Abstract
In Saccharomyces cerevisiae SIR proteins mediate transcriptional silencing, forming heterochromatin structures at repressed loci. Although recruitment of transcription initiation factors can occur even to promoters packed in heterochromatin, it is unclear whether heterochromatin inhibits RNA polymerase II (RNAPII) transcript elongation. To clarify this issue, we recruited SIR proteins to the coding region of an inducible gene and characterized the effects of the heterochromatic structure on transcription. Surprisingly, RNAPII is fully competent for transcription initiation and elongation at the locus, leading to significant loss of heterochromatin proteins from the region. A search for auxiliary factors required for transcript elongation through the heterochromatic locus revealed that two proteins involved in histone H3 lysine 56 acetylation, Rtt109 and Asf1, are needed for efficient transcript elongation by RNAPII. The efficiency of transcription through heterochromatin is also impaired in a strain carrying the K56R mutation in histone H3. Our results show that H3 K56 modification is required for efficient transcription of heterochromatic locus by RNAPII, and we propose that transcription-coupled incorporation of H3 acetylated K56 (acK56) into chromatin is needed for efficient opening of heterochromatic loci for transcription.
In order to produce mRNA, RNA polymerase II (RNAPII) has to contend with chromatin in regulatory and coding regions of genes. Different modifications of histone proteins, rearrangements of nucleosome positioning, and packaging of nucleosomes into higher-order chromatin structures are used to facilitate or repress the accessibility of cellular factors to DNA. Modifications of histone proteins by acetylation and methylation are the most common ways to initiate changes in chromatin structure. In addition to several transcription-coupled modifications of chromatin in active gene loci, nucleosomes can be removed from entire transcribed regions and replaced with new histones after shutdown of transcription (14, 18, 21, 42, 49, 54). A detailed study of nucleosome dynamics in budding yeast has revealed a remarkable replication-independent exchange of histones throughout the genome. While turnover of promoter nucleosomes is detectable regardless of transcriptional activity of the locus, exchange of histones in the coding regions is in good correlation with gene expression level, indicating that turnover of nucleosomes is a rather common feature of ongoing transcription, especially in highly transcribed loci (7).
Gene expression in eukaryotic cells is also controlled by the formation of repressive heterochromatic domains in loci of regulated genes. There are three main regions in the genome of Saccharomyces cerevisiae that are subject to silencing by heterochromatin: telomeres, ribosomal DNA (rDNA) locus, and silent mating type loci (HML and HMR). Heterochromatin at telomeres and HM loci is formed by the complex of silent information regulator (SIR) proteins Sir2, Sir3, and Sir4, which are also required for the maintenance of heterochromatin structure. Disruption of any of these SIR genes greatly reduces recruitment of other Sir proteins to heterochromatic loci and also leads to the loss of telomeric and HM silencing (12, 25, 38, 46). Upon initial recruitment, SIR proteins accumulate and spread along the chromatin. To avoid repression of euchromatic regions in the genome, cells have evolved several mechanisms to restrict the spreading of SIR complex. Those include acetylation and methylation of histones, incorporation of H2A.Z into nucleosomes, and maintenance of highly transcribed nucleosome-free regions in chromatin (37).
For the initiation of SIR protein recruitment and heterochromatin formation in HM loci, specific silencer regions E and I, flanking HMR and HML, are required. Although both E and I regions are involved in full silencing of HM loci, the HMR-E and HML-E silencers are crucial for the recruitment of SIR proteins to chromatin when inserted into different locations in the genome (24, 44).
The classical view of transcriptional silencing by heterochromatin states that the highly condensed structure of chromatin elicits its repressive effects by sterically hindering the access of sequence-specific regulatory factors, required for the binding of transcription machinery, and therefore blocking the whole process (17). Nevertheless, it has been established that repressive heterochromatin in Saccharomyces cerevisiae allows constitutive binding of transcription activators, preinitiation complex (PIC) components, and RNAPII to the promoter regions of repressed genes, indicating that steps other than initial recruitment of transcription factors might be repressed by heterochromatin (43, 44). Indeed, recent data argue that repression of transcription by silenced chromatin is mainly targeting the transition point between RNAPII initiation and elongation by diminishing the recruitment of 5′-capping enzymes and elongation factors (9).
Although the role of heterochromatin in the repression of gene promoters is well characterized, it has remained unclear whether RNAPII that has cleared the promoter and entered the elongation phase of transcription can contend with heterochromatic structures in the coding region. Addressing this question in the present study, we show that elongating RNAPII can efficiently displace SIR proteins from silenced chromatin, and elongation of RNAPII through the coding region of the SIR complex-covered gene locus occurs essentially with the same kinetics as those on the euchromatic template. Additionally our results reveal that histone H3 lysine 56 acetylation is required for efficient elongation of RNAPII through heterochromatic structures in the coding region of the gene.
MATERIALS AND METHODS
Yeast strains.
All Saccharomyces cerevisiae strains used were congenic with strain W303. The GAL-VPS13 (AKY210) strain (49) expressing H3 with a C-terminal E2 tag was used to create the strain AKY362 with a C-terminal triple-E4 tag in Rpb3. GAL-VPS13-HMR-E (AKY354) and GAL-VPS13-HMR-E-0.1kb (AKY359) strains were created by insertion of the HMR-E silencer sequence (nucleotides 292908 to 292388 of chromosome III; Saccharomyces Genome Database) 6,070 or 100 bp downstream of the VPS13 start codon, respectively (see Fig. 1A and 3A). In addition, Sir3 with a C-terminal nine-Myc tag and Rpb3 with a triple-E4 tag in their natural loci were introduced in strains AKY354 and AKY359. Deletion mutant strains with HMR-E sequence at 6 kb in VPS13 were derived from AKY354 using one-step transplacement of the open reading frame (ORF) of genes of interest with kanMX or hphMX markers. H3 mutant K56R and K56Q strains (AKY399 and AKY651, respectively) were created by replacing the HHT2 allele in strain AKY354 with a mutant one in its natural locus. For the rtt109ΔH3K56Q (AKY648) double mutant, the HHT2 allele was replaced with a mutant one in the rtt109Δ strain (AKY380). The GAL-VPS13-term3k-HMR-E strain (AKY612) was created by insertion of the FBA1 termination region (49) 3,220 bp downstream from the VPS13 start codon in strain AKY354 (see Fig. 4A). Deletion mutant strains with wild-type (wt) GAL-VPS13 were derived from AKY362. The H3 mutant K56R strain with wt GAL-VPS13 was created by replacing the HHT2 allele in strain AKY362 with a mutant one in its natural locus. The yeast strains and their genotypes are given in Table 1.
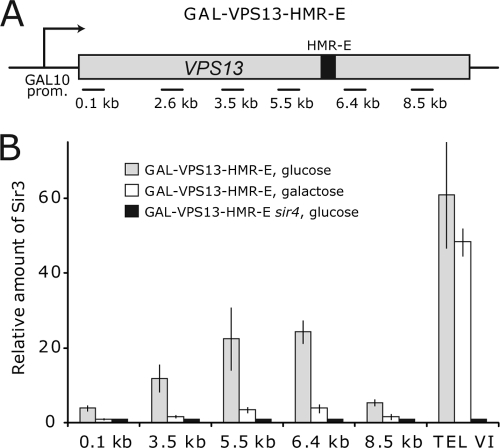
FIG. 1.
Recruitment and displacement of the SIR complex at the coding region of GAL-VPS13-HMR-E. (A) Schematic representation of the GAL-VPS13 locus with HMR-E silencer sequence (black rectangle) inserted at 6 kb from the VPS13 promoter. Horizontal lines beneath the gene region indicate the positions of coding region (0.1-, 2.6-, 3.5-, 5.5-, 6.4-, and 8.5-kb) PCR probes used in ChIP and RT-PCR assays. (B) ChIP assay was used to determine the density of Sir3 protein on the coding region of GAL-VPS13 in GAL-VPS13-HMR-E (AKY354) and GAL-VPS13-HMR-E sir4Δ (AKY355) strains before and after overnight transcription induction. The amount of Sir3 on the right arm of the chromosome VI telomere region is shown as the indicator of SIR complex occupancy on its natural locus. The signal from the FBA1 locus was used to normalize fluctuations in sample handling, and the amount of Sir3 as background signal in the sir4Δ strain was set to 1.
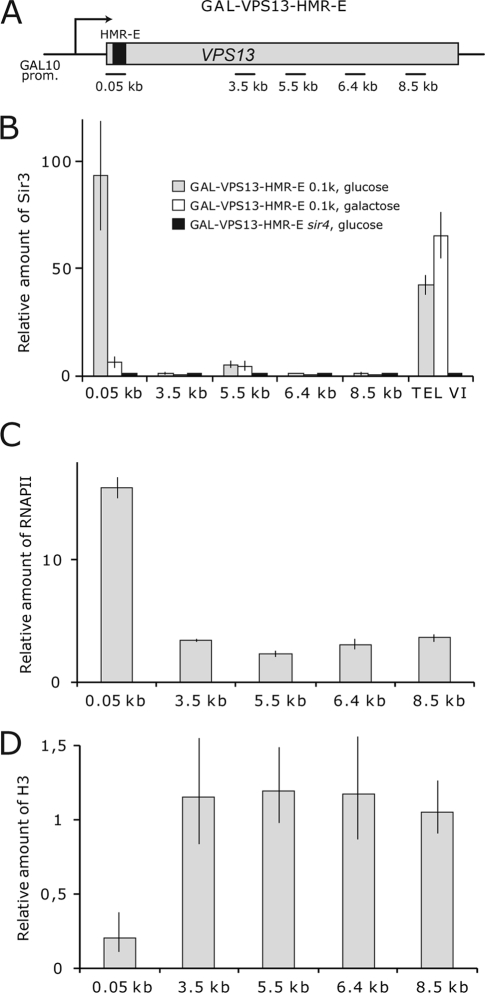
FIG. 3.
Dynamics of the SIR complex, RNAPII, and nucleosomes at the promoter proximal region of GAL-VPS13-HMR-E-0.1kb. (A) Schematic representation of the GAL-VPS13 locus with HMR-E silencer sequence inserted at 0.1 kb from the VPS13 start codon. Horizontal lines beneath the gene region indicate the approximate positions of coding region (0.05-, 3.5-, 5.5-, 6.4-, and 8.5-kb) PCR probes used in ChIP assay. (B to D) ChIP assay was used to determine the density of Sir3 protein (B), RNAPII (C), and histone H3 (D) on the coding region of VPS13 in the GAL-VPS13-HMR-E-0.1kb strain (AKY359). Recruitment of Sir3 is shown before and after overnight transcription induction. The amount of Sir3 on the right arm of the chromosome VI telomere region is shown as the indicator of SIR complex occupancy on its natural locus. The signal from the FBA1 locus was used to normalize fluctuations in sample handling, and the amount of Sir3 as background signal in the sir4Δ strain was set to 1. The bars representing H3 and RNAPII indicate the levels of the respective proteins in galactose-containing medium relative to the levels in cells grown in glucose-containing medium. FBA1 (for RNAPII) and the noncoding region on chromosome VIII (for H3) were used as internal controls.
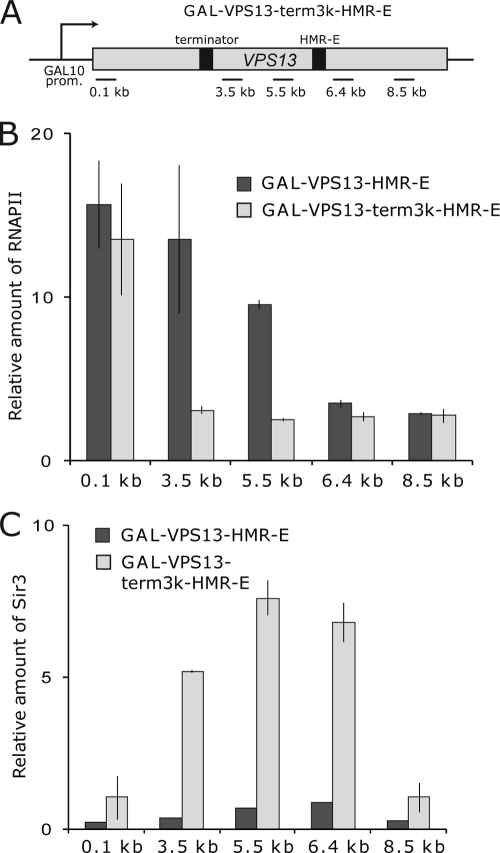
FIG. 4.
Progression of RNAPII through the locus is required for SIR complex displacement from GAL-VPS13-HMR-E. (A) Schematic representation of the GAL-VPS13 locus with FBA1 terminator and HMR-E silencer sequences inserted at 3 kb and 6 kb, respectively. Horizontal lines beneath the gene region indicate the positions of coding region (0.1-, 3.5-, 5.5-, 6.4-, and 8.5-kb) PCR probes used. (B and C) ChIP assay was carried out to determine the density of RNAPII (B) and Sir3 (C) on GAL-VPS13-HMR-E (AKY354) and GAL-VPS13-term3k-HMR-E (AKY612) after transcription induction. The bars indicate the levels of RNAPII in galactose medium relative to the level in cells grown in glucose-containing medium (set as 1) and the level of Sir3 in galactose-containing medium relative to the signal from the coding region of the constitutively expressed FBA1 gene.
TABLE 1.
Names and genotypes of yeast strains used in the study
| Strain | Genotypea | Reference or source |
|---|---|---|
| AKY210 | W303 MATaGAL-VPS13::TRP1 hht1-hhf1Δ::LEU2 HHT2-E2 tag::URA3 | 49 |
| AKY354 | AKY210; Sir3-(9-MYC tag)::TRP1 Rpb3-(3-E4 tag)::SpHIS5 HMR-E silencer sequence at 6 kbp in VPS13 | This study |
| AKY355 | AKY354; sir4Δ::kanMX | This study |
| AKY362 | AKY210; Rpb3-(3-E4 tag)::SpHIS5 | This study |
| AKY372 | AKY354; rpd3Δ::kanMX | This study |
| AKY373 | AKY354; set2Δ::kanMX | This study |
| AKY374 | AKY354; dot1Δ::kanMX | This study |
| AKY377 | AKY354; set1Δ::kanMX | This study |
| AKY378 | AKY354; bre1Δ::kanMX | This study |
| AKY379 | AKY354; gcn5Δ::kanMX | This study |
| AKY380 | AKY354; rtt109Δ::kanMX | This study |
| AKY381 | AKY354; sas2Δ::kanMX | This study |
| AKY383 | AKY354; rsc1Δ::kanMX | This study |
| AKY384 | AKY354; asf1Δ::kanMX | This study |
| AKY398 | AKY354; htz1Δ::kanMX | This study |
| AKY399 | W303 MATaGAL-VPS13::TRP1 hht1-hhf1Δ::LEU2 hht2K56R-E2 tag Sir3-(9-MYC tag)::TRP1 Rpb3-(3-E4 tag)::SpHIS5 HMR-E silencer sequence at 6 kbp in VPS13 | This study |
| AKY605 | AKY362; rtt109Δ::hphMX | This study |
| AKY606 | AKY362; asf1Δ::hphMX | This study |
| AKY607 | AKY362; htz1Δ::hphMX | This study |
| AKY609 | W303 MATaGAL-VPS13::TRP1 Rpb3-(3-E4 tag)::SpHIS5 hht1-hhf1Δ::LEU2 hht2K56R-E2 tag | This study |
| AKY612 | AKY354; FBA1 terminator sequence at 3 kbp in VPS13 | This study |
| AKY646 | AKY354; hst3Δ::kanMX hst4Δ::hphMX | This study |
| AKY648 | AKY651; rtt109Δ::kanMX | This study |
| AKY651 | W303 MATaGAL-VPS13::TRP1 hht1-hhf1Δ::LEU2 hht2K56Q-E2 tag Sir3-(9-MYC tag)::TRP1 Rpb3-(3-E4 tag)::SpHIS5 HMR-E silencer sequence at 6 kbp in VPS13 | This study |
SpHIS5, Schizosaccharomyces pombe HIS5.
ChIP assay.
Cells were grown overnight in yeast extract-peptone (YP) medium containing 2% glucose or galactose as a carbon source before fixation for the chromatin immunoprecipitation (ChIP) assay. For the detection of H3 K56 acetylation dynamics, cells were grown overnight in YP medium containing 2% raffinose, which was then replaced by 2% galactose and glucose for designated periods of time. ChIP assays were performed as described previously (19). Whole-cell extract from 1 × 107 cells was used for ChIP assays with antibodies directed against RNAPII (4H8; Upstate Biotechnology), anti-E4 tag (1E2; Icosagen), anti-Myc tag (9E10; Abcam), anti-E2 tag (5E11; Icosagen), or anti-acetyl-histone H3 Lys56 (Upstate Biotechnology). Coprecipitated DNA was analyzed by quantitative real-time PCR using an ABI Prism 7900HT real-time PCR system under standard conditions (40 cycles; 95°C for 15 s and 60°C for 1 min). Absolute QPCR SYBR green reagents (ABgene) and 5× Hot FIREPol EvaGreen qPCR mix (Solis BioDyne) were used. PCRs were done with primer pairs covering coding region of VPS13, FBA1, the chromosome VI right arm telomere region, and the noncoding region on chromosome VIII. The VPS13 coding region primers were specific for the sequences at 0.1, 3.5, 5.5, 6.4, and 8.5 kb downstream from the start codon of GAL-VPS13. The length of all PCR products was about 160 bp. Exact sequences of all primers are available upon request. In SIR complex removal experiments, samples were normalized to the signal of Sir3 on the constitutively expressed FBA1 gene and further to background signal in the sir4Δ strain. In nucleosome loss experiments, all samples were normalized according to the amount of histone H3 on the noncoding region on chromosome VIII and the amount of RNAPII was normalized to the amount of RNAPII on the constitutively expressed FBA1 gene.
RT-PCR.
For the detection of mRNA, cells were grown overnight in YP medium containing 2% galactose or raffinose. For transcription induction, galactose, with the final concentration of 2%, was added to cells grown in raffinose. Samples were collected before galactose addition and 30, 60, 120, and 180 min after transcription induction. For cell cycle arrest experiments, α-factor mating pheromone (Zymo Research) was used with the final concentration of 100 μM and cell cycle arrest in G1 phase during the experiment was confirmed by fluorescence-activated cell sorting (FACS) analysis. Cells (3 × 107) were collected, and RNA was extracted as described previously (5) with minor modifications. Cells washed with water were resuspended in 600 μl of RNA extraction buffer (10 mM EDTA, 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5% sodium dodecyl sulfate [SDS]), mixed with equal volumes of acid phenol-chloroform (1:1), and lysed at room temperature for 6 min. Glass beads were added, and samples were vortexed at maximum speed for 4 min. Liquid was removed, and phenol-chloroform extraction was performed followed by chloroform extraction. RNA was precipitated with 1/10 volume of 3 M NaOAc and 3 volumes of ice-cold 96% ethanol. Precipitated RNA was washed with 70% ethanol and dissolved in 40 μl RNase-free water. DNase treatment was carried out, and 100 ng of RNA was used for one-step reverse transcriptase PCR (RT-PCR) (48°C for 30 min; 95°C for 5 min; and 20 cycles of 93°C for 3 min, 55°C for 30 s, and 72°C for 30 s). Primers specific for the coding regions of VPS13, FBA1, and GAL10 were used. RT-PCR products were separated on a 2.5% agarose gel stained with ethidium bromide.
RESULTS
Elongating RNAPII displaces SIR proteins from the coding region of the gene.
To study the influence of heterochromatic structures on transcription elongation, we used Saccharomyces cerevisiae strains containing a modified VPS13 gene as our model system. The GAL-VPS13 fusion gene contains the galactose-inducible GAL10 promoter in front of the 9.5-kb-long VPS13 open reading frame, allowing transcription of the gene to be induced or repressed by growing cells in medium containing galactose or glucose, respectively (18). In order to promote SIR complex recruitment and formation of heterochromatic structures in the coding region of the model gene, we inserted an HMR-E silencer into the GAL-VPS13 gene 6 kb downstream of the transcription start site (Fig. 1A). Using chromatin immunoprecipitation (ChIP) and quantitative PCR methods, we then monitored distribution of RNAPII, nucleosomes, and Sir3 protein at different points through the GAL-VPS13-HMR-E gene before and after transcription induction.
In the repressed state, SIR proteins were efficiently recruited to the HMR-E sequence on the GAL-VPS13-HMR-E gene. When bound to HMR-E, SIR complexes spread bidirectionally up- and downstream from the silencer sequence in the wild-type strain where all SIR proteins were present but not in the corresponding sir4Δ strain (Fig. 1B). Spreading of SIR complexes was more prominent toward the 5′ end of the gene, covering at least 3 kb to this side but leaving the promoter and the beginning of the gene free of heterochromatin. The 3′ end of the gene was also essentially free of SIR proteins. This uneven distribution of SIR complex was not unexpected, as the HMR-E silencer also works in an orientation-dependent manner in its native locus (26, 55). Remarkably, upon induction of GAL-VPS13-HMR-E transcription, SIR complexes were removed from the entire locus (Fig. 1B), suggesting that transcriptional activity in the locus was not abolished by heterochromatic structures and indicating that SIR complexes can be removed as a consequence of transcription.
Although the majority of Sir3 protein was found in the open reading frame (ORF) region of the transcriptionally inactive GAL-VPS13-HMR-E locus, small amounts of SIR proteins were also detected in the proximity of the promoter (Fig. 1B). To test whether the GAL-VPS13 promoter can be silenced when large amounts of SIR proteins were recruited directly to the region, we inserted HMR-E sequence into the GAL-VPS13 coding region 100 bp downstream of the beginning of the ORF (see Fig. 3A). As expected, SIR proteins were recruited to the site under repressing conditions, and they were efficiently removed upon activation of the gene (see Fig. 3B). These results show that the GAL-VPS13 promoter is insensitive to SIR-mediated repression, making the GAL-VPS13-HMR-E model gene a suitable experimental system for studies of how heterochromatin affects transcript elongation. These results are also in concordance with previous reports indicating that the GAL1-10 promoter is competent for initiation of transcription of shorter mRNAs when inserted into telomeric heterochromatin (4, 39).
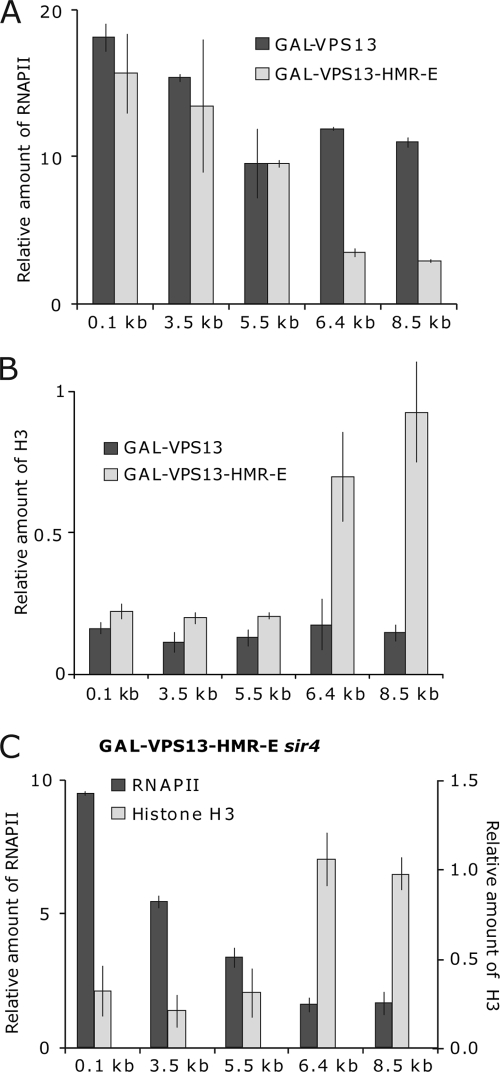
The GAL10 promoter provides a very high-level induction of GAL-VPS13, leading to the removal of nucleosomes from the promoter and coding region of the gene (18, 49). In order to determine whether SIR complexes, as expected, reduce the level of gene induction, we analyzed the density of RNAPII and nucleosomes in GAL-VPS13-HMR-E compared to the unmodified GAL-VPS13 locus. Surprisingly, even though a high level of Sir3p was detected in the GAL-VPS13-HMR-E locus prior to activation, this had no effect on RNAPII density in the region after gene induction. The levels of RNAPII and nucleosomes were similar in the tested strains upstream from the HMR-E insertion site, indicating no major difference in the levels of transcriptional initiation or elongation through this heterochromatic domain. However, neither RNAPII recruitment nor loss of nucleosomes was detected downstream of the HMR-E sequence in the GAL-VPS13-HMR-E strain (Fig. 2A and B). Similar results were observed also in the strain where HMR-E sequence was inserted at 0.1 kb of the GAL-VPS13 gene (Fig. 3C and D). Reverse transcriptase PCR (RT-PCR) experiments also failed to detect VPS13 transcripts beyond the HMR-E sequence, further indicating that although polymerase can traverse the heterochromatin formed upstream of the silencer, it is unable to get through the HMR-E region itself (see Fig. 5; 6.4 kb in the GAL-VPS13-HMR-E strain). Nevertheless, upon galactose induction SIR complexes were largely removed both up- and downstream of the silencer sequence, making us question whether termination of transcription was caused by heterochromatin structure or whether other mechanisms might be responsible. To investigate this further, we carried out similar experiments in a sir4Δ strain which is deficient in heterochromatin and SIR complex formation (12, 25, 32, 35, 38, 46). As shown in Fig. 2C, RNAPII remained unable to transcribe through the HMR-E silencer even in the absence of SIR complexes, strongly suggesting that HMR-E itself contains a signal, for example, an efficient transcription terminator sequence, which causes polymerase to dissociate from the gene. In further support of this idea, nucleosomes were not removed downstream of HMR-E in the sir4Δ strain, indicating that no transcription took place beyond the HMR-E sequence (Fig. 2C). This is in concordance with our previous results showing that the insertion of a strong transcription terminator into the coding region of GAL-VPS13 leads both to dissociation of RNAPII and to lack of nucleosome removal from the region downstream of the terminator sequence (49). The observation that the HMR-E sequence itself is a strong terminator of transcription is interesting but was not the subject of this study and was therefore not pursued further.
FIG. 2.
Transcription-coupled nucleosome removal and RNAPII recruitment at the GAL-VPS13-HMR-E locus. (A and B) ChIP assay was used to determine the density of RNAPII (A) and histone H3 (B) on GAL-VPS13 (AKY210) and GAL-VPS13-HMR-E (AKY354) after transcription induction. (C) Density of RNAPII and nucleosomes in GAL-VPS13-HMR-E sir4Δ strain (AKY355). The bars representing H3 and RNAPII indicate the level of respective proteins in galactose medium relative to the level in cells grown in glucose-containing medium (set as 1). FBA1 (for RNAPII) and the noncoding region on chromosome VIII (for H3) were used as internal controls.
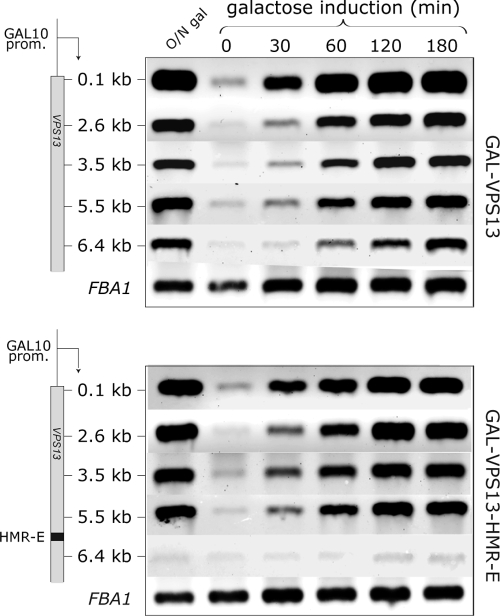
FIG. 5.
Heterochromatic structures do not affect the efficiency of RNAPII-dependent transcription elongation. RNA for one-step RT-PCR analysis was extracted from cells grown overnight in galactose (O/N gal) or raffinose (0) and 30, 60, 120, and 180 min after galactose induction. Different coding region probes (0.1, 2.6, 3.5, 5.5, and 6.4 kb) of GAL-VPS13 (strain AKY210) and GAL-VPS13-HMR-E (strain AKY354) loci are compared. FBA1 serves as an internal control.
Although the displacement of SIR complex from GAL-VPS13-HMR-E was concurrent with the appearance of elongating RNAPII in the region, it remained possible that the process of transcriptional activation at the GAL-VPS13-HMR-E promoter might in itself lead to the disassembly of heterochromatic structure, independently of elongating RNAPII. To clarify this issue, we inserted a powerful transcription terminator sequence into GAL-VPS13-HMR-E 3 kb downstream from the promoter, creating a model gene on which transcription by RNAPII was terminated before it reached the heterochromatic region in the locus (Fig. 4A). As expected (49), RNAPII was detected only upstream from the terminator sequence upon activation (Fig. 4B), showing that the terminator was efficient. Strikingly, SIR complexes located downstream from the terminator were not removed from the locus (Fig. 4C), confirming that elongation through the heterochromatic region is required for displacement of SIR complexes.
The efficiency of RNAPII elongation is not affected by heterochromatin.
Although our results suggested that the presence of the heterochromatic structure did not in itself abolish transcription of the locus, it remained possible that the kinetics of transcription through this region might be significantly slower on a heterochromatic template. For example, DNA replication-dependent removal of SIR complexes might be responsible for the initial displacement of SIR proteins, and only after that, constantly active transcription through the locus might preclude SIR complex re-formation. Therefore, we monitored the accumulation of VPS13 mRNA immediately after the induction of the GAL-VPS13-HMR-E gene. RNA samples were collected 30, 60, 120, and 180 min after galactose induction, and the presence of mRNA was detected by RT-PCR. We observed nearly identical kinetics of VPS13 mRNA accumulation in GAL-VPS13 and GAL-VPS13-HMR-E strains (Fig. 5), indicating that, surprisingly, the SIR complexes in the coding region of the gene did not change the kinetics of gene induction and transcript elongation through the region.
Acetylation of H3 K56 is required for RNAPII elongation through heterochromatin.
Given that histone modifications play an important role in silent chromatin formation, maintenance, and restriction, we asked whether different chromatin modifying factors might be required for elongating RNAPII to open and transcribe SIR complex-covered chromatin in the GAL-VPS13-HMR-E locus. To address this question, we constructed a set of yeast strains with deletions of genes involved in transcription-dependent chromatin dynamics or heterochromatin maintenance and monitored the accumulation of VPS13 mRNA by RT-PCR over a 3-h time course of GAL-VPS13-HMR-E induction. As shown in Fig. 6A, deletion of BRE1 (abolishing transcription-coupled H2B K123 monoubiquitylation and also subsequent methylations of histone H3 residues K4 and K79 [50]); H3 methyltransferase genes SET1, SET2, and DOT1; chromatin remodeling factor gene RSC1; or histone deacetylase gene RPD3 had no inhibitory effect on kinetics of GAL-VPS13-HMR-E induction. We reproducibly detected slightly slower induction of GAL-VPS13-HMR-E mRNA in sas2Δ and gcn5Δ strains (Fig. 6A), confirming the earlier results that both histone acetyltransferases (HATs) play a role in the control of heterochromatin spreading (16, 20, 47). However, severe impairment of GAL-VPS13-HMR-E induction was observed in rtt109Δ, asf1Δ, and htz1Δ strains (Fig. 6A). Rtt109 and Asf1 are both required for H3 K56 acetylation in vivo and in vitro (8, 10, 34, 41, 48). Asf1 is also involved in transcription- and replication-dependent histone dynamics (2, 15, 36). Htz1 is a yeast homolog of mammalian H2A.Z, an alternative form of H2A that is mainly incorporated into nucleosomes located in subtelomeric regions and also in promoter regions of RNAPII-transcribed genes throughout the genome. Subtelomeric Htz1 helps to restrict the spreading of SIR complex, while Htz1 in promoter regions is required for quick opening of chromatin in transcribed loci (6, 23, 29, 30, 53).
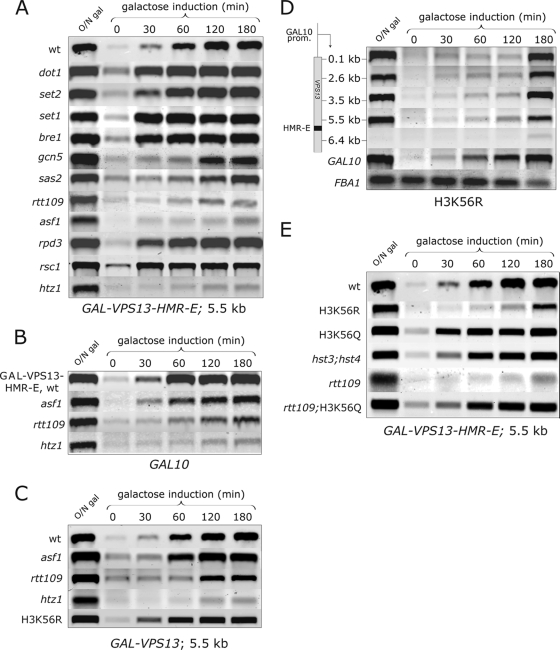
FIG. 6.
Acetylation of lysine 56 on H3 is needed for efficient transcription through the heterochromatic locus. (A) Induction of GAL-VPS13-HMR-E in wild-type (wt) and deletion mutant (dot1Δ, set2Δ, set1Δ, bre1Δ, gcn5Δ, sas2Δ, rtt109Δ, asf1Δ, rpd3Δ, rsc1Δ, and htz1Δ) strains. The amounts of VPS13 mRNA were determined by RT-PCR with a 5.5-kb probe at indicated time points. (B) Transcription induction on the GAL10 coding region in GAL-VPS13-HMR-E, rtt109Δ, asf1Δ, and htz1Δ strains. (C) Deletions of RTT109, ASF1, and HTZ1 and H3 mutation K56R were introduced into the GAL-VPS13 wt strain (AKY362), containing no HMR-E in the locus. The amounts of VPS13 mRNA were determined by RT-PCR with a 5.5-kb probe at indicated time points. (D) Transcription induction of the GAL-VPS13-HMR-E locus in the GAL-VPS13-HMR-E H3 K56R strain (AKY399). (E) Transcription induction of GAL-VPS13-HMR-E at 5.5 kb in wild-type and mutant (H3K56R, H3K56Q, hst3Δ hst4Δ, rtt109Δ, and rtt109Δ H3K56Q) strains. RNA for RT-PCR was collected and analyzed as described for Fig. 5. Equal loadings were confirmed by the FBA1 mRNA amount from each strain (not shown for panels A, B, C, and E).
Although obvious defects in GAL-VPS13-HMR-E induction were detected in rtt109Δ, asfΔ1, and htz1Δ strains, it remained unclear whether the delay of induction was specific for the heterochromatin-covered GAL-VPS13-HMR-E coding region (controlled by the GAL10 promoter) or whether it was due to general defects in the induction of GAL10 promoter-driven transcription in these strains. To clarify the issue, we determined the induction efficiency of the endogenous heterochromatin-free GAL10 gene as an internal control in the same samples. As shown in Fig. 6B, induction of GAL10 was severely delayed in the htz1Δ strain, suggesting that HTZ1 deletion leads to a general delay in the induction of GAL10 promoter-driven genes. Although slightly slower induction of GAL10 was also observed in rtt109Δ and asf1Δ strains, this delay was significantly stronger in the GAL-VPS13-HMR-E locus (compare wt, asf1, and rtt109 rows in Fig. 6A and B), suggesting that Rtt109 and Asf1 become crucial for transcription of a heterochromatic locus.
Alternatively, inefficient transcription of the GAL-VPS13-HMR-E locus in rtt109Δ, asf1Δ, and htz1Δ strains might indicate their inability to transcribe long genes, for example, due to general defects in transcription elongation. To test this possibility, we introduced the same deletions into strains carrying the GAL-VPS13 gene without the HMR-E sequence inserted. Induction of GAL-VPS13 in the asf1Δ strain was indistinguishable from that in the wild type, while deletion of RTT109 led to a slight delay in GAL-VPS13 induction (Fig. 6C). However, the induction defect was considerably stronger in the GAL-VPS13-HMR-E strain, where heterochromatic structures covered the GAL-VPS13 locus (compare rtt109 rows in Fig. 6A and C). Again, a very low level of GAL-VPS13 mRNA was induced in the htz1Δ strain, confirming that the lack of GAL-VPS13-HMR-E induction was not caused specifically by the heterochromatic structures in the locus. The effect of Htz1 observed here was expected, as previous studies have reported slow induction of inducible genes in the htz1Δ strain (1, 40). Interestingly, after overnight growth in galactose-containing medium, GAL-VPS13-HMR-E mRNA was induced in all deletion strains (Fig. 6A, lane O/N gal), indicating that Rtt109, Asf1, and Htz1 were required only for the normal, rapid induction of the heterochromatin-covered gene. It is likely that during the long-term growth under inducing conditions DNA replication-dependent displacement of SIR proteins occurs, and once the SIR proteins are removed, RNAPII gains access to the locus and inhibits re-formation of SIR complexes.
Rtt109 and Asf1 are both involved in acetylation of lysine 56 on histone H3 (8, 10, 34, 41, 48). To investigate the importance of H3 K56 acetylation for efficient transcription of the heterochromatic locus, we also generated a derivate of the GAL-VPS13-HMR-E strain carrying the K56R mutation in histone H3 that mimics the deacetylated state of the residue. Again, transcription in the heterochromatic GAL-VPS13-HMR-E locus was impaired, while induction of the endogenous GAL10 or heterochromatin-free GAL-VPS13 loci was essentially unchanged compared to the wild-type strain (Fig. 6D and C). To explore if constitutive acetylation of H3 K56 also might have an effect on GAL-VPS13-HMR-E transcription, we made two additional strains: one with the deletion of HST3 and HST4 genes, whose products are the major deacetylases of H3 K56 in vivo (3, 27, 52), and another one carrying the K56Q mutant of H3 to mimic the acetylated state of the residue. As expected, GAL-VPS13-HMR-E transcription was not inhibited in these strains (Fig. 6E), and importantly, when the H3 K56Q mutation was combined with the RTT109 deletion, the inhibitory effect of rtt109Δ was reversed (Fig. 6E, compare rtt109 and rtt109;H3K56Q rows). Together, these results indicate that acetylation of lysine 56 on histone H3 plays a key role in efficient transcription through heterochromatic loci.
Acetylation of H3 K56 occurs predominantly in S phase of the cell cycle, where newly synthesized K56-acetylated histones are incorporated into chromatin (28). As acetylation of K56 is required for efficient transcription of a heterochromatic gene, it might be possible that progression through the S phase is also needed for GAL-VPS13-HMR-E transcription. To test this, we compared the induction kinetics of GAL-VPS13 and GAL-VPS13-HMR-E loci in cells arrested in G1 phase by α-factor. As shown in Fig. 7A and B, the two strains had very similar induction patterns of VPS13 mRNA and the overall kinetics of the test gene induction was essentially the same as that in asynchronous cells (Fig. 5). Again, induction of GAL-VPS13-HMR-E in the H3 K56R background was strongly inhibited also in cell cycle-arrested cells (Fig. 7C). These results indicate that cells can transcribe a heterochromatic gene in an H3 K56 acetylation-dependent manner without progressing through the S phase.
FIG. 7.
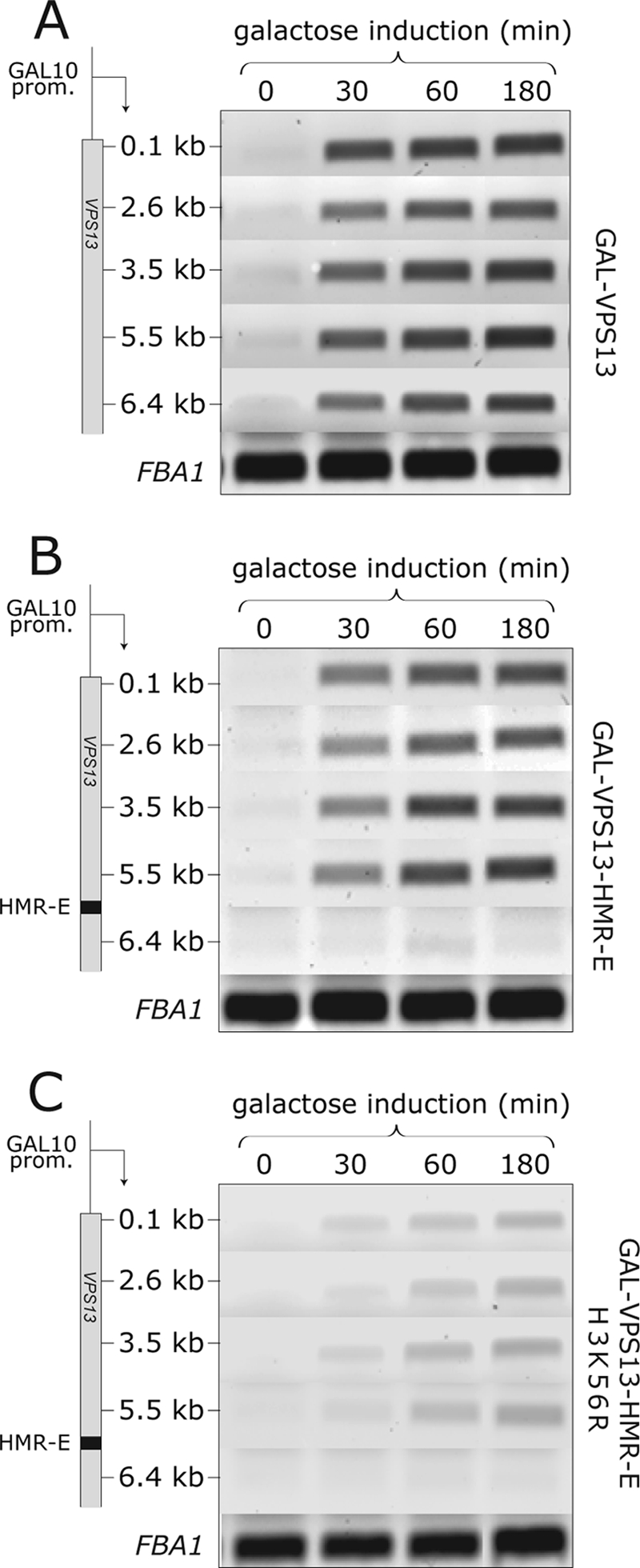
Induction of the GAL-VPS13-HMR-E locus in G1-arrested cells. (A) GAL-VPS13 (AKY362), (B) GAL-VPS13-HMR-E (AKY354), and (C) GAL-VPS13-HMR-E H3 K56R (AKY399) strains were grown overnight in raffinose, arrested in G1 by α-factor, and kept arrested during galactose induction. RNA samples for RT-PCR were extracted from cells grown in raffinose (0) and 30, 60, and 180 min after galactose induction and analyzed as described for Fig. 5. Equal loadings were confirmed by the FBA1 mRNA amount from each strain.
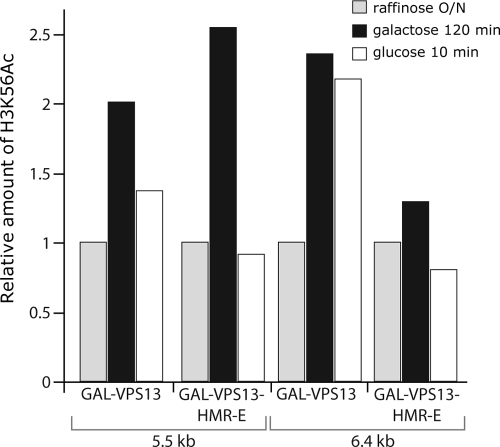
To test whether acetylation of H3 K56 actually occurs in the GAL-VPS13-HMR-E locus during transcription, we compared the presence of H3 acetylated K56 (acK56) in GAL-VPS13 and GAL-VPS13-HMR-E loci before, during, and after transcriptional induction. Acetylated K56 was detected in both GAL-VPS13 and GAL-VPS13-HMR-E strains under induced conditions (Fig. 8, “galactose 120 min”) and also shortly after repression of transcription (Fig. 8, “glucose 10 min”). While the GAL-VPS13 strain contained H3 K56 acetylation at both 5.5-kb and 6.4-kb regions of the locus, the GAL-VPS13-HMR-E strain gained acK56 only at the 5.5-kb region (Fig. 8). These results confirm that H3 acK56 was brought to the locus in a transcription-dependent manner, as the 6.4-kb region of VPS13 was not transcribed in the GAL-VPS13-HMR-E strain (Fig. 2A and 5).
FIG. 8.
Acetylation of H3 K56 in the VPS13 locus. GAL-VPS13 (AKY362) and GAL-VPS13-HMR-E (AKY354) strains were grown in raffinose-YP overnight, induced with galactose for 120 min, and then repressed by glucose for 10 min. The ChIP assay was carried out to determine relative amounts of H3K56 acetylation before and during induction (“raffinose O/N” and “galactose 120 min,” respectively) and after repression (“glucose 10 min”) of transcription at GAL-VPS13 and GAL-VPS13-HMR-E loci. The bars represent H3 K56 acetylation normalized to the overall amount of histone H3 on 5.5-kb and 6.4-kb probes relative to the level of K56 acetylation in cells grown in raffinose-YP medium (set as 1). The signal from the noncoding region on chromosome VIII was used to normalize the samples.
DISCUSSION
Our results reveal that, surprisingly, heterochromatic structures in the coding region of an RNAPII-transcribed gene are permissive for transcript elongation and do not diminish RNAPII transcriptional efficiency. We also show that disruption of histone H3 K56 acetylation, either by mutation of the residue or by the deletion of the H3 K56-specific histone acetyltransferase (HAT) Rtt109 or histone chaperone Asf1, severely impairs the efficiency of RNAPII transcription through heterochromatin.
SIR complexes are removed from the coding region of the gene during RNAPII transcription elongation.
It has been long established that relocation of silencers to other ectopic loci results in the silencing of the adjacent genes (22, 45). The exact mechanisms of transcriptional silencing in yeast are not fully understood; however, it has been shown that promoters embedded into heterochromatin are accessible for binding by basal transcription factors and that those genes are competent for transcription initiation but not for mRNA production (43, 44). These observations were further supported by recent data, suggesting that RNAPII does not switch from transcription initiation to elongation in silenced loci (9), leaving open the question whether RNAPII is capable of elongation on a heterochromatic template at all. To clarify this issue, we induced SIR protein recruitment and heterochromatin formation in the coding region of an inducible model gene and studied how the elongating polymerase can cope with heterochromatic structures in the middle of the gene. We found that the heterochromatic region was efficiently transcribed by RNAPII and that SIR proteins were removed from the gene in a transcription-dependent manner (Fig. 1B, 2, and 4). Surprisingly, transcription was induced with the same kinetics in the heterochromatic locus as in the euchromatic locus, indicating that heterochromatin poses no obvious barrier to transcript elongation by RNAPII (Fig. 5 and 7).
Interestingly, SIR complexes were removed from the entire induced GAL-VPS13-HMR-E locus, despite the fact that transcription of the gene was terminated at the HMR-E sequence and no polymerases were detected beyond the HMR-E site. The HMR-E silencer consists of binding sites for three essential factors—origin recognition complex, Rap1, and Abf1 proteins. All these proteins have affinity for one or more SIR proteins, and all of them are required for the recruitment of the SIR complex to HMR-E (37). We surmise that some or all of these factors might be displaced from DNA by transcribing RNAPII, which in turn disables the efficient recruitment and subsequent spreading of SIR proteins in the locus.
Histone H3 K56 acetylation is needed for effective heterochromatin disruption in the coding regions of genes.
In order to determine which factors are needed for transcription of heterochromatic structures, we analyzed the efficiency of GAL-VPS13-HMR-E mRNA induction in 12 strains with deletions of genes required for different transcription-coupled chromatin modifications. We observed an impairment of transcription induction in three deletion strains. Of those, HTZ1 deletion led to a general delay in galactose induction, while deletions of RTT109 and ASF1 caused slow induction primarily in the heterochromatic locus (Fig. 6). As both Rtt109 and Asf1 are required for H3 K56 acetylation in vivo and in vitro (8, 10, 34, 41, 48), we assumed that this modification was crucial for efficient RNAPII transcription in the presence of SIR proteins. To test it directly, we made a yeast strain carrying the K56R mutation in histone H3 and observed a heterochromatin-specific delay of gene induction similar to those in rtt109 and asf1 strains (Fig. 6D). Moreover, when the H3 K56Q mutation (mimicking the constitutively acetylated state of the residue) was introduced into the test strain, the deletion of RTT109 did not cause the impairment of transcription in the heterochromatic locus (Fig. 6E).
Interestingly, mutation of H3 K56 to either glycine, glutamine, or arginine leads to partial derepression of telomeric and HM mating loci without affecting the binding of Sir proteins to these sites (51). We confirmed that also in our strain background the H3 K56R mutation led to partial derepression of HM silencing, as the K56R strain had about a 10-fold-lower mating efficiency than did its wt counterparts (data not shown). However, the K56R mutation had the opposite effect on gene expression in the VPS13-HMR-E locus. We propose that as the H3 K56 mutations increase chromatin accessibility in telomeric regions (51), this might offer a chance for telomeric promoters to initiate transcription more frequently than in wt cells. However, in the case of GAL-VPS13-HMR-E strains, the GAL10 promoter in front of the model gene remains functional regardless of SIR complex recruitment to the very proximity of the promoter (Fig. 3), and therefore, the possible changes of chromatin structure in the H3 K56R strain cannot make the GAL10 promoter more accessible than it already is. Therefore, the reduced ability of RNAPII to transcribe a heterochromatic locus in rtt109Δ, asf1Δ, and K56R strains most likely reflects the difficulties of RNAPII elongation during transcription. We would also like to emphasize that, in all mutant strains where the slow induction of the GAL-VPS13-HMR-E model gene was detected, the effects were observable only at early time points of gene induction. After overnight induction, all mutants were able to transcribe the model gene as efficiently as did the wt strain (lanes “O/N gal” in Fig. 6A, D, and E). Therefore, the initial activation of the repressed locus might occur during S phase, when SIR proteins are temporarily removed from chromatin, and later on the transcriptional state of the locus might be maintained by RNAPII. This assumption was also supported by the fact that the induction of GAL-VPS13-HMR-E was notably slow in the H3 K56R mutant strain when cells were arrested in G1 (Fig. 7C).
We also studied whether mutations abolishing acetylation of H3 K56 might lead to excessive loading of SIR complexes to the GAL-VPS13-HMR-E locus. However, we did not detect increased recruitment of SIR complexes in rtt109Δ, asf1Δ, and H3 K56R strains compared to wt cells (data not shown). Therefore, the slower induction of the GAL-VPS13-HMR-E gene in these strains was not caused by the larger amount of SIR complexes in the locus.
H3 K56 acetylation is characteristic for newly synthesized histones incorporated into chromatin during S phase, where it is required for proper functioning of the DNA damage checkpoint (28). When DNA synthesis is completed and cells go through G2 and M phases, K56 acetylation is removed by Hst3/4 deacetylases (3, 27, 52). Apparently, acetylation of K56 occurs only before incorporation of H3 into chromatin and histone chaperone Asf1 is required for proper exposition of H3 to Rtt109 acetyltransferase (11, 34, 48). Outside S phase, replication-independent incorporation of H3 acK56 has been reported mainly at promoter nucleosomes but also in the coding regions of highly expressed genes. As H3 K56 acetylation occurs on newly synthesized histones but not on nucleosomes in chromatin, the appearance of acK56 in transcription sites reflects mainly transcription-coupled incorporation of new histones into loci (15, 28, 36, 41). Our results also show that K56-acetylated H3 can be detected in the transcribed but not in the nontranscribed part of the GAL-VPS13-HMR-E gene (Fig. 8), confirming transcription-dependent incorporation of the H3 acK56 into chromatin.
What could be the role of K56 acetylation in transcription-coupled displacement of SIR proteins? Taking into account the results from the current and previous studies, we propose that K56 acetylation might either facilitate removal of nucleosomes during transcription elongation or inhibit reassociation of SIR complex with the transcribed locus. K56 lies in the globular domain of histone H3 extending toward the DNA major groove at the entry and exit points of the nucleosome core particle (13, 33), making modification of this residue a very attractive candidate for regulation of the properties of DNA-nucleosome or nucleosome-nucleosome interactions. Once the elongating RNAPII enters the locus, it will displace nucleosomes from its path and new histones, with K56 acetylation, will be loaded to chromatin as RNAPII moves away (15, 28, 36, 41). This might create a more favorable environment for the next polymerase, as nucleosomes are bound to DNA more loosely due to K56 acetylation and can be removed more easily than in the absence of the modification.
Alternatively, acetylation of H3 K56 might directly inhibit SIR complex binding to the locus and therefore help to open heterochromatic structures. Recent studies have shown that telomeric silencing is disrupted by K56 acetylation and that H3 K56 is maintained in a hypoacetylated state in silenced loci (31, 51, 52). Although a silenced locus might be generally inaccessible due to chromatin-bound SIR proteins, the border of a heterochromatic region is not strictly defined but is likely in a constantly changing equilibrium between SIR protein binding and “free” chromatin. As the original nucleosomes will be replaced by new ones during transcription-coupled reassembly of chromatin (7, 14, 49), they will contain acK56 in histone H3 (15, 36), repelling SIR proteins from the locus. As a result, the border between euchromatin and heterochromatin might be shifted toward the origin of SIR complex spreading and a wider area in the locus will be free of SIR complexes. If acetylation of H3 K56 is abolished by deletion of either ASF1 or RTT109 or by H3 K56R mutation, nucleosomes loaded to the locus do not restrict SIR complex spreading and the opening of the locus is considerably slower than that in wt cells.
Acknowledgments
We thank Jesper Svejstrup, Juhan Sedman, Mart Loog, and Pärt Peterson for critical reading of the manuscript.
This work was supported by the Wellcome Trust International Senior Research Fellowship grant 081756, EMBO Installation grant 1454, and Estonian Science Foundation grant 6604.
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. [DOI] [PubMed] [Google Scholar]
- 3.Celic, I., H. Masumoto, W. P. Griffith, P. Meluh, R. J. Cotter, J. D. Boeke, and A. Verreault. 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr. Biol. 16:1280-1289. [DOI] [PubMed] [Google Scholar]
- 4.de Bruin, D., S. M. Kantrow, R. A. Liberatore, and V. A. Zakian. 2000. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol. 20:7991-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Aguila, E. M., M. B. Dutra, J. T. Silva, and V. M. Paschoalin. 2005. Comparing protocols for preparation of DNA-free total yeast RNA suitable for RT-PCR. BMC Mol. Biol. 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhillon, N., M. Oki, S. J. Szyjka, O. M. Aparicio, and R. T. Kamakaka. 2006. H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 26:489-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman, and O. J. Rando. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405-1408. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, L., and D. S. Gross. 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol. Cell. Biol. 28:3979-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu, and Z. Zhang. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315:653-655. [DOI] [PubMed] [Google Scholar]
- 11.Han, J., H. Zhou, Z. Li, R. M. Xu, and Z. Zhang. 2007. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 282:28587-28596. [DOI] [PubMed] [Google Scholar]
- 12.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyland, E. M., M. S. Cosgrove, H. Molina, D. Wang, A. Pandey, R. J. Cottee, and J. D. Boeke. 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:10060-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamai, A., R. M. Imoberdorf, and M. Strubin. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25:345-355. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan, T., C. L. Liu, J. A. Erkmann, J. Holik, M. Grunstein, P. D. Kaufman, N. Friedman, and O. J. Rando. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4:e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32:370-377. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg, R. D., and Y. Lorch. 1991. Irresistible force meets immovable object: transcription and the nucleosome. Cell 67:833-836. [DOI] [PubMed] [Google Scholar]
- 18.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristjuhan, A., J. Walker, N. Suka, M. Grunstein, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2002. Transcriptional inhibition of genes with severe histone H3 hypoacetylation in the coding region. Mol. Cell 10:925-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristjuhan, A., B. O. Wittschieben, J. Walker, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2003. Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc. Natl. Acad. Sci. U. S. A. 100:7551-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S., and D. S. Gross. 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13:727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo, S., and J. Rine. 1994. Silencers and domains of generalized repression. Science 264:1768-1771. [DOI] [PubMed] [Google Scholar]
- 25.Luo, K., M. A. Vega-Palas, and M. Grunstein. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch, P. J., and L. N. Rusche. 2009. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol. Cell. Biol. 29:43-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maas, N. L., K. M. Miller, L. G. DeFazio, and D. P. Toczyski. 2006. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell 23:109-119. [DOI] [PubMed] [Google Scholar]
- 28.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294-298. [DOI] [PubMed] [Google Scholar]
- 29.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 30.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, A., B. Yang, T. Foster, and A. L. Kirchmaier. 2008. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics 179:793-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. U. S. A. 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozdemir, A., S. Spicuglia, E. Lasonder, M. Vermeulen, C. Campsteijn, H. G. Stunnenberg, and C. Logie. 2005. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 280:25949-25952. [DOI] [PubMed] [Google Scholar]
- 34.Recht, J., T. Tsubota, J. C. Tanny, R. L. Diaz, J. M. Berger, X. Zhang, B. A. Garcia, J. Shabanowitz, A. L. Burlingame, D. F. Hunt, P. D. Kaufman, and C. D. Allis. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. U. S. A. 103:6988-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rine, J., and I. Herskowitz. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116:9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rufiange, A., P. E. Jacques, W. Bhat, F. Robert, and A. Nourani. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27:393-405. [DOI] [PubMed] [Google Scholar]
- 37.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 38.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandell, L. L., D. E. Gottschling, and V. A. Zakian. 1994. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc. Natl. Acad. Sci. U. S. A. 91:12061-12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, J., P. Bajwa, F. C. Johnson, S. R. Bhaumik, and A. Shilatifard. 2006. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281:37270-37274. [DOI] [PubMed] [Google Scholar]
- 42.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403-414. [DOI] [PubMed] [Google Scholar]
- 44.Sekinger, E. A., and D. S. Gross. 1999. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 18:7041-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shei, G. J., and J. R. Broach. 1995. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol. Cell. Biol. 15:3496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 47.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32:378-383. [DOI] [PubMed] [Google Scholar]
- 48.Tsubota, T., C. E. Berndsen, J. A. Erkmann, C. L. Smith, L. Yang, M. A. Freitas, J. M. Denu, and P. D. Kaufman. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25:703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Värv, S., K. Kristjuhan, and A. Kristjuhan. 2007. RNA polymerase II determines the area of nucleosome loss in transcribed gene loci. Biochem. Biophys. Res. Commun. 358:666-671. [DOI] [PubMed] [Google Scholar]
- 50.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 51.Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, B., A. Miller, and A. L. Kirchmaier. 2008. HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol. Biol. Cell 19:4993-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou, Y., Q. Yu, and X. Bi. 2006. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. 26:7806-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]