that binding between 14-3-3τ and p21 is required for 14-3-3τ promotion of p21 proteasomal degradation. We also investigated the role of MDM2 in the in vitro degradation system. Compared with the results for the control, ΔN14-3-3τ, MDM2 did not exert a significant effect on p21 degradation, nor did it have an additive effect with 14-3-3τ in this in vitro degradation system (Fig. 8B). Therefore, 14-3-3τ appears to have a major role in promoting the 20S-mediated degradation of p21 when p21 is incubated with the proteasome. The role of MDM2 might be at the cellular level and therefore could not be demonstrated in this in vitro degradation assay. Since the ubiquitin-independent degradation of p21 involves only free p21 and not CDK-cyclin- or PCNA-complexed p21, we also tested whether 14-3-3τ only promoted the degradation of free p21. As shown in Fig. 8C, the preincubation of p21 with the CDK-cyclin complex blocked its degradation. This result indicates that 14-3-3τ promotes the degradation of the free pool of p21 but not p21 complexed with CDK-cyclin.
FIG. 8.
14-3-3τ promotes 20S-mediated degradation of p21 in vitro. (A) Purified p21 protein was incubated with 20S in the presence or absence of purified 14-3-3τ. The reaction mixtures were incubated at 37°C, and the reactions stopped with SDS loading buffer at the indicated times. The reaction mixtures were then analyzed by immunoblotting. The proteasome inhibitor MG-132 was added as a control reaction. The p21 proteins were detected by Western blot analysis and were quantified by densitometry. The relative amount of remaining p21 following different incubation times compared with that for the control is plotted in the lower panel. (B) Purified p21 protein was incubated with 20S in the presence of purified ΔN14-3-3τ, MDM2, 14-3-3τ, or both 14-3-3τ and MDM2. The p21 degradation reactions were carried out as described for panel A. The graphs in the lower panel were obtained from three independent experiments. (C) The p21 degradation assay was performed in the presence of 14-3-3τ and 20S, as described for panel A, except that one set of p21 proteins was preincubated with the CDK4-cyclin D1 complex before incubation with 14-3-3τ and 20S.
14-3-3τ links tenascin-C signaling to p21.
Given a role of 14-3-3τ in p21 degradation, external stimuli might alter 14-3-3τ levels and, hence, control p21 to regulate cell growth. It has been reported that the 14-3-3τ transcript is induced in response to the signaling of tenascin-C, an extracellular matrix protein involved in tumor initiation and progression (28). Recent studies showed that the expression of tenascin-C in invasion borders of early breast cancer significantly correlates with proliferative activity, a higher risk of distant metastasis, and a local recurrence (18). We therefore tested the effect of tenascin-C on p21 expression in MCF7 breast cancer cells. Consistent with the findings in the published report (28), we observed a higher level of 14-3-3τ expression when MCF7 cells were grown on tenascin-C compared with that in cells grown on fibronectin (Fig. 9A and B). Importantly, p21 expression was reduced when the cells were grown on tenascin-C, and its induction during adriamycin treatment was also repressed by tenascin-C. The regulation of p21 by tenascin-C is mediated by 14-3-3τ, since the depletion of 14-3-3τ blocked the effect of tenascin-C (Fig. 9B). Consistent with a posttranscriptional regulation, the p21 transcript levels remained unchanged between the samples grown on tenascin-C and those grown on fibronectin, despite the reduced p21 protein levels (Fig. 9C). In line with its effect on p21, tenascin-C also abrogated adriamycin-induced G1/S arrest, a known function of p21 (Fig. 9D). Thus, 14-3-3τ can mediate the signaling response from the environmental milieu, such as tenascin-C to p21, and alter the growth properties of cells.
FIG. 9.
Role of 14-3-3τ in tenascin-C signaling. (A) MCF7 cells were grown on plates pretreated with on fibronectin (FN) or tenascin-C (TN) for 24 h. The cell lysates were then immunoblotted. The change in the level of expression on tenascin-C plates relative to that on fibronection plates is shown below each panel. (B and C) MCF7 cells were grown on plates pretreated with fibronectin (F) or tenascin-C (T). On the next day, the cells were infected with Adsi14-3-3τ or control virus for 48 h and then were either left untreated or treated with adriamycin (5 μM) for 18 h. The cells were then harvested for Western blot analysis (B) and quantitative real-time RT-PCR analysis for the p21 transcript (C). The results were normalized to the GAPDH levels. The results derive from triplicate experiments, and P values are based on the paired two-tailed t test. (D) MCF7 cells were grown on plates pretreated with fibronection or tenascin-C for 24 h. The cells were then either left untreated (w/o Adr.) or treated with 5 μM adriamycin (Adr.) for 18 h, followed by performance of the BrdU incorporation assay. The experiment was performed in triplicate. The P values are based on the paired two-tailed t test.
14-3-3τ is frequently overexpressed in breast cancer and is associated with downregulation of p21 and shorter patient survival.
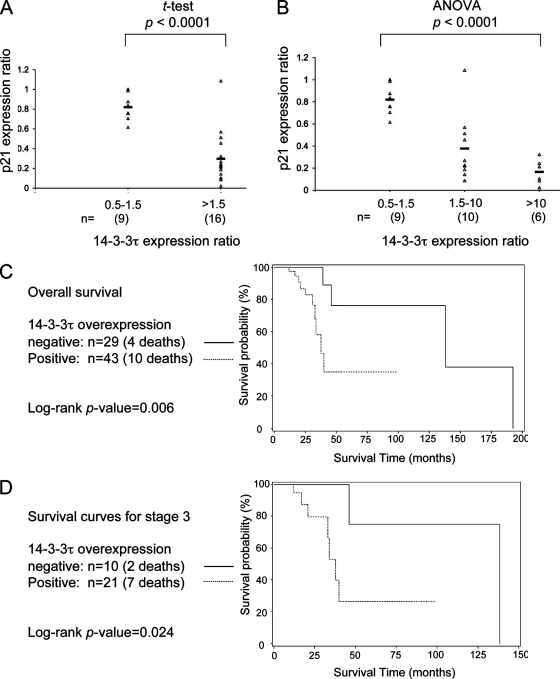
The overexpression of 14-3-3τ has been reported in human lung cancer tissues, although the significance is unclear (34). Given the role of 14-3-3τ in the regulation of p21 degradation, we speculated that the overexpression of 14-3-3τ in cancer cells might lead to the downregulation of p21 expression. To test this hypothesis, we examined the protein and RNA of p21 and 14-3-3τ in 25 pairs of frozen breast tumor tissue samples and their matched healthy breast tissues (see Fig. S6 in the supplemental material). The levels of the p21 and 14-3-3τ proteins were analyzed by immunoblotting and were quantified by densitometry. The protein levels in the tumor tissues were expressed as a ratio relative to the levels in the matched control healthy breast tissues. On the basis of the relative levels of 14-3-3τ in tumors compared to those in their matched controls, we categorized these breast tumors into three groups: those with tumor tissue/healthy tissue expression ratios of 0.5 to 1.5, 1.5 to 10, and >10. As shown in Fig. S6 in the supplemental material, the expression of the p21 protein appears to correlate inversely with that of 14-3-3τ in these breast tumor samples; i.e., 14-3-3τ overexpression is associated with lower levels of p21. The tumor tissue/healthy tissue expression ratios for 14-3-3τ and p21 for each sample were plotted in either two groups of 14-3-3τ expression ratios of 0.5 to 1.5 and >1.5 (Fig. 10A) or in three groups of 14-3-3τ expression ratios of 0.5 to 1.5, 1.5 to 10, and >10 (Fig. 10B). In either case, the inverse correlation between the 14-3-3τ and p21 expression levels was statistically significant (P < 0.0001). Analysis of the RNA showed that higher 14-3-3τ protein levels in tumor tissue (compared with their levels in the matched control tissue) are, in general, correlated with elevated RNA levels; however, lower p21 protein levels are not associated with corresponding changes in the level of its RNA (see Fig. S6B to D in the supplemental material). These results support a role for 14-3-3τ in the regulation of the stability of p21 in breast cancer.
FIG. 10.
Overexpression of 14-3-3τ is associated with the downregulation of p21 and shorter survival times in patients with breast cancer. (A and B) The intensities of p21 on Western blots of the breast tumor tissues and matched healthy tissues were quantified by densitometry. The relative levels of p21 in tumor tissues versus those in healthy control tissues were plotted according to their 14-3-3τ expression ratios between tumor and healthy tissues. (A) The differences in p21 levels were tested between two groups: a group with 14-3-3τ expression ratios of 0.5 to 1.5 and a group with expression ratios of >1.5. The P value was based on the paired two-tailed t test. (B) The differences in p21 levels were tested between three groups: groups with 14-3-3τ expression ratios of 0.5 to 1.5, 1.5 to 10, and >10. The P value was based on analysis of variance (ANOVA). (C) Kaplan-Meier curves of patients' overall survival on the basis of their expression of 14-3-3τ. (D) Kaplan-Meier survival curves of patients with stage 3 breast cancer on the basis of the 14-3-3τ expression status.
To further investigate the clinical significance of 14-3-3τ overexpression, we examined by immunoblotting the expression of 14-3-3τ in frozen breast tumor tissues from a cohort of 79 patients (including the 25 patients described above). A majority of these patients had either stage 2 disease (30 patients) or stage 3 disease (41 patients) (see Tables S1 and S2 in the supplemental material). The levels of 14-3-3τ in tumor tissue were compared with those in the matched healthy breast tissues, when such tissue samples were available (47 patients), or other healthy control breast tissues on the same blots if no matched samples were collected. We found that 14-3-3τ is frequently overexpressed in human breast cancer (47 of 79 samples examined). Although this is a retrospective study, the patient characteristics between the 14-3-3τ-nonoverexpressing group and the 14-3-3τ-overexpressing group were similar by age at the time of diagnosis, race, stage, and Her2/Neu status (see Tables S1 and S2 in the supplemental material). However, patients with 14-3-3τ overexpression tend to have negative estrogen receptor (ER) status (P = 0.026) compared with the ER status of patients without 14-3-3τ overexpression (see Table S2 in the supplemental material). Importantly, patients whose tumors overexpress 14-3-3τ have significantly shorter overall survival times (log-rank test, P = 0.006) (Fig. 10C) and shorter progression-free survival times (P = 0.023) (see Fig. S7 in the supplemental material) compared with the times for those whose tumors do not overexpress 14-3-3τ. The median survival time for patients without overexpression of 14-3-3τ is 138 months (95% confidence interval [CI] = 46, 192) and the median survival time for those with overexpression is 38 months (95% CI = 33, not achievable [NA]). The median progression-free survival time for patients without the overexpression of 14-3-3τ is 107 months (95% CI = 38, 107), and the median progression-free survival time for those with overexpression is 57 months (95% CI = 23, NA). The difference in survival times between women with 14-3-3τ-overexpressing tumors and those with nonoverexpressing tumors remains statistically significant for patients with stage 3 disease (P = 0.024) (Fig. 10D) and lymph node involvement (P = 0.019) (see Fig. S8 in the supplemental material). These data suggest that 14-3-3τ may be a prognostic marker for aggressive breast cancer.
Overexpression of 14-3-3τ abrogates tamoxifen response in MCF7 cells.
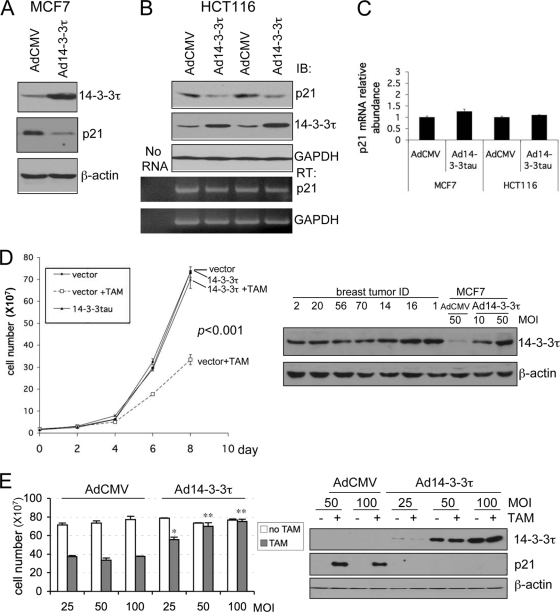
It has been known that p21 is required for a proper response to tamoxifen in patients with breast cancer (2, 6). To further investigate the implications of 14-3-3τ overexpression in breast cancer, we overexpressed 14-3-3τ in an ER-positive breast cancer cell line (MCF7 cells) to see whether it would affect the growth-inhibitory activity of tamoxifen. As expected from its role in p21 degradation, the overexpression of 14-3-3τ decreased the p21 protein levels in MCF7 cells (Fig. 11A). This effect was also seen in other cell lines, such as HCT116 cells, without the alteration of p21 transcript levels (Fig. 11B and C), and in U2OS cells (data not shown). When we examined the tamoxifen response in Ad14-3-3τ-infected and empty vector-infected MCF7 cells, we found that 14-3-3τ alleviated the effect of growth arrest exerted by tamoxifen (Fig. 11D). To correlate the overexpression of 14-3-3τ in MCF7 cells in our experiment with that seen in primary breast tumor tissues, we analyzed their expression levels on the same blot. As shown in the right panels of Fig. 11D, the levels of 14-3-3τ overexpression were very close to those seen in breast cancer tissues, indicating that the inhibition of the effect of tamoxifen by 14-3-3τ overexpression is seen when 14-3-3τ is expressed at levels similar to the levels of expression in primary breast tumor tissues. The abrogative effect on tamoxifen-induced growth arrest by 14-3-3τ occurred in a dose-response manner (Fig. 11E; see also Fig. S9 in the supplemental material). It is also clear that the overexpression of 14-3-3τ abrogated the induction of p21 by tamoxifen (Fig. 11E, right panels). The results of these experiments were analyzed for up to 8 days after tamoxifen treatment. When we carried out similar experiments for up to 11 days after tamoxifen treatment, we obtained the same conclusion (see Fig. S10 in the supplemental material). Since p21 deletion leads only to an increase in the saturation density and not to an increase in the growth rate (9) and since we split and counted the cells before they reached confluence in our experiments, we did not observe a difference in growth curves between control and 14-3-3τ-overexpressing cells.
FIG. 11.
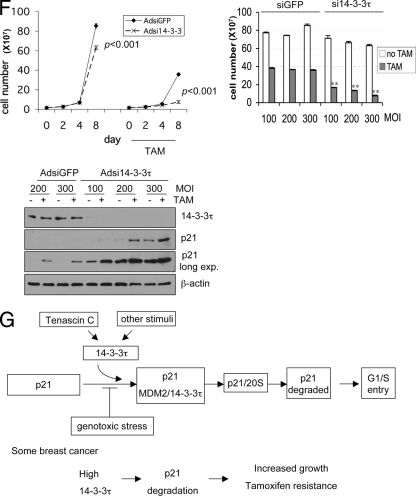
Role for 14-3-3τ in the response to tamoxifen in MCF7 breast cancer cells. (A to C) MCF7 cells (A) or HCT116 cells (B) were infected with AdCMV or Ad14-3-3τ at a multiplicity of infection of 40. Forty-eight hours later, cells were harvested for Western blot and RT-PCR analyses. The results of two independent experiments with AdCMV and Ad14-3-3τ infection are shown (B). (C) Quantitative real-time RT-PCR of p21 transcript levels for MCF7 or HCT116 cells (A and B, respectively). (D) MCF7 cells were infected with AdCMV or Ad14-3-3τ at a multiplicity of infection (MOI) of 50. Two days later, the cells were either treated with tamoxifen (TAM) at 5 μM or a dimethyl sulfoxide vehicle control. The cells were split, reinfected, and counted every 2 days. The data represent the mean cell numbers ± standard deviations for three independent samples for each arm. The difference is significant (P < 0.001), according to the generalized estimating equations (GEE) model. (Right panels) Western blot analysis of cell lysates from the experiment whose results are presented in the left panel of panel D and from breast tumor tissue lysates described in Fig. S6 in the supplemental material. (E) MCF7 cells were infected with AdCMV or Ad14-3-3τ at a multiplicity of infection of 25, 50, or 100 every 2 days and were treated with tamoxifen for 8 days. Cell numbers were counted or analyzed by Western blotting (right panels). *, P ≤ 0.01 compared with the results obtained with AdCMV at the same multiplicity of infection; **, P < 0.001 (t test) compared with the results obtained with AdCMV at the same multiplicity of infection. The results of parallel experiments with tamoxifen treatment for 6 days are presented in Fig. S9 in the supplemental material. (F) MCF7 cells were infected with AdsiGFP or Adsi14-3-3τ at a multiplicity of infection of 100, 200, or 300. Two days later, the cells were treated with either tamoxifen at 5 μM or the dimethyl sulfoxide vehicle control. The cells were split, reinfected, and counted. The data represent the mean the cell numbers ± standard deviations for three independent samples for each arm. (Left panel) The results of infection at a multiplicity of infection of 300. The P values are based on the paired two-tailed t test. (Right panel) The results of infection at a multiplicity of infection of 100 to 300 and following tamoxifen treatment for 8 days. **, P < 0.001 (t test) compared with the results obtained with siGFP at the same multiplicity of infection. (Lower panel) The cell lysates from cells treated with tamoxifen for 8 days were analyzed by Western blotting. (G) A model for the role of 14-3-3τ in the regulation of p21 protein turnover. In response to cellular stimuli such as tenascin-C signaling, 14-3-3τ is induced. Then, 14-3-3τ, along with MDM2, promotes the ubiquitin-independent proteasomal degradation of p21 and induces cell cycle progression. The interaction between 14-3-3τ and p21, however, is inhibited upon genotoxic stress. When 14-3-3τ levels are abnormally elevated in breast cancer, these events may lead to the degradation of p21 and cause resistance to tamoxifen or an enhanced growth property.
We also depleted 14-3-3τ in MCF7 cells and measured the tamoxifen response. Consistent with the data presented earlier for MDA-MB231, U2OS, HEK293, and HCT116 cells (Fig. 1 and 2), 14-3-3τ depletion induced p21 in MCF7 cells and inhibited their growth (Fig. 11F). These effects became more noticeable upon tamoxifen treatment. Together, these results demonstrate that the response to tamoxifen in MCF7 cells depends on the levels of 14-3-3τ and suggest that the overexpression of 14-3-3τ may interfere with the response to tamoxifen in patients with breast cancer.
DISCUSSION
p21 is required for the G1 cell cycle checkpoint; therefore, it is not surprising that the stability of the p21 protein is tightly regulated by multiple regulators. This topic has recently been a subject of intense investigation. In the present study, we uncovered a critical role for 14-3-3τ in the regulation of ubiquitin-independent proteasomal degradation of p21. 14-3-3τ interacts with p21, MDM2, and the C8 α subunit of the 20S proteasome; facilitates their complex formation; and enhances the degradation of the free pool of p21 by the 20S proteasome. In this manner, 14-3-3τ curtails the basal level of p21. This provides an additional layer of regulation to control p21 in response to the environmental milieu. For example, upon genotoxic stress, their interaction can be inhibited, and this may allow p21 to be stabilized. This pathway of regulation is clinically relevant, at least in breast cancer. We show that high levels of 14-3-3τ are associated with smaller amounts of p21 in breast cancer through a posttranscriptional mechanism, likely protein turnover. Importantly, 14-3-3τ overexpression is associated with shorter survival times in patients with breast cancer. We also demonstrate a physiological regulation involving 14-3-3τ and p21 in transmitting the signaling events of tenascin-C. Tenascin-C, an extracellular matrix molecule highly expressed in the microenvironment of most solid tumors, induces 14-3-3τ and downregulates p21. This mechanism may contribute to the known role of tenascin-C in tumor progression (31). Indeed, we show that tenascin-C can mitigate the induction of p21 through 14-3-3τ and abrogate G1 arrest upon adriamycin treatment. The overexpression of 14-3-3τ also inhibits tamoxifen-induced growth arrest in MCF7 cells. These provide a potential mechanism for the association between 14-3-3τ overexpression and shorter survival times in patients with breast cancer. Taken together, these results support a model that p21 protein turnover can be regulated through 14-3-3τ in response to environmental stimuli such as tenascin-C or genotoxic stress (Fig. 11G).
The 26S proteasome in eukaryotic cells is composed of a 20S core and 19S regulatory lids that cap on one or both ends. The 19S multiprotein complex binds to polyubiquitin chains and opens the proteasome core in an ATP-dependent process (16). Structural analysis of the 20S proteasome reveals a cylinder structure consisting of a stack of four rings of hexamers with distal α rings flanking an internal pair of β rings (14). The central axial pore of the cylinder is surrounded by the α rings and is occluded by the N-terminal extensions of some of the α subunits, including α7 (i.e., the C8 subunit). This pore can be opened up upon association with the 19S regulatory complex (24). The 20S proteasome can also recognize the disordered domain of a substrate, and the binding presumably opens the gate to allow threading of the unfolded protein through the channel for endoproteolytic cleavage (27). Given the role of 14-3-3τ in the interaction between p21 and C8 (Fig. 6 and 7) and its ability to directly promote the 20S-mediated degradation of p21 (Fig. 8), we propose that through interactions between 14-3-3τ and both p21 and C8, 14-3-3τ may promote the gating of the 20S proteasome for p21. It is also possible that 14-3-3τ and MDM2 might affect protein folding. Interestingly, Saccharomyces cerevisiae 14-3-3 paralogs BMH1/2 were identified as putative 26S proteasome-interacting proteins by a mass spectrometry approach (15), suggesting an evolutionarily conserved role for 14-3-3 in the regulation of the proteasome function.
Our in vivo data support the published results regarding a role for MDM2 in the proteasomal degradation of p21 (20, 49) and identify 14-3-3τ as an important player in this process. Prior reports did not test MDM2 in the in vitro p21 degradation assay. In this in vitro assay, in which the p21 protein was directly incubated with the 20S proteasome, unlike 14-3-3τ, MDM2 lacked noticeable activity in promoting the 20S-mediated turnover of p21 (Fig. 8B), suggesting a role distinct from that of 14-3-3τ and suggetsing that it is not directly involved in the gating process of the proteasome. The in vitro binding data (Fig. 7A and B) support a role for 14-3-3τ in bridging the interaction between MDM2 and p21. We postulate that after binding with p21, MDM2 might facilitate the translocation of p21 to the proteasome and allow 14-3-3τ to promote the gating of the 20S proteasome for p21. This model can explain why no promoting activity could be detected for MDM2 in the in vitro degradation assay, since p21 was mixed with the 20S proteasome in test tubes. In this regard, MDM2 was recently reported to possess an intrinsic molecular chaperone activity (45). The details on how MDM2 participates in p21 degradation deserve future investigation.
Given the fact that 14-3-3 is involved in multiple survival pathways and increased 14-3-3 expression is seen in some cancers, 14-3-3 is implicated in cancer development (33, 41). Specifically, decreased 14-3-3σ expression through promoter methylation in patients with advanced non-small-cell lung cancer significantly correlates with a longer median survival time (35). On the other hand, the overexpression of 14-3-3σ found in 38.8% of colorectal carcinomas was associated with a significantly decreased survival time (32). Increased 14-3-3β, -γ, and -τ expression was found in lung cancer biopsy specimens (34). However, no clinical correlation has been made for the overexpression of 14-3-3τ in cancers. Here we show that a high level of 14-3-3τ expression was seen in 47 of 79 breast cancer tissue specimens and was associated with poor patient survival. Although our study shows only a trend for an association between 14-3-3τ and high tumor grade, probably due to the limited number of patients in our study, analysis of three breast cancer microarray databases (10, 17, 29) with a total of 694 breast tumor samples by the use of a data-mining tool (ONCOMINE) shows a very significant and consistent association between high levels of 14-3-3τ RNA and high-grade tumors (P = 2.1E-5, 1.6E-9, and 4.9E-9, respectively). Importantly, our data demonstrate a significant correlation between high levels of 14-3-3τ and lower levels of the p21 protein (Fig. 10A and B). Thus, these results are consistent with an oncogenic transformation-promoting property of 14-3-3 and strongly suggest that the downregulation of p21 by 14-3-3τ overexpression may contribute to this activity.
A significant association between 14-3-3τ overexpression and ER-negative status was seen in our series (see Table S2 in the supplemental material). Analysis of data for a total of 445 ER-positive and 167 ER-negative breast tumors from three breast cancer microarray databases (10, 39, 42) via ONCOMINE also shows a strong association between high-level 14-3-3τ expression and negative ER status (P = 4.2E-8, 4.4E-6, and 2E-10, respectively). Even in ER-positive breast cancer, 14-3-3τ overexpression may adversely affect the hormone response, as suggested by the results of the experiments with MCF7 cells (Fig. 11). In the 14-3-3τ-nonoverexpressing group within our series, there were 23 patients who were either ER positive or progestarone receptor (PR) positive (and who had received hormone therapy). Among these patients, three progressed at 32, 36, and 38 months after the initial diagnosis, respectively. There were 28 patients who were either ER positive or PR positive (and who had received hormone therapy) in the 14-3-3τ-overexpressing group. Among those patients, six patients progressed at 9, 11, 17, 19, 23, and 25 months after the initial diagnosis, respectively. The Kaplan-Meier curves of the progression-free survival of these patients are shown in Fig. S11 in the supplemental material. There is a trend favoring the 14-3-3τ-nonoverexpressing group, although the difference has not yet reached statistical significance (log-rank test, P = 0.1035). This result suggests a possible association between 14-3-3τ overexpression and hormonal refractoriness in breast cancer. Whether 14-3-3τ overexpression can lead to resistance to tamoxifen therapy in breast cancer patients, as seen with MCF7 cells, deserves further investigation. Our study implies that 14-3-3τ is a potential novel therapeutic target that may be targeted to overcome hormonal resistance in breast cancer.
Supplementary Material
Acknowledgments
We gratefully acknowledge the UAB Tissue Procurement Facility for providing breast tumor specimens, Stephen W. Michnick for the YFP1 and YFP2 vectors, Wafik El-Deiry for pCEP-p21, Martin Allday and Ruiwen Zhang for GST-C8, Kei-ichi Nakayama for sibling-matched primary Skp2+/+ and Skp2−/− MEFs, Bert Vogelstein for p53+/+ and p53−/− HCT116 cells and p21+/+ and p21−/− HCT116 cells, and James Ou for ts85 cells. We also thank Fannie Lin for valuable discussions and reagents.
The work was supported by grants from NIH (CA 100857) and the U.S. Department of Defense Breast Cancer Research Program (grant W81XWH-09-1-0338). W.-C.L. is a Leukemia and Lymphoma Society scholar.
We all disclose that we have no financial interests that will pose a conflict of interest regarding the work reported in this article.
Footnotes
Published ahead of print on 11 January 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbas, T., U. Sivaprasad, K. Terai, V. Amador, M. Pagano, and A. Dutta. 2008. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22:2496-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abukhdeir, A. M., M. I. Vitolo, P. Argani, A. M. De Marzo, B. Karakas, H. Konishi, J. P. Gustin, J. Lauring, J. P. Garay, C. Pendleton, Y. Konishi, B. G. Blair, K. Brenner, E. Garrett-Mayer, H. Carraway, K. E. Bachman, and B. H. Park. 2008. Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc. Natl. Acad. Sci. U. S. A. 105:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amador, V., S. Ge, P. G. Santamaria, D. Guardavaccaro, and M. Pagano. 2007. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol. Cell 27:462-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom, J., V. Amador, F. Bartolini, G. DeMartino, and M. Pagano. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115:71-82. [DOI] [PubMed] [Google Scholar]
- 5.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278:25752-25757. [DOI] [PubMed] [Google Scholar]
- 6.Cariou, S., J. C. Donovan, W. M. Flanagan, A. Milic, N. Bhattacharya, and J. M. Slingerland. 2000. Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 97:9042-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., L. F. Barton, Y. Chi, B. E. Clurman, and J. M. Roberts. 2007. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 26:843-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., Y. Chi, A. Bloecher, R. Aebersold, B. E. Clurman, and J. M. Roberts. 2004. N-acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1). Mol. Cell 16:839-847. [DOI] [PubMed] [Google Scholar]
- 9.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 10.Desmedt, C., F. Piette, S. Loi, Y. Wang, F. Lallemand, B. Haibe-Kains, G. Viale, M. Delorenzi, Y. Zhang, M. S. d'Assignies, J. Bergh, R. Lidereau, P. Ellis, A. L. Harris, J. G. Klijn, J. A. Foekens, F. Cardoso, M. J. Piccart, M. Buyse, and C. Sotiriou. 2007. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin. Cancer Res. 13:3207-3214. [DOI] [PubMed] [Google Scholar]
- 11.Finley, D., A. Ciechanover, and A. Varshavsky. 1984. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 37:43-55. [DOI] [PubMed] [Google Scholar]
- 12.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40:617-647. [DOI] [PubMed] [Google Scholar]
- 13.Ganguly, S., J. L. Weller, A. Ho, P. Chemineau, B. Malpaux, and D. C. Klein. 2005. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. U. S. A. 102:1222-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groll, M., L. Ditzel, J. Lowe, D. Stock, M. Bochtler, H. D. Bartunik, and R. Huber. 1997. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386:463-471. [DOI] [PubMed] [Google Scholar]
- 15.Guerrero, C., C. Tagwerker, P. Kaiser, and L. Huang. 2006. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol. Cell. Proteomics 5:366-378. [DOI] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Ivshina, A. V., J. George, O. Senko, B. Mow, T. C. Putti, J. Smeds, T. Lindahl, Y. Pawitan, P. Hall, H. Nordgren, J. E. Wong, E. T. Liu, J. Bergh, V. A. Kuznetsov, and L. D. Miller. 2006. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66:10292-10301. [DOI] [PubMed] [Google Scholar]
- 18.Jahkola, T., T. Toivonen, I. Virtanen, K. von Smitten, S. Nordling, K. von Boguslawski, C. Haglund, H. Nevanlinna, and C. Blomqvist. 1998. Tenascin-C expression in invasion border of early breast cancer: a predictor of local and distant recurrence. Br. J. Cancer 78:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jascur, T., H. Brickner, I. Salles-Passador, V. Barbier, A. El Khissiin, B. Smith, R. Fotedar, and A. Fotedar. 2005. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol. Cell 17:237-249. [DOI] [PubMed] [Google Scholar]
- 20.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22:6365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, Y., S. X. Zeng, X. X. Sun, H. Lee, C. Blattner, Z. Xiao, and H. Lu. 2008. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol. Cell. Biol. 28:1218-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerppola, T. K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y., N. G. Starostina, and E. T. Kipreos. 2008. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22:2507-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohler, A., P. Cascio, D. S. Leggett, K. M. Woo, A. L. Goldberg, and D. Finley. 2001. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell 7:1143-1152. [DOI] [PubMed] [Google Scholar]
- 25.Lau, J. M., X. Jin, J. Ren, J. Avery, B. J. DeBosch, I. Treskov, T. S. Lupu, A. Kovacs, C. Weinheimer, and A. J. Muslin. 2007. The 14-3-3tau phosphoserine-binding protein is required for cardiomyocyte survival. Mol. Cell. Biol. 27:1455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X., L. Amazit, W. Long, D. M. Lonard, J. J. Monaco, and B. W. O'Malley. 2007. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell 26:831-842. [DOI] [PubMed] [Google Scholar]
- 27.Liu, C. W., M. J. Corboy, G. N. DeMartino, and P. J. Thomas. 2003. Endoproteolytic activity of the proteasome. Science 299:408-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, D., M. Brown-Luedi, and R. Chiquet-Ehrismann. 2003. Tenascin-C signaling through induction of 14-3-3 tau. J. Cell Biol. 160:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, L. D., J. Smeds, J. George, V. B. Vega, L. Vergara, A. Ploner, Y. Pawitan, P. Hall, S. Klaar, E. T. Liu, and J. Bergh. 2005. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U. S. A. 102:13550-13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishitani, H., Y. Shiomi, H. Iida, M. Michishita, T. Takami, and T. Tsurimoto. 2008. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283:29045-29052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orend, G., and R. Chiquet-Ehrismann. 2006. Tenascin-C induced signaling in cancer. Cancer Lett. 244:143-163. [DOI] [PubMed] [Google Scholar]
- 32.Perathoner, A., D. Pirkebner, G. Brandacher, G. Spizzo, S. Stadlmann, P. Obrist, R. Margreiter, and A. Amberger. 2005. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 11:3274-3279. [DOI] [PubMed] [Google Scholar]
- 33.Porter, G. W., F. R. Khuri, and H. Fu. 2006. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin. Cancer Biol. 16:193-202. [DOI] [PubMed] [Google Scholar]
- 34.Qi, W., X. Liu, D. Qiao, and J. D. Martinez. 2005. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int. J. Cancer 113:359-363. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez, J. L., R. Rosell, M. Taron, M. Sanchez-Ronco, V. Alberola, R. de Las Penas, J. M. Sanchez, T. Moran, C. Camps, B. Massuti, J. J. Sanchez, F. Salazar, and S. Catot. 2005. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival. The Spanish Lung Cancer Group. J. Clin. Oncol. 23:9105-9112. [DOI] [PubMed] [Google Scholar]
- 36.Remy, I., A. Montmarquette, and S. W. Michnick. 2004. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6:358-365. [DOI] [PubMed] [Google Scholar]
- 37.Seimiya, H., H. Sawada, Y. Muramatsu, M. Shimizu, K. Ohko, K. Yamane, and T. Tsuruo. 2000. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5:403-410. [DOI] [PubMed] [Google Scholar]
- 39.Sotiriou, C., P. Wirapati, S. Loi, A. Harris, S. Fox, J. Smeds, H. Nordgren, P. Farmer, V. Praz, B. Haibe-Kains, C. Desmedt, D. Larsimont, F. Cardoso, H. Peterse, D. Nuyten, M. Buyse, M. J. Van de Vijver, J. Bergh, M. Piccart, and M. Delorenzi. 2006. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98:262-272. [DOI] [PubMed] [Google Scholar]
- 40.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzivion, G., V. S. Gupta, L. Kaplun, and V. Balan. 2006. 14-3-3 proteins as potential oncogenes. Semin. Cancer Biol. 16:203-213. [DOI] [PubMed] [Google Scholar]
- 42.van de Vijver, M. J., Y. D. He, L. J. van't Veer, H. Dai, A. A. Hart, D. W. Voskuil, G. J. Schreiber, J. L. Peterse, C. Roberts, M. J. Marton, M. Parrish, D. Atsma, A. Witteveen, A. Glas, L. Delahaye, T. van der Velde, H. Bartelink, S. Rodenhuis, E. T. Rutgers, S. H. Friend, and R. Bernards. 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347:1999-2009. [DOI] [PubMed] [Google Scholar]
- 43.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187-5190. [PubMed] [Google Scholar]
- 44.Wang, B., K. Liu, F. T. Lin, and W. C. Lin. 2004. A role for 14-3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J. Biol. Chem. 279:54140-54152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wawrzynow, B., A. Zylicz, M. Wallace, T. Hupp, and M. Zylicz. 2007. MDM2 chaperones the p53 tumor suppressor. J. Biol. Chem. 282:32603-32612. [DOI] [PubMed] [Google Scholar]
- 46.Yaffe, M. B. 2002. How do 14-3-3 proteins work?—gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513:53-57. [DOI] [PubMed] [Google Scholar]
- 47.Yang, S. Z., and S. A. Abdulkadir. 2003. Early growth response gene 1 modulates androgen receptor signaling in prostate carcinoma cells. J. Biol. Chem. 278:39906-39911. [DOI] [PubMed] [Google Scholar]
- 48.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc. Natl. Acad. Sci. U. S. A. 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Z., H. Wang, M. Li, S. Agrawal, X. Chen, and R. Zhang. 2004. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 279:16000-16006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.