Abstract
Small heterodimer partner (SHP) is an epigenetically regulated nuclear transcriptional repressor that suppresses the development of liver cancer by inhibiting cellular growth. Here we report a novel cytoplasmic function of SHP through its regulation of mitochondrial activity. SHP is a pivotal cell death receptor that targets mitochondria, where it binds with Bcl-2, disrupts Bcl-2/Bid interaction, and induces cytochrome c release. The apoptosis inducer AHPN {retinoid 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid} acts by regulating SHP gene expression and promotes the translocation of SHP from the nucleus to the mitochondria. Induction of apoptosis by SHP activation inhibits peritoneal pancreatic tumor growth. Our findings provide for the first time a mechanism by which SHP regulates cell survival, namely, by controlling mitochondrial function via modulating the activity of Bcl-2 through AHPN-mediated or AHPN-independent action. Thus, SHP regulates a mechanism by which apoptotic signals can mediate local control of mitochondrial function and apoptosis, which in turn may limit tumorigenesis.
Resistance to apoptosis is an important characteristic of human cancers (1, 11). Apoptosis is a distinct form of programmed cell death that is best defined morphologically by nuclear and cell fragmentation (16). Apoptosis plays a major role in liver disease, and altered regulation of apoptosis has been shown to be associated with the pathogenesis of human hepatocellular carcinoma (HCC) and dysplasia (4, 10, 18).
Mitochondria are crucial cellular organelles that regulate apoptosis (27). Apoptosis can be executed through either the intrinsic mitochondrion-dependent pathway or the extrinsic mitochondrion-independent pathway (13, 20). The intrinsic pathway is triggered by a variety of stressors and is mediated through cleavage and activation of the Bcl-2 family protein BID (21). Cleaved BID translocates to mitochondria and interacts with other Bcl-2 family proteins, which disrupt the mitochondrial transmembrane potential (Δψ) through pore-forming proapoptotic factors leading to leakage of cytochrome c (5). Cytochrome c interacts with the adapter protein Apaf-1, dATP, and caspase-9 to subsequently activate caspase-3, leading to cell death (22). These events can be promoted by other mitochondrial factors, including apoptosis-inducing factor (AIF), smac/Diablo, endonuclease G, and Omi/HtrA (6, 20, 38, 39, 41). The extrinsic pathway is mediated through binding of cognate ligands to their death receptors, including CD95 (Fas/APO-1), TNF-R1, and the TRAIL receptors DR4 and DR5 (2, 35). This results in direct cleavage and activation of procaspases-8 to further activate executor caspase-3 and regulate target proteins involved in apoptosis (36). Several studies have reported translocation of selected proteins, including the orphan nuclear receptor TR3 (26), to mitochondria during apoptosis, where they exert regulatory effects on apoptosis (3).
Small heterodimer partner (SHP, NR0B2) is an atypical orphan nuclear receptor that trans-represses its target genes in several metabolic pathways (, 33, 43). SHP is an epigenetically regulated transcriptional repressor that suppresses the development of liver cancer in both humans and mice. Epigenetic gene silencing disrupts SHP function by downregulating its expression in human HCC (12). SHP also plays a role in the maintenance of cellular proliferation that is associated with the development of hepatoma in mice (46). In this regard, SHP inhibits gene transcription of the key cell cycle regulator cyclin D1, thus controlling the G1/S transition.
The synthetic retinoic acid receptor γ (RARγ) agonist AHPN {retinoid 6-[3-(1-adamantyl)-4-hydroxyphenyl]-2-naphthalene carboxylic acid, also called CD437} induces apoptosis in cancer cells independently of the RAR interaction (47). The AHPN derivative 3-Cl-AHPC {4-[3′-(1-adamantyl)-4′-hydroxyphenyl]-3-chlorocinnamic acid} does not transactivate RARs but induces apoptosis by increasing epidermal growth factor receptor (EGFR) proteolysis (9) or by activating NF-κB (7). Although numerous potential pathways have been suggested, a common biochemical basis for the mechanism of their apoptotic action remains unclear. Moreover, interpreting the findings of AHPN-induced apoptosis is confounded due to diverse responses resulting from different genetic pathways in cells of different lineages (47).
It has recently been shown that AHPN and 3-Cl-AHPC can bind to SHP protein, which forms a complex with Sin3A (8). This raises an interesting question whether SHP is an active component of apoptosis signaling. In this study, we investigated the underlying mechanism and the function of SHP in regulating apoptosis. We show that SHP is a potent activator of apoptosis by directly targeting mitochondria. AHPN and 3-Cl-AHPC induced SHP mRNA expression and SHP protein translocation from the nucleus to mitochondria, where it interacted with Bcl-2 and disrupted Bcl-2/Bid interaction, which induced cytochrome c release and activated apoptosis. Our findings provide for the first time a mechanism by which SHP regulates cell survival, namely, by controlling mitochondrial function by modulating the activity of Bcl-2.
MATERIALS AND METHODS
Animals.
SHP+/+ (wild type [WT]), SHP−/− (SKO), SHP nontransgenic control (NC), and hepatocyte-specific SHP transgenic (STG) mice have been described previously (46). Primary mouse embryonic fibroblasts (MEFs) were isolated and immortalized as described previously (46). AHPN and 3-Cl-AHPC were emulsified in 25% cremophor EL-75% water and were given by oral or intraperitoneal (i.p.) injection. A dose of 10 to 20 mg/kg body weight was given each day for 1 to 7 days. Protocols for animal use were approved by the Institutional Animal Care and use Committee at the University of Utah.
Chemicals and antibodies.
AHPN and monoclonal antibodies to Flag M2 (catalog number F1804) and β-actin (catalog number A3853) were purchased from Sigma (Sigma Chemical Co., St. Louis, MO). Polyclonal antibodies to poly(ADP-ribose) polymerase (PARP) (catalog number 9532), Bcl-2 (catalog number 2870s), Hsp60 (catalog number 4870s), and hepatocyte nuclear factor 4 alpha (HNF4α) (catalog number 3113s) were obtained from Cell Signaling (Cell Signaling Technology, Inc., Danvers, MA). Monoclonal antibodies to cytochrome c (catalog number 556433) and Bcl-2 (catalog number sc-7382) were purchased from BD Pharmingen (BD Biosciences, San Jose, CA) and Santa Cruz (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), respectively. Rabbit anti-human SHP was purchased from MBL (catalog number LS-A5411; MBL International Corporation, MA). MitoTracker Red 580 was purchased from Molecular Probes, Inc. (Eugene, OR). Caspase-3 inhibitor (Z-DEVD-FMK; catalog number FMK004), caspase-8 inhibitor (Z-IETD-FMK; catalog number FMK007), and caspase-9 inhibitor (Z-LEHD-FMK; catalog number FMK008) were purchase from R & D (R & D Systems, Minneapolis, MN). Annexin V-phycoerythrin (PE) apoptosis detection kit I was obtained from BD Pharmingen (catalog number 559763; BD Biosciences, San Jose, CA). d-Luciferin was purchased from Xenogen (Xenogen Corporation, Alameda, CA). Flag-SHP plasmid was obtained from Timothy F. Osborne, and hemagglutinin (HA)-HNF4α was obtained from Akiyoshi Fukamizu. LRH-1 and Bcl-2 small interfering RNAs (siRNAs) were purchased from Thermo Scientific Dharmacon RNAi Technologies (NR5A2 ON-TARGET plus SMART pool [L-003430-00-0005] and Bcl-2 ON-TARGET plus SMART pool [L-003307-00-0005]).
Anti-Fas antibody injections and histologic examination.
Two-month-old nontransgenic littermates (NC) or transgenic mice (STG) bred on a C57BL/6 × SJL background were injected i.p. with 10 μg of an affinity-purified hamster monoclonal antibody against mouse Fas antigen (Jo2) diluted in 100 μl of a 0.9-g/liter NaCl solution. Mice were euthanized at specific times after treatment. Tissues were harvested immediately after death, fragments of tissues were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and embedded in paraffin, and 5-μm sections were stained with hematoxylin and eosin (H&E).
The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed using the In Situ Cell Death Detection Kit, Fluorescein (catalog number 11684795910; Roche Applied Science, Mannheim, Germany), following the manufacturer's instructions. In brief, 5-μm cryopreserved tissue sections were fixed in 4% paraformaldehyde for 20 min at room temperature (RT), washed with PBS, and permeabilized with freshly prepared 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice. After washing with PBS, the slides were overlaid with 100 μl of TUNEL reaction mixture, according to the manufacturer's instructions, and incubated for 1 h at 37°C. Finally, cells were washed in PBS, and coverslips were mounted onto slides with Prolong Gold antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole) (catalog number p36931; Molecular Probes, Inc., Eugene, OR). Cells were imaged using an Olympus AX70 fluorescence microscope. The magnifications of the photographs of the histology are ×20.
Apoptosis analyses.
For UV-induced apoptosis, subconfluent fibroblasts growing in the logarithmic phase were subjected to UVC (40 J/m2) irradiation. Cells were fixed, and apoptotic cells were identified by TUNEL assays (Roche). To examine tumor necrosis factor alpha (TNF-α)-induced apoptosis, cells were treated with 10 μg/ml cycloheximide (CHX) plus 30 ng/ml TNF-α at the indicated times and examined for apoptotic cell death by annexin V staining. For AHPN- and 3-Cl-AHPC-induced apoptosis, cells were treated with both agents for 24 h at different doses as indicated. For annexin V staining, cells were washed twice with cold PBS and resuspended in binding buffer provided in the annexin V-PE apoptosis detection kit I at a concentration of 106 cells/ml. A total of 105 cells were then stained with 5 μl annexin V-PE and 5 μl 7-aminoactinomycin D (7-AAD) and incubated for 15 min at RT in the dark. A total of 30,000 cells were acquired by flow cytometry (FACSCalibur; Becton Dickinson) and analyzed with CellQuest software (BD Bioscience).
Immunofluorescence and confocal laser scanning microscopy.
Cells were cultured overnight on coverslips and were treated with various agents as required. Cells were washed twice with PBS, fixed in 3.7% paraformaldehyde (in PBS) for 5 min at RT, and then permeabilized in chilled methanol for 3 min at RT. After washing with PBS, the cells were incubated with blocking solution containing 1% bovine serum albumin (BSA)-PBS for 1 h at RT. To identify SHP protein, cells were incubated first with anti-SHP IgG antibody (3 μg/ml; MBL) diluted in 1% BSA-PBS for 2 h at RT and then with the corresponding Alexa Fluor 488 goat anti-rabbit IgG (1:200; Molecular Probes) as the secondary antibody. To identify Bcl-2, cells were incubated with anti-Bcl-2 IgG antibody (2 μg/ml; Santa Cruz), followed by the corresponding Alexa Fluor 350 goat anti-mouse IgG (1:200; Molecular Probes). To localize mitochondria, cells were incubated with MitoTracker Red 580 at 200 nM in the dark for 20 min at RT. Cells were then washed in PBS, and coverslips were mounted onto slides with Prolong Gold antifade reagent (p36930; Molecular Probes Inc., Eugene, OR). For each sample, confocal images were collected in the fluorescence mode, followed by electronic merging of the images using a confocal microscope (Olympus IX81; Olympus America Inc., Melville, NY).
Western blotting.
Cells grown in 100-mm culture dishes were washed three times with ice-cold phosphate buffered-saline (PBS) and collected in radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris [pH 8.0], 1% NP-40, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) with protease inhibitors (catalog number 78410; Thermo Fisher Scientific Inc., Rockford, IL). After a 30-min incubation on ice, cell lysates were sonicated and centrifuged at 14,000 × g for 15 min at 4°C, and the clear supernatants were saved as whole-cell lysates. To isolate the mitochondrion-enriched fraction, cells were washed with PBS, using the mitochondrion isolation kit's protocol (catalog number 89874; Thermo Fisher Scientific Inc., Rockford, IL). For nuclear fractionation, the nuclear and cytoplasmic extraction kit (catalog number 78833; Thermo Fisher Scientific Inc., Rockford, IL) was used. Protein concentrations were determined using the DC protein assay (catalog number 500-0112; Bio-Rad, Richmond, CA). Cell lysates (20 to 40 μg) were resolved by SDS-PAGE and transferred to nitrocellulose membranes according to standard procedures. Membranes were washed in Tris-buffered saline containing 0.05% Tween 20 (TBST), blocked for 1 h with TBST containing 5% nonfat milk, and then incubated with primary antibodies at a 1:1,000 dilution in TBST containing 5% nonfat milk overnight at 4°C. Membranes were then washed with TBST before incubation with horseradish peroxidase-conjugated secondary antibody (3 μg/liter in TBST containing 5% nonfat milk) for 1 h at RT. Membranes were washed four times with TBST before antibody binding was visualized with SuperSignal West Femto maximum-sensitivity substrate (catalog number 34095; Thermo Fisher Scientific Inc., Rockford, IL) according to the manufacturer's protocol. Equal loading of whole lysate protein, nuclear protein, and mitochondrial protein was verified with β-actin, PARP, and Hsp60, respectively.
Co-IP.
For coimmunoprecipitation (co-IP), 1% NP-40 lysis buffer was used instead of RIPA buffer. Protein concentrations of the lysates were quantified by DC protein assay (catalog number 500-0112; Bio-Rad, Richmond, CA). Two hundred micrograms of mitochondrion-enriched lysate was incubated overnight with 1 μg anti-Bcl-2 antibody (Cell Signaling) with agitation at 4°C. The resulting immune complex was precipitated by adding 30 μl Dynabeads M-280 sheep anti-rabbit IgG (SKU number 112-03D; Invitrogen Dynal As, Oslo, Norway) with agitation at 4°C for 2 h, washed four times with the lysis buffer, subjected to 10% SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was probed with the anti-Flag antibody (1:1,000; Sigma).
Colorimetric assay of caspase activities.
Measurements were performed using a kit for each caspase (catalog numbers K106-100, K113-100, and K119-100; BioVision, Inc., Mountain View, CA), in accordance with the manufacturer's instructions. The kits detect specific colorimetric substrates which, when cleaved, increase light absorption at 405 nm. Briefly, cells were cultured overnight on 60-mm culture dishes and then treated with various agents as required. Cells were resuspended in cell lysis buffer provided in the kits and incubated on ice for 10 min, and then cell lysates were centrifuged at 10,000 × g for 1 min at 4°C to collect the supernatants. Protein concentrations were determined using the DC protein assay (catalog number 500-0112; Bio-Rad, Richmond, CA). Two hundred micrograms of protein for each assay was incubated with reaction buffer and substrate at 37°C for 2 h and then measured in a microtiter plate reader.
Athymic mouse pancreatic cancer xenograft studies.
Athymic 5-week-old female nu/nu mice were injected intraperitoneally (i.p.) with 106 BxPc-3 cancer cells for 1 week. Mice were then randomized to oral treatment with either AHPN (10 mg/kg body weight), 3-Cl-AHPC (20 mg/kg body weight), or an equal volume of vehicle (cremophor EL) daily for 5 days. A mixture of 10 μl/mg of firefly d-luciferin (15 mg/ml) (Xenogen), 200 μl ketamine HCl, and 20 μl xylazine (100 mg/ml) was injected i.p. immediately prior to bioluminescent imaging to anesthetize the mice and provide a substrate for the luciferase-expressing cancer cells. The peritoneal tumor implants were imaged with the Xenogen bioluminescent imaging system. The tumor implants for each mouse were verified by microscopy and histopathology.
Mitochondrial respiration assay.
Mitochondrial oxygen consumption and ATP production were measured using techniques described previously (31). Isolation of mitochondria was performed in livers from 10-week-old male mice. Mice were killed by cervical dislocation. The livers were immediately placed in ice-cold STE buffer (250 mM sucrose, 5 mM Tris, 2 mM EGTA at pH 7.4) and cut into small pieces. After homogenization four times, samples were centrifuged at 1,000 × g for 3 min and the pellet discarded. The resultant supernatant was then centrifuged three times at 12,000 × g for 10 min. The isolated mitochondria were resuspended in 500 μL STE buffer, and the concentration of mitochondria was determined using the Bradford protein assay (500-0203; Bio-Rad, Hercules, CA). Rates of oxygen consumption by mitochondria were measured with a FOXY-R-AF probe (Ocean Optics Inc., Dunedin, FL). Measurements were performed with two independent substrates in respiration buffer containing 120 mM KCl, 5 mM KH2PO4, 1 mM EGTA, 1 mg/ml BSA, and 3 mM HEPES, pH 7.2, at 25°C:. Substrates used were (i) 0.02 mM palmitoyl-carnitine and 5 mM malate or (ii) 5 mM succinate and 0.01 mM rotenone. Oxygen consumption rates were expressed as nmol of O2·min−1·mg dry weight−1. Respiratory parameters were defined as follows. Basal respiration rates before the addition of ADP (V0) were defined as state 2. Maximal ADP (1 mmol/liter)-stimulated respiration rates (VADP) were defined as state 3, and respiration rates in the absence of ADP phosphorylation and measured in the presence of 1 μg/ml oligomycin (Voligomycin) were termed state 4. Respiration assays were performed in triplicate. The ATP concentration was determined by a bioluminescence assay based on the luciferin/luciferase reaction with the ATP assay kit (ThermoLabsystems).
Detection of ROS.
Production of reactive oxygen species (ROS) was measured using 2′,7′-dichlorofluorescein diacetate (H2DCFDA) (Sigma, D6883). Huh7 cells were grown in 60-mm dishes and loaded with 20 μM H2DCFDA for 2 h at 37°C. After washing twice with ice-cold PBS, cell fluorescence was immediately analyzed using a FACScan flow cytometer (Becton-Dickinson Immunocytometry Systems, San Jose, CA).
Biochemical analysis of Flag-SHP localization in mitochondria. (i) Subcellular fractionations.
Twenty micrograms of Flag-SHP plasmid DNA was transfected into Huh7 cells in a 15-cm dish. The transfected cells were harvested and disrupted in isotonic buffer (PBS containing 0.2 M mannitol, 0.07 M sucrose, and 1 mM EDTA) containing protease inhibitors (catalog number 78410; Thermo Fisher Scientific Inc., Rockford, IL), followed by centrifugation at 800 × g at 4°C for 10 min to obtain postnuclear supernatant (PNS). PNS was centrifuged at 17,000 × g at 4°C for 10 min to obtain the mitochondrion-enriched precipitate fraction (P1). The supernatant was centrifuged at 170,000 × g at 4°C for 30 min to separate the microsome-enriched precipitate (P2) and supernatant (S) fractions. The subcellular fractions were separated by SDS-PAGE and then analyzed by Western blotting.
(ii) Alkaline treatment of mitochondria.
One hundred micrograms of mitochondrial protein was prepared from the Huh7 cells expressing Flag-SHP protein and treated with 100 mM Na2CO3 in 10 times the volume of mitochondrial suspension for 1 h on ice. The reaction mixtures were centrifuged at 170,000 × g at 4°C for 30 min to separate the precipitate and supernatant fractions. The fractions were subjected to SDS-PAGE followed by Western blot analysis.
(iii) Trypsin protection assay.
The mitochondrion were treated with either H2O or 2% Triton X-100 in 10 times the volume of mitochondrial suspension on ice for 1 h and then treated with 50 μg/ml trypsin on ice for 1 h. The reaction mixtures were separated by SDS-PAGE and then analyzed by Western blotting.
Statistical analysis.
All the experiments were repeated at least three times, and error bars represent the standard errors of the means (SEM). Statistical analyses were carried out using Student's unpaired t test; a P value of <0.01 was considered statistically significant.
RESULTS
SHP activates apoptotic signaling in mouse hepatocytes.
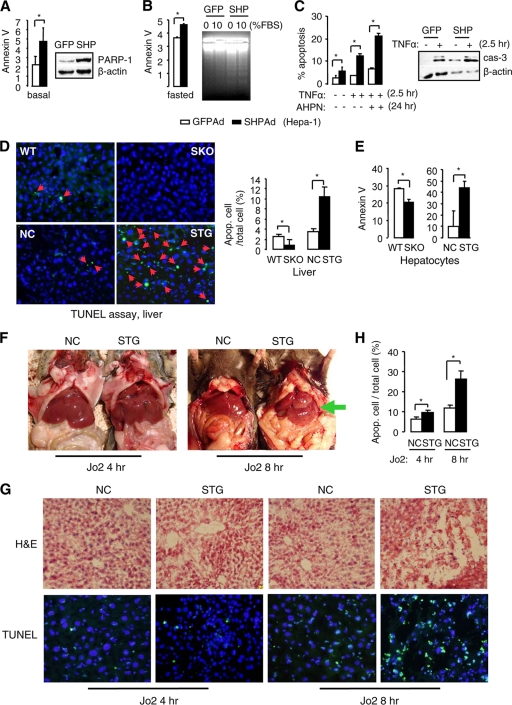
To determine the apoptotic effect of SHP, we first tested the mouse HCC Hepa-1 cell line, which has lost endogenous SHP expression (37, 46). Overexpression of SHP resulted in a marked increase in the percentage of apoptotic cells as examined by annexin V staining (Fig. 1A, left) and a robust induction of the cleaved form of the poly(ADP-ribose) polymerase (PARP-1) protein as determined by Western blotting (right). PARP-1 functions as a DNA damage sensor and is activated in response to DNA injury (42). SHP overexpression also enhanced the sensitivity of cells to starvation-induced apoptosis as assayed by annexin V staining (Fig. 1B, left) and DNA fragmentation (right). Although treatment with tumor necrosis factor alpha (TNF-α) induced apoptosis in green fluorescent protein (GFP)-adenovirus-infected cells, the effect was more dramatic in the presence of SHP, as confirmed by annexin V staining (Fig. 1C, left) and by increased cleavage of caspase-3 protein (right). In addition, cells overexpressing SHP exhibited higher sensitivity to AHPN-induced apoptosis than controls (Fig. 1C, left). Thus, SHP overexpression alone was sufficient to induce apoptosis in the absence of apoptotic stressors. SHP also enhanced the apoptotic effects of TNF-α and AHPN.
FIG. 1.
Overexpression of SHP induces hepatocyte apoptosis. (A) Hepa-1 cells were transduced with GFP or SHP adenovirus for 2 days. The percentage of apoptotic cells was analyzed by annexin V staining (left), and PARP-1 protein cleavage was analyzed by Western blotting (right). Data are represented as mean ± SEM (*, P < 0.01). (B) Hepa-1 cells were serum starved with 0.01% serum for 72 h. Left, annexin V staining of the apoptotic cells (*, P < 0.01). Right, DNA fragmentation analysis of cells without or with SHP overexpression. (C) Annexin V staining of the apoptotic cells treated with TNF-α or AHPN (left) and Western blotting of cleaved caspase-3 protein (right). Data are represented as mean ± SEM (*, P < 0.01). (D) TUNEL assays of liver sections from wild-type (WT), SHP−/− (SKO), SHP nontransgenic (NC), and hepatocyte specific SHP-transgenic (STG) mice (left). Hepatocytes apoptosis was significantly higher in STG mice than in NC mice (right). Mice were analyzed at 10 to 12 months of age. Data are represented as mean ± SEM (*, P < 0.01). (E) Analysis of apoptosis in hepatocytes from 2-month-old WT, SKO, NC, and STG mice by annexin V staining (n = 3). Data are represented as mean ± SEM (*, P < 0.01). (F) Gross morphology of livers from NC and STG mice treated with Jo2 antibody (10 μg/25 g/mouse). (G) H&E (top) and TUNEL (bottom) staining of liver sections from NC and STG mice treated with Jo2 antibody. (H) Statistical analysis (mean ± SEM; *, P < 0.01) of the number of apoptotic hepatocytes from NC and STG mice treated with Jo2 antibody, as determined by TUNEL assays in panel G. A total of six fields from each group (n = 3) were counted.
To further evaluate the contribution of SHP in regulating apoptotic signaling in vivo, we performed TUNEL assays of liver sections from wild-type (WT), SHP−/− (SKO), and hepatocyte-specific SHP-transgenic (STG) mice and their nontransgenic littermate controls (NC). Ten- to 12-month-old mice were used because older SKO mice showed marked alterations of hepatocyte proliferation and developed HCC (46). Deletion of SHP decreased apoptotic hepatocytes in SHP−/− livers, whereas overexpression of SHP in STG livers induced robust apoptosis (Fig. 1D, red arrowheads). In addition, in primary hepatocytes isolated from 2-month-old mice, the number of apoptotic cells was consistently lower in SKO mice and higher in STG mice compared to their controls (Fig. 1E).
Death receptor-mediated hepatocyte apoptosis is implicated in a wide range of liver diseases, including cancer. Fas (also known as APO-1/CD95) is an apoptosis-signaling receptor molecule that is highly expressed in the liver, and in vivo administration of the agonistic anti-Fas Jo2 antibody in mice induces severe apoptosis in the liver (28, 34). Because older STG mice showed increased apoptosis in their livers, we used 2-month-old STG mice to determine the sensitivity of the mice to Jo2-induced apoptosis. Four hours after Jo2 injection, the gross appearance of the liver did not differ in both types of mice (Fig. 1F, left). However, histologic lesions of the STG liver after antibody injection showed more extensive hemorrhage and apoptosis than in control mice (Fig. 1G, left four panels). By 8 h following Jo2 administration, massive hemorrhagic destruction of the liver was observed in STG mice by gross appearance (Fig. 1F, right, green arrow) and by histology (Fig. 1G, right four panels), which was present to a much lesser extent in control mice. The hemorrhagic lesions were characterized by widespread hepatocyte destruction. Cytoplasmic swelling of most surviving hepatocytes in STG mice was accompanied by frequent nuclear chromatin condensation, indicative of apoptosis. Quantitation of apoptotic nuclei showed a significant increase in Jo2-treated STG mice relative to similarly treated controls (Fig. 1H). In contrast, SKO primary hepatocytes showed a decreased response to Jo2-induced cell death compared to controls (data not shown). These data demonstrate that mice overexpressing SHP in hepatocytes are hypersensitive to Fas-induced hepatocyte apoptosis.
Deletion of SHP diminishes apoptotic signaling in mouse fibroblasts.
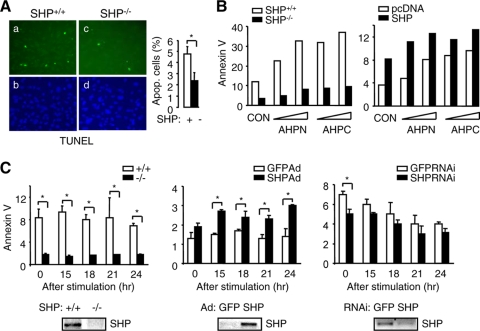
To determine if a similar proapoptotic effect of SHP exists in other cell systems, we examined the sensitivity of mouse embryonic fibroblasts (MEFs) lacking SHP to UV irradiation, which induces mitochondrion-mediated apoptosis. DAPI and TUNEL staining of the nuclei revealed that SHP−/− cells displayed greater viability after stimulation by UV and were markedly less sensitive to UV-induced apoptosis than SHP+/+ cells (Fig. 2A). In addition, apoptosis induced by AHPN and 3-Cl-AHPC was significantly attenuated in SHP−/− relative to SHP+/+ cells (Fig. 2B, left), and the sensitivity of SHP−/− cells to both agents was partially restored by SHP reexpression (Fig. 2B, right, and data not shown). When cells were synchronized in G0 and subsequently released into the cell cycle, apoptosis rates were consistently lower in SHP−/− than in control cells (Fig. 2C, left). Overexpression of SHP by adenovirus in SHP−/− cells induced apoptosis (Fig. 2C, middle), and knockdown of SHP by siRNA in SHP+/+ cells decreased apoptosis (Fig. 2C, right). The modest effect of SHP knockdown was due to the low basal level of SHP in MEFs. Thus, SHP appeared to function as an active component of the apoptotic signaling system in MEFs.
FIG. 2.
AHPN induces apoptosis through SHP in fibroblasts. (A) Micrographs of DAPI (panels b and d) and TUNEL (panels a and c) staining of immortalized SHP+/+ and SHP−/− MEF cells exposed to UV (left). Statistical results represent means ± SEMs of apoptotic cell counts from six different fields (right) (*, P < 0.01). (B) Left, responses of SHP+/+ and SHP−/− MEFs to AHPN (1 or 10 μM)- or 3-Cl-AHPC (10 or 100 μM)-induced apoptosis. Right, SHP−/− MEFs were transfected with pcDNA3 or SHP expression plasmid and then treated with AHPN or 3-Cl-AHPC. Cells were analyzed by annexin V staining. CON, control (0.1% dimethyl sulfoxide [DMSO]). The experiments were repeated three times (triplicate plates/time) with similar results. One representative result is shown. (C) The MEF cells were synchronized at G0 and released into the cell cycle, and apoptotic cells were determined. Left, percentage of apoptotic cells in SHP+/+ and SHP−/− cells; middle, reexpression of SHP in SHP−/− cells by adenovirus (Ad); right, knockdown of SHP in SHP+/+ cells by RNAi. Bottom, semiquantitative PCR analysis of SHP mRNA expression in SHP+/+ and SHP−/− MEFs (left), SHP−/− MEFs that overexpressed SHP adenovirus (middle), and SHP+/+ MEFs that were infected with SHP siRNA (right). Data are represented as mean ± SEM (*, P < 0.01).
AHPN and 3-Cl-AHPC induce SHP gene expression via LRH-1.
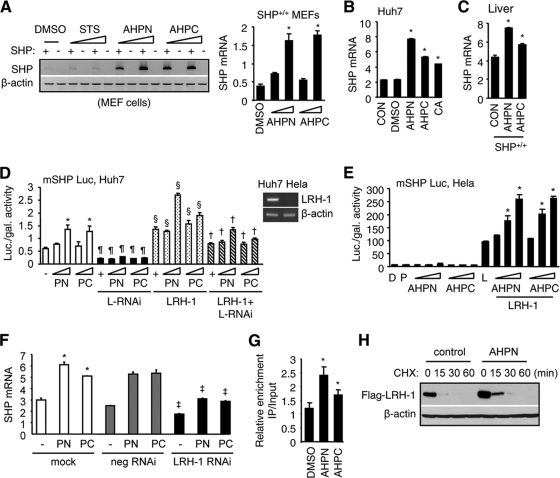
AHPN and 3-Cl-AHPC are retinoid-like agonists that bind to SHP protein (8). However, it remains to be determined if they regulate SHP expression at the transcriptional level. We first determined if APHN and 3-Cl-AHPC induced SHP mRNA expression in MEFs. AHPN and 3-Cl-AHPC significantly induced SHP mRNA levels in a dose-dependent manner in SHP+/+ MEFs (Fig. 3A), which was not seen with staurosporine (STS), another initiator of apoptosis, suggesting a specific effect of both agents on activating SHP expression.
FIG. 3.
AHPN and 3-Cl-AHPC activate SHP gene expression via LRH-1. (A) Semiquantitative (left) and real-time PCR (right) analyses of SHP mRNAs in SHP+/+ and SHP−/− MEFs treated with AHPN (1 or 10 μM) or 3-Cl-AHPC (10 or 100 μM). Staurosporine (0.1 or 10 μM) showed no effect and thus served as a negative control. (B) Real-time PCR analysis of SHP expression in Huh7 cells. CON, nontreated cells; DMSO, 0.1%; AHPN, 1 μM; 3-Cl-AHPC, 10 μM. Cholic acid (CA) (100 μM) served as a positive control. Data are represented as mean ± SEM (*, P < 0.01). (C) Real-time PCR analysis of hepatic SHP mRNAs in SHP+/+ mice following i.p. administration of AHPN (10 mg/kg/mouse) or 3-Cl-AHPC (10 mg/kg/mouse) for 3 days. Data are represented as mean ± SEM (*, P < 0.01). (D) Transient-transfection assays of mouse SHP (mSHP) promoter luciferase (Luc) reporter in Huh7 cells. Luc activities were determined and normalized to β-galactosidase (gal) activities. PN, AHPN (1 or 10 μM); PC, 3-Cl-AHPC (10 or 100 μM); L-RNAi, LRH-1-RNAi. *, significant difference compared to the group (lane 1) without AHPN and 3-Cl-AHPC treatment (P < 0.01); ¶, significant difference compared to each corresponding group without L-RNAi (white bars); §, significant difference compared to each corresponding group without (white bars) and with (black bars) L-RNAi; †, significant difference compared to each corresponding group with L-RNAi (black bars) and LRH-1 (stippled bars). Top right, reverse transcription-PCR (RT-PCR) analysis of LRH-1 mRNA in Huh7 and HeLa cells. (E) Transient-transfection assays of mouse SHP (mSHP) promoter luciferase (Luc) reporter in HeLa cells. D, DMSO; P, pcDNA3; L, LRH-1. Doses, AHPN, 0.1, 1, and 10 μM; 3-Cl-AHPC, (1, 10, and 100 μM. Data are represented as mean ± SEM (*, P < 0.01 versus L). (F) Real-time PCR analysis of SHP mRNA in Huh7 cells transfected with negative (neg) control or LRH-1 RNAi in the presence of AHPN (PN) and 3-Cl-AHPC (PC). *, significant difference compared to nontreated group; ‡, significant difference compared to each corresponding “mock” group. (G) ChIP assays to determine the enhanced recruitment of LRH-1 to the endogenous hSHP promoter by AHPN (10 μm) and 3-Cl-AHPC (100 μm) in Huh7 cells (*, P < 0.01 versus DMSO). (H) Western blotting to determine the effect of AHPN (1 μm) on LRH-1 protein degradation in the presence of the protein synthesis inhibitor cycloheximide (CHX) (50 μm) in Huh7 cells.
Due to the lack of specific antibodies against the mouse SHP, human hepatoma Huh7 cells were further tested for this and subsequent studies because a specific human SHP antibody is available for analysis of the cellular distribution of hSHP by immunohistochemistry (12). Using human Huh7 cells would also allow us to test if the findings in mice are applicable to humans. As expected, the mRNA expression of SHP was upregulated by AHPN and 3-Cl-AHPC, as well as by cholic acid (CA), in Huh7 cells (Fig. 3B); the latter was known to induce SHP expression and thus served as a positive control. In agreement with these in vitro findings, in vivo administration of AHPN and 3-Cl-AHPC in mice increased SHP expression in SHP+/+ (Fig. 3C), but not in SHP−/− (not shown) liver. It was noted that the efficacy of 3-Cl-AHPC to induce SHP expression was weaker than that of AHPN in both Huh7 cells and mouse liver.
In Huh7 cells which express high endogenous levels of liver receptor homolog 1 (LRH-1, NR5A2) (Fig. 3D, top right), AHPN and 3-Cl-AHPC activated the SHP promoter in a dose-dependent manner in the absence of LRH-1 cotransfection (white bars) but also significantly enhanced the transactivation of the SHP promoter by exogenously expressed LRH-1 (stippled bars). The effects of AHPN and 3-Cl-AHPC were via LRH-1 because they were markedly decreased by knockdown of LRH-1 using siRNA (black and hatched bars) but not by a negative-control siRNA (not shown). On the other hand, HeLa cells have low levels of endogenous LRH-1 (Fig. 3D, top right), and AHPN and 3-Cl-AHPC did not show activation of the SHP promoter without LRH-1 cotransfection but were able to enhance the effect of ectopically expressed LRH-1 (Fig. 3E). In contrast, STS showed no such effect on SHP promoter activity (data not shown), consistent with the results that it did not induce SHP (Fig. 3A). In addition, the activation effects of AHPN and 3-Cl-AHPC were not seen with retinoic acid receptors RAR/RXR (data not shown) and hepatocyte nuclear factor 4 alpha (HNF4α, NR2A1) (data not shown); both are known transactivators of the SHP promoter, further indicating the specific effects of AHPN and 3-Cl-AHPC on LRH-1 activation of the SHP promoter. In agreement with these observations, both the endogenous SHP and SHP induced by AHPN and 3-Cl-AHPC were decreased by LRH-1 RNAi (Fig. 3F).
The proximal promoter region of human SHP contains three LRH-1 response elements (LRE) (nucleotides [nt] −100 to −109, −305 to −314, and −338 to −346). Next, the effects of AHPN and 3-Cl-AHPC on the recruitment of LRH-1 to the endogenous hSHP promoter were determined in Huh7 cells using chromatin immunoprecipitation (ChIP) assays (Fig. 3G). Huh7 cells were transfected with a Flag-LRH1 expression vector, and LRH-1 co-IP on the hSHP promoter of Huh7 cells was determined using an anti-Flag antibody with specific primers covering the three LREs in the hSHP promoter. We observed an enhanced recruitment of LRH-1 to the hSHP promoter by both agonists (Fig. 3G). In contrast, primers located 4 kb upstream of the promoter did not produce PCR products (data not shown). Because AHPN did not induce LRH-1 mRNA expression (data not shown) or show transcriptional activation on Gal4-LRH-1 in the mammalian one-hybrid assay (data not shown), which indicated that it did not bind to LRH-1, we reasoned that AHPN may enhance the transactivation of the SHP promoter by LRH-1 via increasing LRH-1 protein stability. The degradation rate of LRH-1 was monitored after the Huh7 cells were treated with the protein synthesis inhibitor cycloheximide (CHX) to block protein synthesis. The half-life of LRH-1 was about 15 min after CHX treatment, which extended to 30 min in the presence of AHPN (Fig. 3H). The increased basal level of LRH-1 by AHPN before CHX treatment (0 min) was consistent with its decreased degradation rate (Fig. 3H). These data suggest that AHPN and 3-Cl-AHPC function as potent activators of SHP gene transcription by enhancing LRH-1-mediated SHP expression.
SHP shuttles between nucleus and mitochondria.
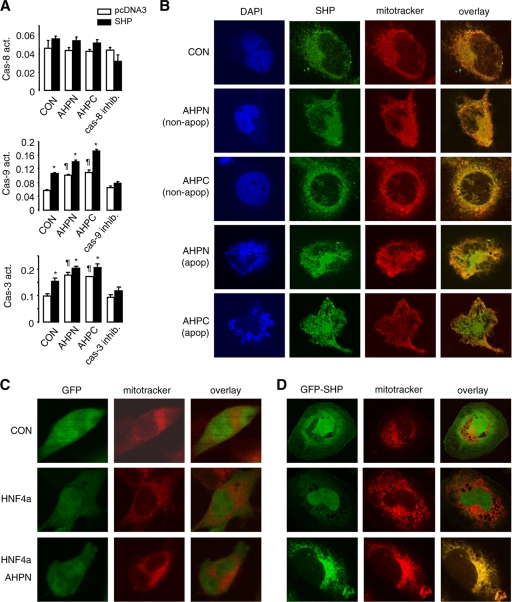
Thus far we have shown that SHP not only activated apoptosis but also enhanced apoptosis induced by AHPN and 3-Cl-AHPC and by the death receptor ligands TNF-α and Jo2. To elucidate the mechanism by which SHP activates apoptosis, we examined the activities of several caspases. Caspase-8 is the first cleaved caspase in the death receptor-mediated extrinsic pathway (5). Overexpression of SHP in Huh7 cells led to moderate activation of caspase-8 activity, but AHPN and 3-Cl-AHPC did not increase caspase-8 activity in nontransfected cells (Fig. 4A, top). Caspase-9 (Fig. 4A, middle) and caspase-3 (Fig. 4A, bottom, and data not shown) are cleaved mainly during mitochondrially mediated apoptosis, and their activities were independently increased by SHP and by AHPN and 3-Cl-AHPC, respectively. Thus, we hypothesized that the major proapoptotic effect of SHP was through its action on mitochondria.
FIG. 4.
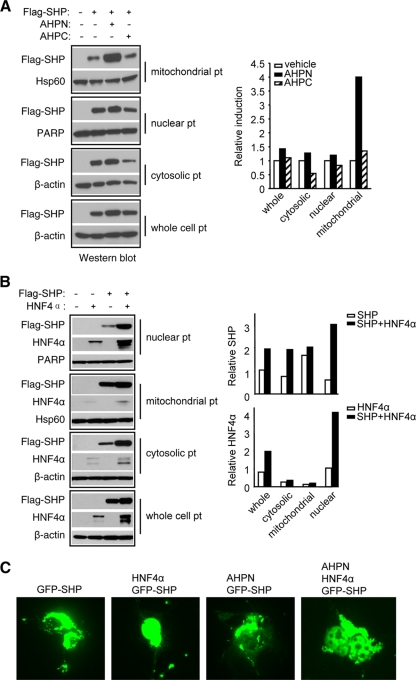
AHPN and HNF4α induce SHP trafficking between mitochondria and nucleus. (A) Caspase-8, -9, and -3 activities in Huh7 cells transfected with pcDNA3 (empty bars) or SHP (filled bars) in the absence (CON, 0.1% DMSO) or presence of AHPN (1 μM) or 3-Cl-AHPC (10 μM). Data are represented as mean ± SEM. *, P < 0.01 for SHP versus pcDNA3; ¶, P < 0.01 for AHPN or 3-Cl-AHPC versus pcDNA3. (B) Micrographs of DAPI, SHP, and MitoTracker staining of Huh7 cells. Human SHP protein was detected by immunocytochemistry using specific hSHP antibodies. CON, 0.1% DMSO; AHPN, 1 μM; 3-Cl-AHPC, 10 μM. (C) GFP control adenoviruses were cotransfected with pcDNA3 (CON) or HNF4α expression vector in Huh7 cells without or with AHPN (1 μM) treatment. Subcellular localization of GFP was observed by confocal laser scanning microscopy. (D) GFP-SHP adenoviruses were cotransfected with pcDNA3 (CON) or HNF4α expression vector in Huh7 cells without or with AHPN (1 μM) treatment. Subcellular localization of GFP-SHP was observed by a confocal laser scanning microscopy.
We determined the endogenous SHP subcellular localization in Huh7 cells by immunocytochemistry using specific hSHP antibodies (12). Unexpectedly, the SHP protein was located in a cytoplasmic compartment that colocalized with the mitochondrial marker MitoTracker under normal culture conditions (Fig. 4B, row 1). For cells that have not undergone apoptosis induced by AHPN and 3-Cl-AHPC, as indicated by intact nuclei, the SHP cellular distribution appeared to be mainly located in mitochondria, similar to the case for control cells (rows 2 and 3). When cells underwent apoptosis as evidenced by condensed nuclei, a diffuse distribution pattern of SHP throughout the cells was observed (rows 4 and 5), suggesting that SHP was released into the cytosol during apoptosis.
Because of the localization of endogenous SHP in mitochondria, it was technically difficult to ascertain if AHPN or 3-Cl-AHPC played a role in nucleus-to-mitochondrion shuttling of SHP. It has been reported that HNF4α possesses a nuclear localization signal (NLS) and is always located in the nucleus in the HIT-T15 pancreatic β-cell line (17). In contrast, SHP lacks such a signal and resides primarily in the cytoplasm in the absence of HNF4α in COS-7 cells, but it can be transported to the nucleus by overexpression of HNF4α (29). Thus, we overexpressed an SHP-GFP fusion protein in Huh7 cells and examined the effects of HNF4α and AHPN on the exogenous SHP cellular distribution.
In cells infected with GFP alone (Fig. 4C), the GFP signal was located in both the cytoplasm and nucleus (top), and its distribution pattern was altered neither by HNF4α coexpression (middle) nor by AHPN treatment (bottom). In GFP-SHP-infected cells, GFP-SHP appeared to reside throughout the cytoplasm and nucleus, with part of the cytoplasmic signal colocalizing with MitoTracker (Fig. 4D, top). Exogenous coexpression of a wild-type HNF4α induced redistribution of most of the GFP-SHP into the nucleus (middle). In contrast, treatment with AHPN resulted in SHP cytoplasmic translocation which overlapped exclusively with MitoTracker (bottom). Thus, APHN was able to induce the nucleus-to mitochondrion translocation of SHP.
AHPN and HNF4α affect SHP cellular trafficking and protein stability.
To further confirm that AHPN and 3-Cl-AHPC affected mitochondrial translocation of SHP, we examined SHP protein levels by Western blotting by overexpressing a Flag-SHP expression plasmid in Huh7 cells and examined SHP protein levels using anti-Flag antibodies. Flag-SHP was observed in all cellular fractions, and its accumulation was significantly increased by AHPN in the mitochondrial fraction (Fig. 5A). Interestingly, cytosolic or nuclear SHP levels were not decreased. 3-Cl-AHPC treatment slightly increased the level of SHP protein in mitochondria, with a concomitant decrease in the cytosol and nucleus.
FIG. 5.
AHPN and HNF4α regulate SHP protein cellular translocation. (A) Left, Western blots of SHP protein expression in Huh7 cells transfected with a Flag-SHP vector or treated with AHPN (1 μM) and 3-Cl-AHPC (10 μM). We overexpressed the cells with a Flag-SHP expression vector and examined the level of SHP protein with an anti-Flag antibody. pt, protein. Right, band intensities were measured by densitometry, and the intensities relative to that of the vehicle (set as 1) were plotted. (B) Left, Western blots of SHP protein expression in COS-7 cells treated with AHPN (1 μM) and 3-Cl-AHPC (10 μM) after transfection with a Flag-SHP or HNF4α plasmid. Right, band intensities were measured by densitometry and normalized to each corresponding internal control. (C) GFP-SHP plasmid was cotransfected with HNF4α expression vector in COS-7 cells without or with AHPN treatment, and subcellular localization of GFP-SHP was observed by microscopy.
We next determined if HNF4α could increase SHP nuclear protein levels. Huh7 cells express abundant HNF4α protein exclusively in the nucleus, but with two different sizes that are smaller than the wild-type HNF4α (not shown). To eliminate the interference of those unknown forms of HNF4α on SHP nuclear translocation, we used COS-7 cells, which have undetectable endogenous HNF4α (29), to express a wild-type HNF4α. The nuclear Flag-SHP was markedly increased in cells that were cotransfected with an HNF4α expression vector compared to cells transfected with the SHP vector alone (Fig. 5B). Interestingly, the mitochondrial SHP level was not decreased, and cytosolic SHP was rather increased. On the other hand, HNF4α protein, as visualized by anti-HNF4α antibodies, was increased in nuclear lysate isolated from SHP coexpression cells, which was consistent with its increased level in whole-cell lysate. This indicates that the interaction between SHP and HNF4α may facilitate translocation of both proteins to the nucleus. When COS-7 cells were transfected with a GFP-SHP plasmid, the majority of GFP signal was located in the cytoplasmic compartment, but it was largely concentrated to the nucleus in the presence of HNF4α (Fig. 5C). GFP-SHP signal remained in the cytosol in cells treated with AHPN alone, which was redistributed, although not completely, to the nucleus with HNF4α overexpression. Taken together with the observation in Fig. 4D, the results suggest that there exists a competition between AHPN and HNF4α to traffic SHP between mitochondria and nucleus.
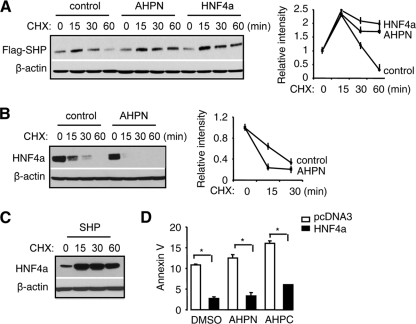
To address the question of whether SHP protein stability was altered by AHPN and HNF4α, we treated Huh7 cells with the protein synthesis inhibitor cycloheximide (CHX) after Flag-SHP overexpression. The level of Flag-SHP started to decrease by 30 min of CHX treatment and was further decreased by 60 min (Fig. 6A). In contrast, Flag-SHP protein remained at constant levels until 60 min in the presence of AHPN and HNF4α, suggesting that it had a lower degradation rate. At 15 min of CHX treatment relative to 0 min in all groups, Flag-SHP levels were increased, indicating incomplete blocking of protein synthesis by CHX at this time point. An opposite effect of AHPN on HNF4α half-life was observed, where it facilitated the rate of HNF4α degradation (Fig. 6B). On the other hand, SHP overexpression efficiently blocked HNF4α degradation, as shown by the accumulation of HNF4α in the presence of CHX (Fig. 6C). Thus, the observations that SHP was not depleted from the nucleus by AHPN (Fig. 5A) and that it was not depleted from mitochondria and was increased in the cytoplasm by HNF4α (Fig. 5B) may be in part associated with the increased SHP stability. The increased total HNF4α in whole-cell lysate by SHP may be associated with the decreased HNF4α degradation.
FIG. 6.
AHPN regulates SHP and HNF4α protein stability. (A) Western blotting to determine the effects of AHPN (1 μM) and HNF4α on SHP protein degradation in the presence of the protein synthesis inhibitor cycloheximide (CHX) (50 μM) in Huh7 cells. The relative intensity of bands is presented on the right. (B) Western blots to determine the effect of AHPN (1 μM) on HNF4α protein degradation in the presence of the protein synthesis inhibitor cycloheximide (CHX) (50 μM) in Huh7 cells. (C) Western blots to determine the effect of SHP on HNF4α protein degradation in the presence of the protein synthesis inhibitor cycloheximide (CHX) (50 μM) in Huh7 cells. (D) Annexin V staining of Huh7 cells transfected with pcDNA3 (empty bars) or HNF4α (filled bars) in the absence (DMSO, 0.1%) or presence of AHPN (1 μM) or 3-Cl-AHPC (10 μM). Data are represented as mean ± SEM (*, P < 0.01).
If SHP mitochondrial targeting is crucial in activating apoptosis, it would be expected that overexpressing HNF4α would decrease apoptosis by sequestering SHP in the nucleus. As expected, overexpression of HNF4α significantly reduced basal and attenuated AHPN- and 3-Cl-APHC-induced apoptosis (Fig. 6C). Currently we do not have a satisfactory explanation for the cytoplasmic localization of the endogenous SHP in Huh7 cells, which should be determined in future studies. Nevertheless, ectopic expression of a functional wild-type HNF4α efficiently resulted in SHP trafficking to the nucleus, which can then be shuttled back to mitochondria by AHPN.
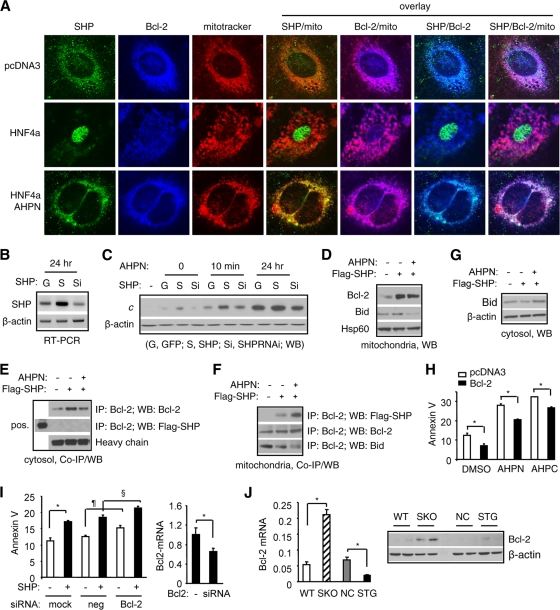
SHP colocalizes and interacts with Bcl-2 in mitochondria to induce cytochrome c release.
To determine how SHP activates mitochondrial apoptotic pathways, we analyzed the interaction of SHP with the well-characterized antiapoptotic protein B-cell lymphoma protein 2 (Bcl-2) (32). We hypothesized that SHP activates apoptosis through an interaction with Bcl-2 in mitochondria to disrupt its association with other proapoptotic proteins and block its antiapoptotic effect. We performed immunocytochemistry analysis of Huh7 cells using SHP and Bcl-2 antibodies and MitoTracker dye. In control pcDNA3-transfected cells, the SHP signal largely overlapped with mitochondrial Bcl-2 (Fig. 7A, row 1). Consistent with previous results, overexpression of HNF4α induced SHP translocation to the nucleus, which was not affected by 30 min of AHPN treatment (row 2). However, treatment with AHPN for 24 h resulted in SHP trafficking back to mitochondria and colocalizing with Bcl-2. AHPN did not induce marked changes in the levels of Bcl-2 or on Bcl-2 cellular distribution.
FIG. 7.
SHP colocalizes and interacts with Bcl-2 in mitochondria to induce cytochrome c release. (A) Immunocytochemical analysis of SHP and Bcl-2 colocalization in mitochondria of Huh7 cells using specific hSHP and Bcl-2 antibodies. (B) Semiquantitative PCR analysis of SHP mRNAs in Huh7 cells with SHP overexpression (S) or knockdown (Si). (C) Western blotting to determine cytochrome c (cyto c) release from mitochondria by SHP overexpression or knockdown without or with AHPN treatment in Huh7 cells. (D) Western blots to determine mitochondrial Bcl-2 and Bid accumulation by SHP overexpression in Huh7 cells. (E and F) Coimmunoprecipitation (co-IP) and Western blotting (WB) to determine SHP and Bcl-2 protein interaction in cytoplasm (E) and SHP and Bcl-2 or Bcl-2 and Bid protein interaction in mitochondria (F). Huh7 cells were overexpressed with a Flag-SHP expression vector in the absence or presence of AHPN (1 μM). Specific Flag, Bcl-2, or Bid antibodies were used in co-IP and WB. (G) Western blotting to determine cytosolic Bid accumulation by SHP overexpression in Huh7 cells. (H) Annexin V staining of Huh7 cells transfected with a control (empty bars) or Bcl-2 (filled bars) in the absence (DMSO, 0.1%) or presence of AHPN (5 μM) or 3-Cl-AHPC (50 μM). (I) Annexin V staining of Huh7 cells transfected with control (neg) or Bcl-2 siRNA in the absence or presence of SHP. Right, real-time PCR analysis of Bcl-2 mRNA in Huh7 cells transfected with Bcl-2 siRNA. Data are represented as mean ± SEM. (J) Left, real-time PCR analysis of hepatic Bcl-2 mRNA in wild-type (WT), SHP knockout (SKO), nontransgenic (NC), and liver SHP transgenic (STG) mice. Right, Western blotting of hepatic Bcl-2 protein in WT, SKO, NC, and STG mice.
Consistent with SHP mitochondrial localization, overexpression of SHP (Fig. 7B) induced cytochrome c release (Fig. 7C, lane 3), which was inhibited by SHP siRNA (lane 4). Short-term treatment with AHPN (10 min) enhanced both basal and SHP-induced cytochrome c release (lane 5 versus 2 and lane 6 versus 3), and the effect was attenuated in SHP RNAi-infected cells (lane 7). After 24 h of treatment, AHPN increased both basal and SHP-induced cytochrome c release (lanes 8 and 9).
Despite the colocalization in the mitochondria, it remained to be determined if SHP protein directly interacted with Bcl-2 protein on the mitochondrial membrane. We overexpressed a Flag-SHP protein in Huh7 cells and examined cells for a direct interaction of SHP with Bcl-2. Interestingly, SHP overexpression increased both mitochondrial (Fig. 7D) and cytosolic (Fig. 7E) Bcl-2 protein accumulation, whereas no enhanced effect was observed with AHPN treatment. Although Bcl-2 was readily detected in the cytosolic fraction of the cells, SHP was not observed to interact with Bcl-2 in the cytoplasm (Fig. 7E). Analysis of proteins purified from mitochondria revealed a direct interaction between SHP and Bcl-2 in cells overexpressing SHP (Fig. 7F, row 1, lane 2), which was strongly enhanced by APHN (lane 3).
We next asked how the SHP-Bcl-2 interaction resulted in cytochrome c release. We hypothesized that the binding of SHP to Bcl-2 disrupted the interaction of Bcl-2 with other proapoptotic proteins that and the release of the proapoptotic protein(s) resulted in cytochrome c release. We examined proapoptotic proteins Bax and Bok and the BH3-only proteins Bad, Bik, Bid, Puma, and Bim. Bid was coimmunoprecipitated with Bcl-2 in mitochondria (Fig. 7F, row 3, lane 1). The Bid/Bcl-2 mitochondrial interaction was markedly decreased in SHP-overexpressing cells (lane 2) and was further reduced in AHPN-treated cells (lane 3). The levels of Bid protein in mitochondria and cytoplasm were not affected by elevated SHP levels, whereas they were decreased in mitochondria (Fig. 7D, row 2) but increased in cytoplasm in cells treated with AHPN (Fig. 7G). In contrast, such effects of SHP and AHPN were not observed for other Bcl-2-interacting proapoptotic proteins (data not shown). Taking the results together, because the binding of Bcl-2 to Bid in mitochondria inhibits Bid's ability to activate apoptosis, the interaction of SHP with Bcl-2 disrupts the Bcl-2/Bid complex and prevents Bcl-2 from inhibiting Bid's proapoptotic effect.
If the sequestration of Bcl-2 by SHP leads to an increase of cytochrome c release, then overexpression of Bcl-2 may inhibit the effects of AHPN and 3-Cl-AHPC on apoptosis. As expected, ectopic expression of Bcl-2 attenuated the apoptotic effects of AHPN and 3-Cl-AHPC (Fig. 7H). In contrast, knockdown of Bcl-2 using siRNA enhanced basal as well as SHP-induced apoptosis (Fig. 7I). On the other hand, Bcl-2 expression was increased in SHP knockout mice but decreased in SHP transgenic mice, further indicating the involvement of Bcl-2 in SHP-mediated apoptosis (Fig. 7J).
Because AHPN was shown to target nuclear receptor TR3 (Nur77) to mitochondria in prostate and colon cancer cells (19), we asked if Nur77 is involved in SHP-mediated apoptosis in Huh7 cells. Nur77 was barely detectable in Huh7 and BxPc cells or induced by AHPN and 3-Cl-AHPC (data not shown), suggesting that it is unlikely to be involved in SHP-mediated apoptosis in those two cell lines. Interestingly, genes encoding a group of mitochondrial chaperon proteins were induced by AHPN and 3-Cl-AHPC (data not shown). It remains to be determined which chaperon protein plays an important role in SHP mitochondrial targeting.
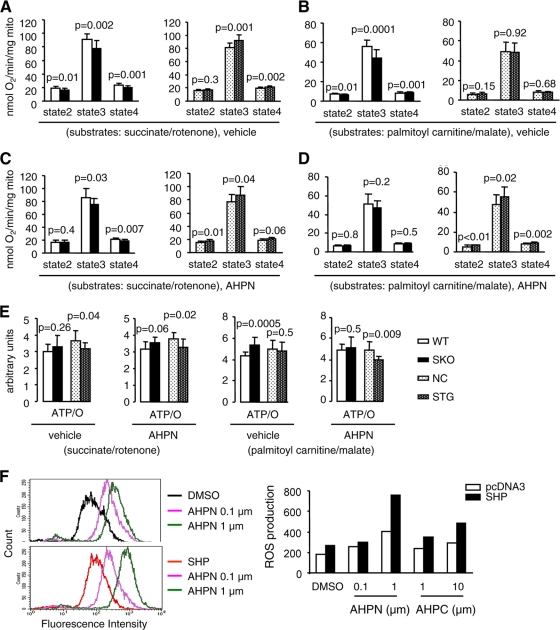
SHP modulates mitochondrial respiration and ROS production.
We sought to determine if SHP-mediated mitochondrial apoptotic signaling was associated with changes in mitochondrial oxygen consumption and energetics. Mitochondria were isolated from primary hepatocytes of WT, SKO, NC, and STG mice under normal conditions or treated with vehicle or AHPN for 1 day. Respiratory parameters were defined as follows. Basal respiration rates before the addition of ADP (V0) were defined as state 2. Maximally ADP-stimulated respiration rates (VADP) were defined as state 3, and respiration rates in the absence of ADP phosphorylation (measured in the presence of the ATP synthase inhibitor oligomycin [Voligomycin]) were termed state 4. Studies were performed with two substrates. Succinate in the presence of the complex I inhibitor rotenone reflects activity of the flavoprotein-dependent succinate dehydrogenase (complex II). Palmitoyl carnitine (PC) respiration reflects activity of the beta oxidation pathways and delivery of reducing equivalents to complex II and complex I (via the tricarboxylic acid [TCA] cycle).
Relative to those in littermate controls, state 3 mitochondrial respiration rates were lower in SKO mice in the presence of either succinate or PC (Fig. 8A and B). The reduction in succinate-supported respiration persisted in APHN-treated animals; however, respiration rates with PC were no longer reduced in AHPN-treated SKO mice (Fig. 8C and D). In contrast, overexpression of SHP was associated with increased succinate-supported respiration rates in both the presence and absence of AHPN treatment (Fig. 8A and B). Although basal rates of PC respiration were unchanged in STG mice, there was a further increase in PC-supported respiration following AHPN treatment (Fig. 8C and D). Examination of ATP/O ratios (a general measure of mitochondrial coupling) revealed that mitochondria from SKO mice were more coupled (increased ATP/O ratios) in untreated mice (with PC) and following treatment with AHPN (with succinate) (Fig. 8E). In contrast, in STG mice ATP/O ratios with succinate in untreated mice and with PC in vehicle- and AHPN-treated mice were reduced, suggesting mitochondrial uncoupling (data not shown). Increased state 4 respirations rates in STG mice with succinate (with or without AHPN) and PC (with AHPN) are also consistent with mitochondrial uncoupling (data not shown).
FIG. 8.
SHP regulates mitochondrial respiration and coupling in hepatocytes. (A to D) Mitochondrial respiratory rates of isolated hepatic mitochondria from 10-week-old WT, SHP KO (SKO), NC, SHP transgenic (STG) livers using succinate (5 mM)-rotenone (10 μM) or palmitoyl-carnitine (20 μM)-malate (5 mM) as substrates (n = 5 mice, triplicate assays of liver mitochondria isolated from each mouse liver). The mice were treated with vehicle (A and B) or APHN (C and D) (10 mg/kg of body weight) for 1 day by oral feeding. Data are represented as mean ± SEM. (E) ATP/O (ATP/state3) ratios with succinate-rotenone and palmitoylcarnitine-malate as substrates in isolated liver mitochondria from WT, SKO, NC, and STG mice treated with vehicle or AHPN. Data are represented as mean ± SEM. (F) Detection of reactive oxygen species (ROS) in Huh7 cells overexpressing an SHP expression plasmid in the absence (DMSO) or presence of AHPN (0.1 μM or 1 μM) or 3-Cl-AHPC (1 μM or 10 μM) for 24 h. The experiments were repeated twice (triplicate plates/time) with similar results. One representative result is shown.
The basis for increased uncoupling of STG mitochondria could reflect increased oxidative stress that could render them more prone to apoptosis. We examined the rate of reactive oxygen species (ROS) production in Huh7 cells. Expression of SHP increased cellular ROS production, which was further enhanced by AHPN or 3-Cl-AHPC in a dose-dependent manner (Fig. 8F).
Thus, coupled mitochondrial respiration in SHP-deficient hepatocytes is associated with resistance to apoptosis in SKO livers. In contrast, increased oxygen consumption and mitochondrial uncoupling in SHP-overexpressing liver cells might contribute to the activation of apoptosis or be a marker of increased apoptosis susceptibility in STG mice. Future studies will determine if increases in SHP expression modulate mitochondrial inner membrane potential and if these changes might contribute to cytochrome c release.
Activation of SHP by AHPN inhibits tumor growth.
We next asked if activation of apoptosis via induction of SHP by AHPN or 3-Cl-AHPC repressed tumor growth in vivo. We evaluated two pancreatic cancer cell lines, BxPc-3 and SU8 and one colon cancer cell line, HT29, which were stably transduced with a retrovirus containing pLuc-puro, a constitutive luciferase reporter. Implantation of these cells into the peritoneums of nude mice generates a peritoneal tumor model. The luciferase reporter in the cells permits in vivo monitoring of tumor growth using the Xenogen bioluminescent imaging system. Huh7 cells could not be used for this study, because they are not suitable for use as a peritoneal tumor model in vivo.
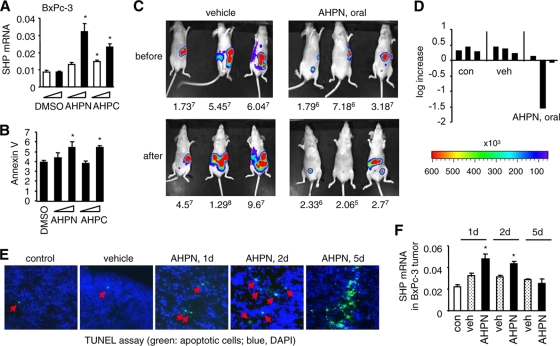
We first determined the effect of AHPN or 3-Cl-AHPC on SHP expression in cultured BxPc-3, SU8, and HT29 cells in vitro. The expression of SHP was significantly induced in all three cell lines treated with AHPN or 3-Cl-AHPC (Fig. 9A and data not shown). We chose to use BxPc-3 cells for subsequent in vivo studies. Induction of SHP by AHPN and 3-Cl-AHPC paralleled the activation of apoptosis (Fig. 9B).
FIG. 9.
Induction of SHP by AHPN inhibits tumor growth through activation of apoptosis. (A) Real-time PCR analysis of SHP expression in pancreatic cancer BxPc-3 cells treated with AHPN (0.1 and 1 μM) or 3-Cl-AHPC (1 and 10 μM). Data are represented as mean ± SEM (*, P < 0.01). (B) Annexin V staining of BxPc-3 cells treated with AHPN (0.1 or 1 μM) or 3-Cl-AHPC (1 or 10 μM). Data are represented as mean ± SEM (*, P < 0.01). (C) Bioluminescent detection of peritoneal tumor implants. Adult athymic female mice were injected i.p. with 106 BxPc-3 cancer cells, and the tumor implantation was examined 1 week later (top row, before). The mice then received vehicle (veh) or AHPN (oral) daily for 5 days (10 mg/kg of body weight) (bottom row, after). Mice were euthanized before and after the treatment and the tumor implants visualized by bioluminescence and confirmed by histopathology. The number below each mouse represents tumor images detected (photons/s/cm2/steridian). (D) The log increase represents the rate of peritoneal tumor growth before and after the treatment with AHPN. Each bar corresponds to each individual mouse in panel C. (E) TUNEL staining of peritoneal pancreatic tumors to examine the apoptotic cell death. (F) Real-time PCR analysis of SHP expression in peritoneal pancreatic tumors. Data are represented as mean ± SEM (*, P < 0.01).
We then determined the effect of AHPN or 3-Cl-AHPC on peritoneal BxPc-3 tumor growth in vivo. Nude mice received an i.p. injection of BxPc-3 cells, and the peritoneal tumor implants were imaged with the Xenogen bioluminescent imaging system 1 week later (Fig. 9C, top row). The mice then received vehicle (bottom row, left) or AHPN by oral feeding (right) every day for 5 days, and the tumor implants were imaged at the end of the feeding. Feeding with AHPN resulted in an average negative tumor growth rate (Fig. 9D) compared to that in the vehicle-treated nontreated groups. Activation of apoptosis in tumors was observed from day 1 of AHPN treatment, and they became massive by day 5 (Fig. 9E). 3-Cl-AHPC given by oral feeding showed less of an effect in inhibiting tumor growth than AHPN, although the decreased tumor growth was associated with marked induction of apoptosis (data not shown).
The levels of SHP expression were dramatically increased in tumors collected from the mice from day 1 of AHPN treatment (Fig. 9F). Unexpectedly, SHP levels were decreased by day 5 of AHPN feeding. Because SHP is able to repress its own expression, we reasoned that the decreased SHP level in tumors upon long-term AHPN feeding was due to a self-inhibitory mechanism. To test this hypothesis, SHP promoter reporter activity was assayed in HEK293 cells that lacked endogenous SHP as determined by quantitative PCR (not shown). LRH-1 transactivated the SHP promoter, which was inhibited by coexpression of SHP (data not shown). AHPN and 3-Cl-AHPC markedly enhanced LRH-1 activation of the SHP promoter, which was completely blocked by addition of SHP (data not shown). In addition, SHP protein was shown to have a short half-life, and SHP protein ubiquitination may also contribute to the downregulation of SHP (25). Thus, despite SHP downregulation by the end of AHPN feeding, apoptotic cell death was already induced and appeared to be sufficient to inhibit tumor growth. Taken together, the data demonstrated that induction of SHP by AHPN serves as a potent inhibitor of tumor growth.
DISCUSSION
Many studies have reported the apoptotic activity of AHPN or 3-Cl-AHPC in various different tumor cells, but a mechanism has been elusive. In this study, we have identified a mechanism of their action mediated by the SHP gene. The mechanism of AHPN-mediated apoptosis depends in part on its regulation of SHP. AHPN-induced apoptosis through SHP appears to be via distinct but synergistic mechanisms: (i) activation of transcription of the SHP gene in the nucleus and SHP protein translocation to the mitochondria and (ii) enhanced SHP/Bcl-2 interaction in mitochondria to disrupt Bcl-2/Bid interaction, leading to cytochrome c release. The finding provides new insight into SHP-dependent apoptotic signaling and may have important clinical implications.
Inhibition of SHP expression largely diminished but did not completely eliminate the proapoptotic effect of AHPN. This suggests that alternative mechanisms for AHPN-mediated apoptosis in an SHP-independent manner may exist. A report by Li et al. shows that in prostate and lung cancer cells, AHPN targets nuclear receptor TR3 (Nur77) to mitochondria (19), where TR3 interacts with Bcl-2 to convert Bcl-2 from an antiapoptotic to a proapoptotic molecule (23). However, Nur77 was not expressed or induced by AHPN and 3-Cl-AHPC in Huh7 and BxPc cells, suggesting that at least in those two cell types, SHP-induced apoptosis mediated by both agonists is independent of the AHPN-TR3 pathway. We show that the interaction between SHP and Bcl-2 disrupts the Bcl-2/Bid complex, thus preventing Bcl-2 from inhibiting Bid's proapoptotic effector. This provides a molecular basis that explains SHP's induction of cytochrome c release.
Interestingly, it was shown that SHP repressed the transactivation function of Nur77 and physically associated with Nur77 in the nucleus (45). How the interaction of SHP and Nur77 in the nucleus affects the function of both receptors in mitochondria remains unknown. We speculate that AHPN may play a role in dissociating SHP and Nur77 in the nucleus. AHPN would cause SHP mitochondrial targeting and release SHP-mediated retention of Nur77 in the nucleus, thereby promoting Nur77 mitochondrial translocation. Our data elucidate a common molecular basis for AHPN-mediated apoptosis through SHP in both mouse and human hepatoma cells and MEFs. It is likely that genetic differences in other tumor cells may require different signaling molecules that may determine their apoptotic fate in response to manipulation of SHP.
Huh7 cells easily undergo cell death in culture and are more sensitive to death stimulus-induced apoptosis than other HCC cells, in which SHP is localized in the nucleus, suggesting that the mitochondrial location of SHP sensitizes Huh7 cells to apoptosis. On the other hand, in the absence of apoptotic stimuli, overexpression of SHP alone is sufficient to trigger cellular apoptosis and induce mitochondrial cytochrome c release. Thus, SHP appears to function as a death receptor independent of external apoptotic stimulating agents, with a direct action in mitochondria. The present finding shows that SHP acts through a transcription-independent mechanism involving translocation to mitochondria and leading to cytochrome c release. The identification of SHP subcellular localization and function is of significance, because it may regulate activities of other known transcription factors, such as c-Myc and c-Jun, which mediate both cell death and proliferation (15, 24). The identity of endogenous signaling molecules that are involved in SHP mitochondrion targeting is currently unknown. Identifying such molecules will allow better understanding of the molecular and genetic program governing SHP-induced apoptosis.
Swales et al. reported that the nuclear receptor farnesoid X receptor (FXR), an SHP transcriptional activator, is present in human breast cancer cells and that activation of FXR by its ligands induces cell apoptosis and the expression of SHP mRNA (40). However, it remains unknown how FXR activation triggers apoptosis. Based on our studies, one potential explanation is that FXR activation induces SHP and SHP induces apoptosis. In contrast to the report by Swales et al., another report shows that FXR activation protects hepatocytes from apoptosis (44). In the latter study, it is unclear what role SHP plays. It is possible that FXR-SHP-dependent and -independent mechanisms may control apoptotic signaling in a tissue-specific manner. Our study shows that activation of SHP triggers massive apoptosis in a variety of cells and tissue types originating from both mouse and human, and thus these findings are generally applicable and are not limited to specific cells in a single species.
In summary, using a series of in vivo and in vitro experimental approaches, our study presents a novel cytoplasmic function of SHP through its regulation of mitochondrial activity, which is mediated by its subcellular localization. We provide convincing evidence that SHP is a critical component of mitochondrial apoptotic signaling and mediates the proapoptotic effects of AHPN. We have established a role for SHP in induction of apoptosis via disruption of the function of mitochondria through an interaction with Bcl-2 to release Bid and cause cytochrome c release. AHPN attenuated tumor growth through induction of SHP in an in vivo peritoneal tumor model, suggesting that activation of SHP may be applied to a variety of human cancers to suppress tumor growth.
Acknowledgments
We thank Curt Hagedorn for critically reading the manuscript. We thank Timothy F. Osborne (UCI) and Akiyoshi Fukamizu (University of Tsukuba) for kindly providing the expression plasmids. We thank Phillip Gray for helping with the Xenogen bioluminescent imaging system.
We acknowledge the use of core facilities (Cell Imaging Core and Flow Cytometry Core) supported by grant P30 CA042014 to the Huntsman Cancer Institute. This work is in part supported by National Institutes of Health grants DK080440 to L.W. and HL73167 to E.D.A.
No conflicts of interest exist for any of the authors.
Y.Z., J.S., K.P., and G.V. conducted the experiments; S.K. provided the BxPc-3, SU8, and HT29 cell lines; E.D.A. oversaw the mitochondrial function assays and edited the manuscript; and L.W. directed the experiments, analyzed the data, and wrote the manuscript.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Abou-Alfa, G. K. 2006. Hepatocellular carcinoma: molecular biology and therapy. Semin. Oncol. 33:S79-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnhart, B. C., E. C. Alappat, and M. E. Peter. 2003. The CD95 type I/type II model. Semin. Immunol. 15:185-193. [DOI] [PubMed] [Google Scholar]
- 3.Brenner, C., and G. Kroemer. 2000. Apoptosis. Mitochondria—the death signal integrators. Science 289:1150-1151. [DOI] [PubMed] [Google Scholar]
- 4.Bursch, W., M. Chabicovsky, U. Wastl, B. Grasl-Kraupp, K. Bukowska, H. Taper, and R. Schulte-Hermann. 2005. Apoptosis in stages of mouse hepatocarcinogenesis: failure to counterbalance cell proliferation and to account for strain differences in tumor susceptibility. Toxicol. Sci. 85:515-529. [DOI] [PubMed] [Google Scholar]
- 5.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 6.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 7.Farhana, L., M. I. Dawson, and J. A. Fontana. 2005. Apoptosis induction by a novel retinoid-related molecule requires nuclear factor-kappaB activation. Cancer Res. 65:4909-4917. [DOI] [PubMed] [Google Scholar]
- 8.Farhana, L., M. I. Dawson, M. Leid, L. Wang, D. D. Moore, G. Liu, Z. Xia, and J. A. Fontana. 2007. Adamantyl-substituted retinoid-related molecules bind small heterodimer partner and modulate the Sin3A repressor. Cancer Res. 67:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farhana, L., M. I. Dawson, Y. Huang, Y. Zhang, A. K. Rishi, K. B. Reddy, R. S. Freeman, and J. A. Fontana. 2004. Apoptosis signaling by the novel compound 3-Cl-AHPC involves increased EGFR proteolysis and accompanying decreased phosphatidylinositol 3-kinase and AKT kinase activities. Oncogene 23:1874-1884. [DOI] [PubMed] [Google Scholar]
- 10.Grasl-Kraupp, B., B. Ruttkay-Nedecky, L. Müllauer, H. Taper, W. Huber, W. Bursch, and R. Schulte-Hermann. 1997. Inherent increase of apoptosis in liver tumors: implications for carcinogenesis and tumor regression. Hepatology 25:906-912. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 12.He, N., K. T. Park, Y. X. Zhang, J. S. Huang, S. Lu, and L. Wang. 2008. Epigenetic inhibition of nuclear receptor SHP is associated with and regulates hepatocellular carcinoma growth. Gastroenterology 134:793-802. [DOI] [PubMed] [Google Scholar]
- 13.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J. S., J. Iqbal, P. Saha, J. Liu, L. Chan, M. Hussain, D. D. Moore, and L. Wang. 2007. Molecular characterization of the role of orphan receptor SHP in development of fatty liver. Hepatology 46:147-157. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, G. L., and K. Nakamura. 2007. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim. Biophys. Acta 1773:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krysko, D. V., T. Vanden Berghe, K. D'Herde, and P. Vandenabeele. 2008. Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods 44:205-221. [DOI] [PubMed] [Google Scholar]
- 17.Laine, B., J. Eeckhoute, L. Suaud, I. Briche, H. Furuta, G. I. Bell, and P. Formstecher. 2000. Functional properties of the R154X HNF-4alpha protein generated by a mutation associated with maturity-onset diabetes of the young, type 1. FEBS Lett. 479:41-45. [DOI] [PubMed] [Google Scholar]
- 18.Ledda-Columbano, G. M., H. Shinozuka, S. L. Katyal, and A. Columbano. 1996. Cell proliferation, cell death and hepatocarcinogenesis. Cell Death Differ. 3:17-22. [PubMed] [Google Scholar]
- 19.Li, H., S. K. Kolluri, J. Gu, M. I. Dawson, X. Cao, P. D. Hobbs, B. Lin, G. Chen, J. Lu, F. Lin, et al. 2000. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science 289:1159-1164. [DOI] [PubMed] [Google Scholar]
- 20.Li, L. Y., X. Luo, and X. Wang. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95-99. [DOI] [PubMed] [Google Scholar]
- 21.Li, P., D. Nijhawan, and X. Wang. 2004. Mitochondrial activation of apoptosis. Cell 116:S57-S59. [DOI] [PubMed] [Google Scholar]
- 22.Li, P., D. Nijhawan, I. Budihardjo, S. M. Srinivasula, M. Ahmad, E. S. Alnemri, and X. Wang. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479-489. [DOI] [PubMed] [Google Scholar]
- 23.Lin, B., S. K. Kolluri, F. Lin, W. Liu, Y. H. Han, X. Cao, M. I. Dawson, J. C. Reed, and X. K. Zhang. 2004. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell 116:527-540. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, N., S. S. Kim, and L. Z. Penn. 2006. The Oscar-worthy role of Myc in apoptosis. Semin. Cancer Biol. 16:275-287. [DOI] [PubMed] [Google Scholar]
- 25.Miao, J., Z. Xiao, D. Kanamaluru, G. Min, P. M. Yau, T. D. Veenstra, E. Ellis, S. Strom, K. Suino-Powell, H. E. Xu, and J. K. Kemper. 2009. Bile acid signaling pathways increase stability of small heterodimer partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 23:986-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moll, U. M., N. Marchenko, and X. K. Zhang. 2006. p53 and Nur77/TR3—transcription factors that directly target mitochondria for cell death induction. Oncogene 25:4725-4743. [DOI] [PubMed] [Google Scholar]
- 27.Oberst, A., C. Bender, and D. R. Green. 2008. Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ. 15:1139-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogasawara, J., R. Watanabe-Fukunaga, M. Adachi, A. Matsuzawa, T. Kasugai, Y. Kitamura, N. Itoh, T. Suda, and S. Nagata. 1993. Lethal effect of the anti-Fas antibody in mice. Nature 364:806-809. [DOI] [PubMed] [Google Scholar]
- 29.Ogata, M., T. Awaji, N. Iwasaki, S. Miyazaki, G. I. Bell, and Y. Iwamoto. 2002. Nuclear translocation of SHP and visualization of interaction with HNF-4alpha in living cells. Biochem. Biophys. Res. Commun. 292:8-12. [DOI] [PubMed] [Google Scholar]
- 30.Omura, T. 1998. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 123:1010-1016. [DOI] [PubMed] [Google Scholar]
- 31.O'Neill, B. T., J. Kim, A. R. Wende, H. A. Theobald, J. Tuinei, J. Buchanan, A. Guo, V. G. Zaha, D. K. Davis, J. C. Schell, et al. 2007. A conserved role for phosphatidylinositol 3-kinase but not Akt signaling in mitochondrial adaptations that accompany physiological cardiac hypertrophy. Cell Metab. 6:294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ow, Y. L., D. R. Green, Z. Hao, and T. W. Mak. 2008. Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9:532-542. [DOI] [PubMed] [Google Scholar]
- 33.Park, Y. J., M. Qatanani, S. S. Chua, J. L. LaRey, S. A. Johnson, M. Watanabe, D. D. Moore, and Y. K. Lee. 2008. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology 47:1578-1586. [DOI] [PubMed] [Google Scholar]
- 34.Peter, M. E., and P. H. Krammer. 2003. The CD95 (APO-1/Fas) DISC and beyond. Cell Death Differ. 10:26-35. [DOI] [PubMed] [Google Scholar]
- 35.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, et al. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz, D. R., and W. J. Harrington, Jr. 2003. Apoptosis: programmed cell death at a molecular level. Semin. Arthritis Rheum. 32:345-369. [DOI] [PubMed] [Google Scholar]
- 37.Song, G. S., and L. Wang. 2008. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRγ. Nucleic Acids Res. 36:5727-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, et al. 1999. Molecular characterization of mitochondrial apoptosis inducing factor. Nature 97:441-446. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki, Y., Y. Imai, H. Nakayama, K. Takahashi, K. Takio, and R. Takahashi. 2001. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8:613-621. [DOI] [PubMed] [Google Scholar]
- 40.Swales, K. E., M. Korbonits, R. Carpenter, D. T. Walsh, T. D. Warner, and D. Bishop-Bailey. 2006. The farnesoid X receptor is expressed in breast cancer and regulates apoptosis and aromatase expression. Cancer Res. 66:10120-10126. [DOI] [PubMed] [Google Scholar]
- 41.Verhagen, A. M., P. G. Ekert, M. Pakusch, J. Silke, L. M. Connolly, G. E. Reid, et al. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43-53. [DOI] [PubMed] [Google Scholar]
- 42.Virág, L., and C. Szabó. 2002. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 54:375-429. [DOI] [PubMed] [Google Scholar]
- 43.Wang, L., J. Liu, P. Saha, J. S. Huang, L. Chan, B. Spiegelman, and D. D. Moore. 2005. The orphan nuclear receptor SHP regulates PGC-1α expression and energy production in brown adipocytes. Cell Metab. 2:227-238. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y. D., F. Yang, W. D. Chen, Y. Huang, L. Lai, B. M. Forman, and W. Huang. 2008. Farnesoid X receptor protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo. Mol. Endocrinol. 22:1622-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo, M. G., Y. G. Yoo, H. S. Choi, Y. K. Pak, and M. O. Lee. 2005. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol. Endocrinol. 19:950-963. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y. X., P. Xu, K. T. Park, Y. H. Kim, D. D. Moore, and L. Wang. 2008. Orphan receptor SHP suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology 48:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, X., and R. A. Spanjaard. 2003. The apoptotic action of the retinoid CD437/AHPN: diverse effects, common basis. J. Biomed. Sci. 10:44-49. [DOI] [PubMed] [Google Scholar]