Abstract
The c-Jun NH2-terminal kinase (JNK) is implicated in proliferation. Mice with a deficiency of either the Jnk1 or the Jnk2 genes are viable, but a compound deficiency of both Jnk1 and Jnk2 causes early embryonic lethality. Studies using conditional gene ablation and chemical genetic approaches demonstrate that the combined loss of JNK1 and JNK2 protein kinase function results in rapid senescence. To test whether this role of JNK was required for stem cell proliferation, we isolated embryonic stem (ES) cells from wild-type and JNK-deficient mice. We found that Jnk1−/− Jnk2−/− ES cells underwent self-renewal, but these cells proliferated more rapidly than wild-type ES cells and exhibited major defects in lineage-specific differentiation. Together, these data demonstrate that JNK is not required for proliferation or self-renewal of ES cells, but JNK plays a key role in the differentiation of ES cells.
The c-Jun NH2-terminal kinase (JNK) is a member of the mitogen-activated protein (MAP) kinase group of signaling proteins. JNK is encoded by two ubiquitously expressed genes (Jnk1 and Jnk2) and by a third gene (Jnk3) that is selectively expressed in neurons (14). Gene disruption studies demonstrate that mice without Jnk1 or Jnk2 are viable, but compound deficiency of both Jnk1 and Jnk2 causes early embryonic lethality (14). Murine embryonic fibroblasts (MEFs) isolated from Jnk1−/− Jnk2−/− mice exhibit a severe growth retardation phenotype (54). The markedly reduced growth of Jnk1−/− Jnk2−/− MEFs is consistent with the finding that JNK is critically required for the regulation of AP1-dependent gene expression (56) that is implicated in cellular proliferation (26). Thus, Jnk1−/− Jnk2−/− MEFs express low levels of AP1 proteins (e.g., c-Jun and JunD) and exhibit marked defects in AP1 target gene expression (34, 56). This loss of AP1 function is mediated, in part, by reduced phosphorylation of the activation domain of Jun family proteins and ATF2 (56).
More recent studies using a conditional gene ablation strategy have demonstrated that compound JNK deficiency causes rapid senescence (12). This conclusion was confirmed by using chemical genetic analysis with MEFs isolated from mice with a germ line mutation that sensitizes JNK to inhibition by a predesigned small-molecule drug (12, 25). This form of senescence was found to be p53 dependent (12) and resembles the p53-dependent senescence of c-Jun−/− MEFs (49). These data indicate that JNK plays a critical role in cellular proliferation. Indeed, it is possible that the p53-dependent senescence observed in JNK-deficient cells may contribute to aging. This is because altered p53 function is established to be an important determinant of early aging (36, 55). Importantly, this role of p53 in aging appears to be distinct from p53-mediated tumor suppression and DNA damage responses (21, 39, 43).
One aspect of the aging process is a reduction in the regenerative capacity of stem cells (50). Indeed, it has been established that altered p53 activity associated with aging causes decreased stem cell function (8, 18, 42) and that disruption of the p53 pathway can increase stem cell function (1). Since JNK can influence p53-dependent senescence (12), these data indicate that JNK may be important for stem cell proliferation and self-renewal potential.
Embryonic stem (ES) cells proliferate and are capable of both self-renewal and differentiation to multiple cell types. Indeed, murine ES cells can differentiate to create all tissues within a mouse. The profound growth retardation and rapid p53-dependent senescence of Jnk1−/− Jnk2−/− MEFs (12) suggests that JNK may play a critical role in the normal function of ES cells, including self-renewal and differentiation potential. The purpose of the present study was to test this hypothesis. Our approach was to isolate ES cells from wild-type and JNK-deficient mice. We demonstrate that JNK is not required for self-renewal or the proliferation of ES cells. However, JNK is required for ES cell differentiation.
MATERIALS AND METHODS
Mouse studies.
Jnk1−/− mice (16) and Jnk2−/− mice (60) on a C57BL/6J genetic background were described previously. C57BL/6J mice and C57BL/6J-scid (B6.CB17-prkdcscid/SzJ) mice were obtained from the Jackson Laboratories. These mice were housed in a facility that is accredited by the American Association for Laboratory Animal Care and the studies were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School.
Genotype analysis.
Jnk1 and Jnk2 genotypes were examined by PCR analysis of genomic DNA (16, 60). Sex determination of ES cells was performed by PCR amplification of genomic DNA to detect the presence of X and Y chromosomes (46).
ES cell culture.
Blastocysts (embryonic day 3.5 [E3.5]) were isolated (47) and transferred to 24-well tissue culture dishes with a feeder cell layer of primary mouse embryo fibroblasts (MEFs) inactivated with mitomycin C (Sigma) in Dulbecco modified Eagle medium (DMEM; Invitrogen), 15% fetal bovine serum (Atlanta Biologicals), 2 mM glutamine, 1 mM sodium pyruvate, 100 μM nonessential amino acids, 0.1 mM β-mercaptoethanol (Invitrogen), and 1,000 U of leukemia inhibitory factor (LIF) (Chemicon)/ml. Five days after plating, the inner cell mass was treated with trypsin and harvested, and replated on feeder cell layers in 24-well dishes. ES cell colonies were replated on feeder cell layers every 2 to 3 days.
Retroviral transduction studies were performed by using the vector pMSCV-Flag-JNK1-IRES-Puro (31). ES cells were plated on puromycin resistant RJ feeder layers (provided by Stephen N. Jones, University of Massachusetts Medical School) in ES cell growth medium (48 h) prior to infection. The medium was replaced at 24 h postinfection with fresh medium supplemented with 2 μg of puromycin/ml. Positive ES cell clones were screened for expression of the JNK1 polypeptide by immunoblot analysis.
Embryoid bodies (EBs) were cultured by plating single cells in suspension using DMEM supplemented with 10% fetal bovine serum (Invitrogen), 10% newborn calf serum (Invitrogen), 1% horse serum (Invitrogen), and 2 mM glutamine using uncoated petri dishes (8 days) and then 0.1% gelatin-coated dishes (7 days). The number of cells was measured by transferring 100 EBs to 1 ml of phosphate-buffered saline (PBS) in one well of a 24-well culture plate. These EBs were incubated with trypsin and dissociated to a single-cell suspension, and the number of cells was measured by using a hemacytometer.
Teratomas.
ES cell-derived teratomas were prepared by subcutaneous injection of 4 × 106 ES cells into the flanks of C57BL/6J-scid mice. The mice were euthanized at 3 weeks postinjection. The teratomas were removed, washed in PBS, fixed (14 h) in 4% paraformaldehyde, and processed for histology.
Cardiomyocyte differentiation in vitro.
ES cell-derived cardiomyocytes were prepared in vitro (61). The ES cells were cultured using dishes coated with 0.1% gelatin (3 days) in DMEM supplemented with 10% fetal bovine serum (Invitrogen), 10% newborn calf serum (Invitrogen), 1% horse serum (Invitrogen), 2 mM glutamine, 0.1 mM β-mercaptoethanol, 1,000 U of LIF/ml, and 0.15 μg of Noggin-Fc (R&D Systems)/ml. Single cells were cultured in the same medium without LIF and β-mercaptoethanol (1 day) and then in the same medium without LIF (Chemicon), β-mercaptoethanol, and Noggin-Fc (7 days) in suspension to form EBs using uncoated petri dishes. The EBs were transferred to gelatin-coated tissue dishes and cultured for 16 days.
Immunohistochemistry.
Paraformaldehyde-fixed tissue was processed, embedded in paraffin, and 4-μm sections were stained with hematoxylin and eosin (H&E). Sections were also stained with antibodies to insulin (Dako), α-fetoprotein (Meridian Life Science), troponin I (Santa Cruz), keratin 5/8 (Lab Vision), desmin (Dako), Nestin (Chemicon), and the cytokeratin Endo-A (TROMA-1; Developmental Studies Hybridoma Bank, University of Iowa) using indirect immunoperoxidase detection (58). Alkaline phosphatase activity was measured using cells fixed with 4% paraformaldehyde at room temperature (1 h) using the Red Alkaline phosphatase substrate kit I (Vector). TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assays were performed by using an in situ cell death detection kit (Roche).
Immunofluorescence.
Staining was performed using coverslips washed in PBS, incubated in 90% methanol containing 5% acetic acid at −20°C (5 min), washed with PBS, and then blocked by incubation with PBS supplemented with 1% skimmed milk for 1 h at room temperature and incubated with antibodies to SSEA-1 (Developmental Hybridoma Cell Bank, University of Iowa), Oct-4 (Chemicon), or FGF-4 (Santa Cruz, CA) in PBS containing 1% skimmed milk at 4°C (14 h). The cells were washed and then incubated with secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 546 (Molecular Probes) at 37 °C (1 h). Washed slides were mounted and examined by using a Leica SP2 laser-scanning confocal fluorescence microscope.
Bromodeoxyuridine (BrdU) incorporation was examined by addition of 10 μM BrdU (Sigma) to the EB culture medium for 24 h. EB were fixed with 4% paraformaldehyde at 4°C (14 h) and embedded in OCT compound. Frozen sections (4 μm) were prepared, stained with a fluorescein isothiocyanate-conjugated antibody to BrdU (BD-Pharmingen), and examined by fluorescence microscopy (59).
Electron microscopy.
Cells were fixed with 1.25% glutaraldehyde (30 min) at room temperature and with 2.5% glutaraldehyde in cacodylate buffer (14 h) at 4°C. The cells were then postfixed with 1% (wt/vol) osmium tetraoxide in PBS, dehydrated, and embedded in Lx 112/Araldite 502 epoxy resin. Ultrathin sections were mounted on copper support grids in serial order, contrasted with lead citrate and uranyl acetate, and examined on a Philips CM 10 transmission electron microscope (20).
Immunoblot analysis.
Cell extracts were prepared by using Triton lysis buffer (20 mM Tris [pH 7.4], 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 10 μg of aprotinin and leupeptin/ml). Extracts (50 μg) were examined by immunoblot analysis. The blots were probed with antibodies to JNK (BD-Pharmingen), p38, ERK1, ERK2 (Santa Cruz), and α-tubulin (Sigma). Immunocomplexes were detected by enhanced chemiluminescence (NEN).
RNA analysis.
Total RNA was prepared by using an RNeasy lipid tissue minikit (Qiagen) and was treated with DNase (Sigma). First-strand cDNA was synthesized by using an iScript cDNA synthesis kit (Bio-Rad). PCR analysis was performed using amplimers (Table 1).
TABLE 1.
Amplimers used in this study
| Gene | Amplimer pair sequences (5′-3′) |
|
|---|---|---|
| Amplimer 1 | Amplimer 2 | |
| Anf | GGGGGTAGGATTGACAGGAT | CAGAGTGGGAGAGGCAAGAC |
| Apoa2 | CACTGCTGGTAACCATCTG | TCGAGGTTCATTAAACTGCT |
| BMHC | CTACAGGCCTGGGCTTACCT | TCTCCTTCTCAGACTTCCGC |
| Bmp4 | GTTTCCTCTTCAACCTCAGC | TATACGGTGGAAGCCCTGTT |
| Brachyury | CTCACCAACAAGCTCAATGG | ACAGCTATGAACTGGGTCTC |
| Emx2 | AGCTTTTAAGGCTAGAGCAC | CCTGAGTTTCCGTAAGACTG |
| Flk1 | GGCGGTGGTGACAGTATCTT | CTCGGTGATGTACACGATGC |
| Gata4 | TGTCAATTGTGGGGCCATGTC | GTGCCCCAGCCTTTTACTTTG |
| Hnf1 | GAAAGCAACGGGAGATCCTCCGAC | TAGGCATCCATGGCCAGCTTCTGC |
| Hnf3 | TGGTCACTGGGGACAAGGGAA | GCAACAACAGCAATAGAGAAC |
| Keratin17 | GTAGTGCTCTTGGGAGCAACAG | AGAACCAGTCTTCGGCATCCTT |
| MEF2c | ACTGGGAAACCCCAATCTTC | ATCAGACCGCCTGTGTTACC |
| Mlv2v | AAAGAGGCTCCAGGTCCAAT | CCTCTCTGCTTGTGTGGTCA |
| Nkx2.5 | TGCAGAAGGCAGTGGAGCTGGACAAGCC | TTGCACTTGTAGCGACGGTTCTGGAACCAG |
| Nodal | TGGAGCGCATTTGGATGGAGAC | TACTGCTTGGGGTAGATGATCC |
| Otx1 | TTCACGCGCTCACAGCTGGA | ACCAAACCTGGACTCTGGAC |
| Pax2 | TGCCGAATACAAGCGACAGAAC | TGGAAGACATCGGGATAGGAAG |
| Sox17 | GCCAAAGACGAACGCAAGCGG | TCATGCGCTTCACCTGCTTG |
| Transferrin | ATTCTCAAAGTGGCACAGGAACAC | GGGTTCTTTCCTTCGGTGTTATCC |
| Wnt1 | TCTTCGGCAAGATCGTCAACCG | AAGGTTCATGAGGAAGCGTAGG |
Statistical analysis.
Values are expressed as the mean ± the standard deviation. Comparisons among values for all groups were determined using a Student t test.
RESULTS
Isolation of JNK-deficient ES cells.
We prepared E3.5 blastocysts from C57BL/6J mice and isolated ES cells in culture using a feeder layer of inactivated MEFs. Studies using mice without or with targeted disruption of the Jnk1 and Jnk2 genes led to the isolation of wild-type ES cell clones and also JNK-deficient ES cell clones lacking expression of Jnk1 and/or Jnk2. These clones were screened for the presence of X and Y chromosomes, and 18 male (XY) ES cell clones were selected for further study.
Representative ES cell clones were expanded in culture and examined by immunoblot analysis. JNK isoforms (46 and 55 kDa) were detected in lysates prepared from wild-type ES cells (Fig. 1A). Reduced expression of 46- and 55-kDa JNK isoforms was detected in lysates prepared from Jnk1−/− and Jnk2−/− ES cells, respectively. In contrast, no JNK was detected in lysates prepared from compound mutant Jnk1−/− Jnk2−/− ES cells (Fig. 1A). Control studies demonstrated that these ES cells did not express different levels of other MAP kinases (ERK1/2 and p38α).
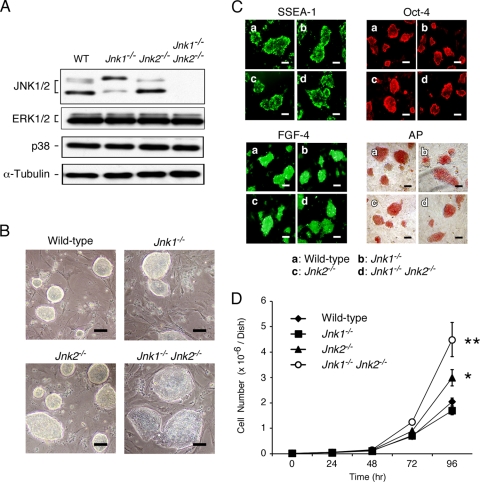
FIG. 1.
Isolation of JNK-deficient ES cells. (A) Wild-type, Jnk1−/−, Jnk2−/−, and Jnk1−/− Jnk2−/− ES cells were examined by immunoblot analysis with antibodies to JNK1/2, ERK1/2, p38 MAPK, and α-tubulin. (B) Wild-type and JNK-deficient ES cells grown on a layer of feeder cells (92 h) were examined by phase-contrast microscopy. Scale bar, 40 μm. (C) ES cells were stained for alkaline phosphatase activity and with antibodies to SSEA-1, Oct-4, and FGF-4. Scale bar, 40 μm. (D) Wild-type and JNK-deficient ES cells (104 cells) were plated in six-well gelatin-coated dishes. The number of cells on different days was measured. The data are shown as means ± the standard deviations (SD) (n = 3). Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01).
The wild-type and JNK-deficient ES cells grow as colonies on feeder layers. No morphological differences between wild-type and JNK-deficient ES cell colonies were detected, although the compound mutant Jnk1−/− Jnk2−/− ES cells formed larger colonies than did the other ES cells (Fig. 1B). All of the ES cells we isolated expressed phenotypic markers that characterize an undifferentiated state (Fig. 1C), including alkaline phosphatase, SSEA-1, Oct-4, and FGF-4 (28). Together, these data indicate that JNK deficiency does not prevent the isolation of ES cells that express markers of the undifferentiated state.
JNK is not essential for ES cell proliferation.
Compound JNK deficiency in MEFs causes severe growth retardation and early senescence (12, 54). We therefore examined the growth of wild-type and JNK-deficient ES cells grown as colonies. The proliferation of wild-type and Jnk1−/− ES cells was similar (Fig. 1D). In contrast, Jnk2−/− ES cells grew more rapidly than wild-type ES cells and compound mutant Jnk1−/− Jnk2−/− ES cells exhibited markedly increased proliferation compared to wild-type ES cells (Fig. 1D). The high proliferation rate of Jnk1−/− Jnk2−/− ES cells suggests that the normal function of JNK may be to limit ES cell proliferation and most likely accounts for the observation that these cells form large colonies on feeder cell layers (Fig. 1B). These data demonstrate that JNK is not required for ES cell proliferation. This observation contrasts with the finding that JNK deficiency causes rapid senescence in MEFs (12, 54).
JNK is required for the normal development of ES cell-derived EBs.
To examine the properties of wild-type and JNK-deficient ES cells, we investigated the ability of these ES cells to form EBs. Wild-type ES cells, Jnk1−/− ES cells, and Jnk2−/− ES cells formed EBs that increased in size (Fig. 2A) and cell number (Fig. 2B) during culture in vitro. The Jnk1−/− EBs, and especially the Jnk2−/− EBs, formed larger luminal spaces than wild-type EBs (Fig. 2A). However, the proliferation of wild-type, Jnk1−/−, or Jnk2−/− cells in the EBs was similar (Fig. 2B). This finding contrasts with the observation that undifferentiated Jnk2−/− ES cell colonies proliferated more rapidly than wild-type or Jnk1−/− ES cell colonies in culture (Fig. 1D).
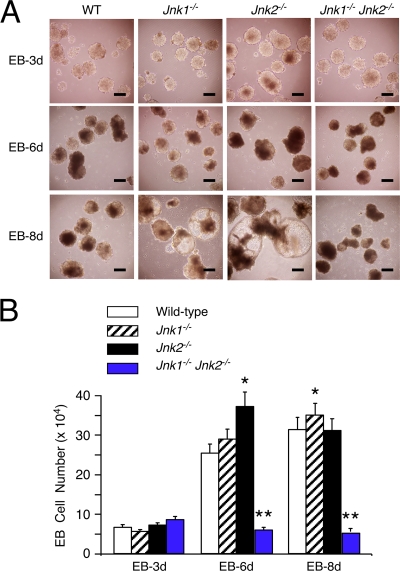
FIG. 2.
Effect of JNK deficiency on the formation of EBs. (A) EBs at days 3, 6, and 8 were examined by phase-contrast microscopy. Scale bar, 250 μm. (B) The number of cells per EB was measured. The data are presented as the means ± the SD (n = 100). Statistically significant differences are indicated (*, P < 0.05; **, P < 0.001).
The compound mutant Jnk1−/− Jnk2−/− ES cells also formed EBs (Fig. 2A). These structures lacked the large luminal spaces found in Jnk1−/− and Jnk2−/− EBs (Fig. 2A) and the number of cells within the compound mutant Jnk1−/− Jnk2−/− EBs did not increase during culture in vitro (Fig. 2B). Thus, while JNK is not required for ES cell proliferation (Fig. 1D), JNK is required for proliferation of cells that have initiated a differentiation program (Fig. 2B). This stage-specific role for JNK correlates with a change in the rapid cell cycles of ES cells to the slower cell cycles of differentiated cells that include fully formed gap (G1/G2) phases.
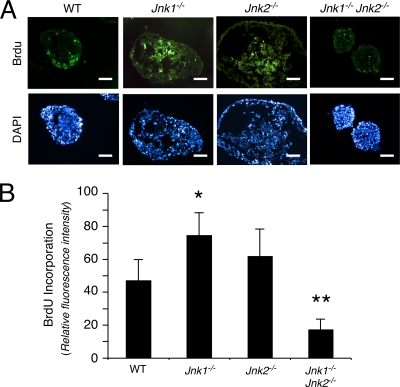
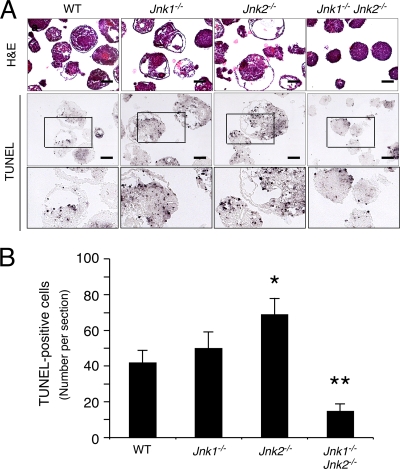
Previous studies have implicated JNK in both proliferation and death (14). The effect of compound JNK deficiency to prevent the accumulation of cells within EBs may therefore be caused by changes in either cellular proliferation or death. To test this hypothesis, we examined proliferation in EBs by investigating the incorporation of BrdU into DNA (Fig. 3) and cell death by TUNEL assays (Fig. 4). These studies demonstrated that the compound mutant Jnk1−/− Jnk2−/− EBs exhibited decreased DNA synthesis and also decreased apoptosis. The failure of compound mutant Jnk1−/− Jnk2−/− EBs to grow (Fig. 2B) is therefore accounted for by decreased proliferation rather than increased cell death.
FIG. 3.
Effect of JNK deficiency on proliferation. (A) Day 7 EBs were pulse-labeled with BrdU and fixed. Frozen sections were stained with DAPI (4′,6′-diamidino-2-phenylindole) and an antibody to BrdU and then examined by immunofluorescence analysis. Scale bar, 150 μm. (B) The mean fluorescence intensity of the BrdU immunofluorescence was measured by using ImageJ software and is presented as the means ± the SD (n = 100 EBs). Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01).
FIG. 4.
Effect of JNK deficiency on apoptosis. (A) Day 7 EBs were fixed and embedded in paraffin. Sections were stained with H&E and examined by TUNEL assay. Scale bar, 250 μm. A higher-magnification image of a selected region (indicated with a box) of the TUNEL assay is also shown. (B) The number of TUNEL-positive cells in sections of EBs was measured and is presented as the mean ± the SD (n = 100 EBs). Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01).
Role of JNK in the expression of endodermal lineage genes in vitro.
Development of the endoderm is one of the earliest steps in embryonic development and genes associated with endodermal differentiation (e.g., Apoa2, Hnf1, Hnf3, Sox17, and Transferrin) are expressed during the formation of ES cell-derived EBs. Transferrin and Hnf3 are essential for differentiation of the visceral endoderm (7, 17) and show a similar pattern of expression in EBs formed by wild-type and JNK-deficient ES cells (Fig. 5A). Similarly, JNK deficiency did not cause major changes in the expression of Apoa2, a gene that is critically required for liver development (53). However, we found JNK-dependent differences in the expression of Sox17 and Hnf1, genes that are required for differentiation of definitive endoderm (27) and visceral endoderm (5), respectively. Thus, Sox17 expression was downregulated in late-stage EBs formed by JNK1/2-deficient ES cells compared to wild-type ES cells (Fig. 5A). In contrast, the expression of Hnf1 was increased at the early stages of EB formation by JNK1/2-deficient ES cells compared to wild-type ES cells (Fig. 5A). Together, these data indicate a nonessential role of JNK for endodermal differentiation, but JNK may influence this developmental program.
FIG. 5.
Effect of JNK deficiency on differentiation-associated gene expression in vitro. Semiquantitative reverse transcription-PCR (RT-PCR) analysis was performed to detect the expression of mRNA associated with differentiation to endoderm (A), mesoderm (B), and ectoderm (C). Total RNA from undifferentiated ES cells (un-diff) or various stages of EB development and differentiation between days 3 and 15 (EB-3d to EB-15d) was examined. The amplimers used for this analysis are listed in Table 1. Gapdh gene expression was analyzed as an internal control. The results are representative of three independent experiments.
Role of JNK in the expression of mesodermal lineage genes in vitro.
Members of the transforming growth factor-β superfamily, including Bmp4 and Nodal, are implicated in the induction of mesoderm and gastrulation (4). Nodal was expressed at similar levels in wild-type and JNK1/2-deficient EBs, but some variations in Bmp4 expression were detected (Fig. 5B). These data suggest that the initial development of mesoderm may be JNK independent. Nevertheless, Brachyury, a gene that is critical for mesoderm development (22), was poorly expressed by EBs formed by compound mutant Jnk1−/− Jnk2−/− ES cells (Fig. 5B). This observation suggests that JNK may play an important role in the development of mesodermal lineages.
To further examine the JNK dependence of mesodermal lineage gene expression, we examined genes related to cardiomyogenesis, including Flk1, Gata4, Nkx2.5, Mef2c, β-Mhc, Mlv2v, and Anf. These genes play important roles in the specification of cardiac progenitor cells and cardiac development (6, 9, 19, 24, 29, 33, 35, 44, 45, 51). Expression of Gata4 and Mef2c transcription factor mRNA was markedly reduced in JNK1/2-deficient EBs at early stages of differentiation compared to wild-type EBs (Fig. 5B). At late stages of differentiation, very low levels of Flk1, Mef2c, β-Mhc, Mlv2v, and Anf gene expression were detected in EBs formed by compound mutant Jnk1−/− Jnk2−/− ES cells (Fig. 5B). These data strongly support the conclusion that the JNK pathway is important for mesodermal differentiation, including the development of cardiac cells in vitro.
Role of JNK in the expression of ectodermal lineage genes in vitro.
We examined whether JNK deficiency affected the development of ectoderm in ES cell-derived EBs. Keratin17 is a marker for epidermal cells (41), and its expression was modestly decreased in EBs formed by compound mutant Jnk1−/− Jnk2−/− ES cells compared to wild-type ES cells at late stages of development (Fig. 5C). These data suggest that JNK is not critically required for ectodermal differentiation.
Compound deficiency of Jnk1 and Jnk2 in embryos causes early death associated with defects in neurogenesis (32). We therefore examined the expression of genes related to early neurogenesis in wild-type and JNK-deficient EBs. The expression of Emx2, a gene that is essential for forebrain development (37), was decreased in JNK1/2-deficient EBs compared to wild-type EBs (Fig. 2D). Expression of Otx1, a marker for early neuroectodermal development that is expressed at the 1-3 somite stage in the anterior neuroectoderm of mouse embryos (10), was increased in early stage JNK1/2-deficient EBs (Fig. 5C). It has been established that Wnt1 and Pax2 regulate mesencephalon and metencephalon development (38), and the expression of these genes was markedly downregulated in EBs formed by compound mutant Jnk1−/− Jnk2−/− ES cells compared to wild-type ES cells (Fig. 5C). Together, these findings support the conclusion that JNK contributes to ectoderm development and indicate that JNK-deficient ES cells exhibit abnormal neurogenesis in vitro.
Role of JNK in differentiation in vivo.
In vitro analysis of ES cells indicates that JNK can influence differentiation (Fig. 5). Marker gene expression analysis demonstrated that JNK deficiency caused large defects in the development of mesoderm and more limited defects in ectodermal and endodermal development in vitro (Fig. 5). Methods that can be used to study differentiation in vivo include the analysis of mutant and mosaic mice. However, these approaches were not feasible for the analysis of JNK function because Jnk1−/− Jnk2−/− mice die during early embryonic development (32) and mosaic mice prepared by injection of Jnk1−/− Jnk2−/− ES cells into wild-type blastocysts exhibit low levels of chimerism (15). Therefore, to test the role of JNK in differentiation in vivo, we examined the formation of ES cell-derived teratomas, including tissues formed by all three germ cell layers (Fig. 6 and 7).
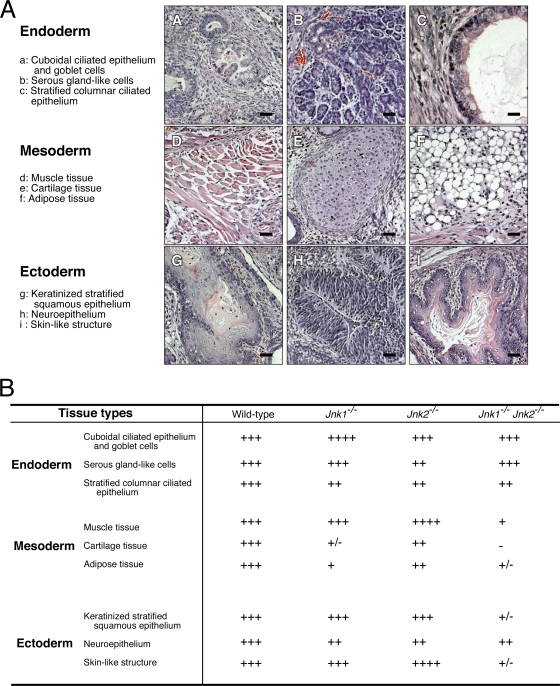
FIG. 6.
Effect of JNK deficiency of ES differentiation in vivo. (A) Subcutaneously injected ES cells form teratomas in congenic mice. H&E-stained sections of teratomas formed by wild-type ES cells show the formation of endodermal (a to c), mesodermal (d to f), and ectodermal (g to i) tissues. Scale bar, 60 μm. (B) Comparison of teratomas derived from wild-type and JNK-deficient ES cells. Different tissues within sections of teratomas were identified and the area within each section was determined by using ImageJ software. The relative expression of each tissue within teratomas formed by ES cells with different genotypes is presented with a scale from “−” (lowest) to “++++” (highest). This scale is defined in Table 2.
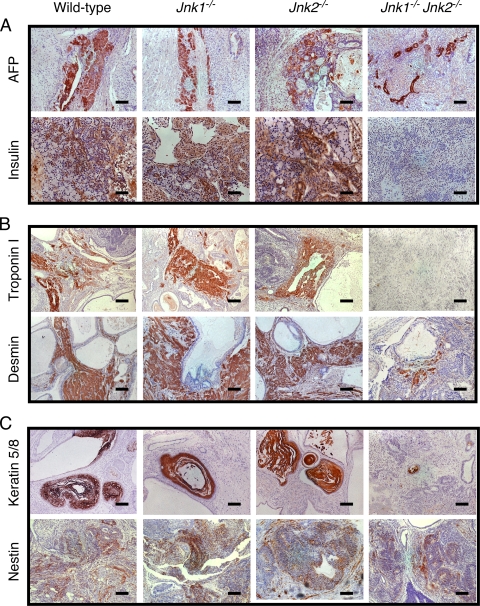
FIG. 7.
Defective differentiation of JNK-deficient ES cells in vivo. Sections of teratomas formed by wild-type and JNK-deficient ES cells were examined by staining with H&E. (A) Hepatocytelike cells (upper panel) and β-like cells (lower panel) of endodermal origin were detected by staining with antibodies to α-fetoprotein (AFP) and insulin. Scale bars: upper, 250 μm; lower, 125 μm. (B) Cardiomyocytelike cells (upper panel) and striated-muscle-like cells (lower panel) of mesodermal origin were detected with antibodies to troponin I and desmin. Scale bars, 250 μm. (C) Keratinocytelike cells (upper panel) and primitive neuroepitheliumlike cells (lower panel) of ectodermal origin were detected with antibodies to keratin5/8 and nestin. Scale bars, 250 μm.
Histological analysis of sections prepared from teratomas demonstrated the presence of endodermal tissues (Fig. 6A). Stratified columnar ciliated epithelium originates from the endoderm (28) and a modest reduction in the amount of this tissue was detected in JNK-deficient teratomas (Fig. 6B). However, JNK deficiency was found to cause no difference in tissues with serous gland-like cells or cuboidal ciliated epithelium (Fig. 6B). Immunohistochemical staining of tissue sections for the cytokeratin Endo-A (an endodermal marker) with the TROMA1 antibody (30) did not reveal differences between wild-type and JNK-deficient teratomas (data not shown). However, in contrast to wild-type teratomas, decreased numbers of hepatocytelike cells (that stain with an antibody to α-fetoprotein) and no insulin-positive pancreatic β-like cells were detected in compound mutant Jnk1−/− Jnk2−/− teratomas (Fig. 7A). These data indicate that JNK is not essential for endodermal development, but JNK may influence differentiation of some specific cell types derived from the endoderm.
Tissues with a mesodermal origin in wild-type and JNK-deficient teratomas, including muscle, cartilage, and adipose tissue, were examined. Major reductions in the amount of all three tissues were detected in JNK-deficient teratomas compared to wild-type teratomas (Fig. 6B). Immunohistochemical staining of tissue sections using antibodies to cardiac muscle troponin I and striated muscle desmin indicated the presence of both cardiomyocytelike and striated-muscle-like cells in wild-type teratomas, but no cardiac muscle tropinin I and significantly decreased striated muscle desmin were detected in compound mutant Jnk1−/− Jnk2−/− teratomas (Fig. 7B). These data indicate that mesoderm development is markedly impaired by JNK deficiency in vivo.
We examined tissues with an ectodermal origin in wild-type and JNK-deficient teratomas, including keratinized stratified squamous epithelium, neuroepithelium, and skinlike structures. Studies of compound mutant Jnk1−/− Jnk2−/− teratomas demonstrated reduced amounts of all three ectodermal tissues compared to wild-type teratomas (Fig. 6B). Immunohistochemical staining of tissue sections using an antibody to keratin 5/8, a marker for epidermal skin cells, demonstrated markedly reduced expression in compound mutant Jnk1−/− Jnk2−/− teratomas compared to wild-type teratomas (Fig. 7C). A small decrease in Nestin expression (a marker of primitive neuroectoderm) was also detected in compound mutant Jnk1−/− Jnk2−/− teratomas compared to wild-type teratomas (Fig. 7C). Together, these data indicate that JNK can play an important role in ectoderm development in vivo.
JNK is not required for ES cell self-renewal.
Studies of ES cells in vitro and in vivo demonstrate that JNK deficiency causes lineage-specific defects in differentiation (Fig. 5 to 7). These defects in differentiation may result from either a role of JNK in the differentiation program or because JNK-deficient ES cells have lost their differentiation potential due to a failure of self-renewal. To distinguish between these possibilities, we performed complementation analysis using recombinant JNK expressed in compound mutant Jnk1−/− Jnk2−/− ES cells. The rationale for these studies was that if the ES cells exhibited a failure of self-renewal, genetic complementation would not lead to restoration of differentiation potential. In contrast, if the JNK-deficient ES cells underwent self-renewal and JNK was required for differentiation, genetic complementation would restore differentiation potential.
We examined ES cells using a Noggin-induced differentiation protocol that leads to the formation of cardiomyocytes (61). Indeed, spontaneous beating of cell sheets was reproducibly detected within 6 days after attachment of wild-type, Jnk1−/−, and Jnk2−/− EBs in culture (Fig. 8A; see also Videos S1 to S3 in the supplemental material). Within 16 days after attachment, spontaneous beating of cells with a distinct muscle morphology was observed (Fig. 8A; see also Videos S5 to S7 in the supplemental material). Spontaneous beating is a property of both cardiomyocytes and smooth muscle cells. To confirm that these cultures contain cardiomyocytes (61), we examined the differentiated cells by transmission electron microscopy. Sarcomeric and Z-bands structures were detected consistent with the presence of cardiomyocytes (Fig. 8B).
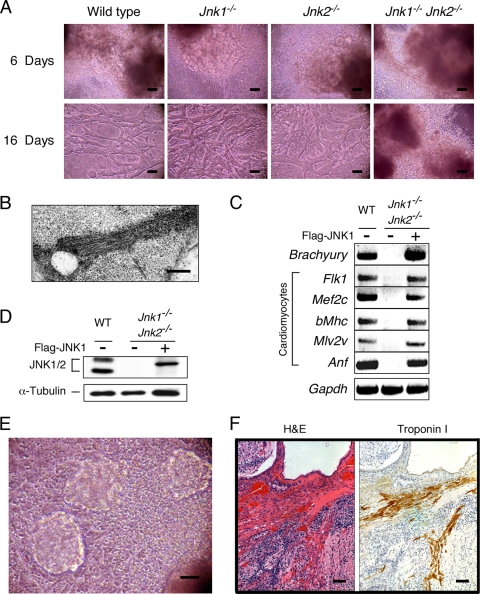
FIG. 8.
JNK is not required for ES cell self-renewal. (A) ES cell-derived cardiomyocytes after 6 or 16 days of differentiation were examined by phase-contrast microscopy. Scale bar, 20 μm. The images represent single frames of movies (see Videos S1 to S8 in the supplemental material). (B) Cardiomyocytes derived from wild-type ES cells were examined by transmission electron microscopy. Sarcomeric and Z-band structures were detected. Scale bar, 1 μm. (C) Complementation analysis demonstrates that the expression of Flag-JNK1 in in Jnk1−/− Jnk2−/− ES cells restored the expressed of mesodermal differentiation markers in EBs at 15 days. Total RNA was isolated and mRNA expression was examined by semiquantitative RT-PCR analysis. (D) Flag-JNK1 was expressed in Jnk1−/− Jnk2−/− ES cells by retroviral transduction. The amount of JNK and α-tubulin in ES cell lysates was examined by immunoblot analysis. (E) Beating ES cell-derived cardiomyocytes in cultures of Jnk1−/− Jnk2−/− ES cells that express Flag-JNK1 were detected by phase-contrast microscopy. Scale bar, 20 μm. The image represents a single frame of a movie (see Video S9 in the supplemental material). (F) Sections of teratomas derived from Jnk1−/− Jnk2−/− ES cells that express Flag-JNK1 demonstrate the presence of cardiomyocytelike structures stained with H&E (left) and an antibody to troponin I (right). Scale bar, 250 μm.
No spontaneously beating cell sheets or muscle were detected in cultures of compound mutant Jnk1−/− Jnk2−/− cells (Fig. 8A; see also Videos S4 and S8 in the supplemental material). Indeed, gene expression analysis demonstrated that the mesoderm marker Brachyury and the cardiomyocyte markers Anf, Flk1, Mef2c, β-Mhc, and Mlv2v expressed in wild-type cultures were not expressed in cultures of compound mutant Jnk1−/− Jnk2−/− cells (Fig. 8C). These data confirm that JNK is essential for the formation of cardiomyocytes.
Complementation tests were performed using retroviral transduction of epitope-tagged JNK1α1 in compound mutant Jnk1−/− Jnk2−/− ES cells. Immunoblot analysis of cell lysates demonstrated that the 46- and 55-kDa JNK isoforms present in wild-type ES cells were absent in JNK-deficient ES cells (Fig. 8D). However, epitope-tagged JNK1α1 was detected in JNK-deficient ES cells transduced with the JNK1α1 retroviral vector (Fig. 8D). The expression of JNK1α1 in compound mutant Jnk1−/− Jnk2−/− ES cells restored the wild-type ES cell phenotype, including gene expression (Anf, Brachyury, Flk1, Mef2c, β-Mhc, and Mlv2v) and the development of beating cardiomyocytes in culture (Fig. 8E; see Video S9 in the supplemental material). Furthermore, analysis of teratomas formed by the complemented JNK-deficient ES cells demonstrated the restoration of cardiomyocyte development in vivo (Fig. 8F). Together, these data confirm that JNK is required for the development of cardiomyocytes and demonstrate that JNK is not required for ES cell self-renewal.
DISCUSSION
Studies of primary MEFs demonstrate that Jnk1−/− MEFs proliferate more slowly than wild-type MEFs and that Jnk2−/− MEFs proliferate more rapidly than wild-type MEFs (54). This observation has been interpreted to indicate that JNK1 and JNK2 may have opposite roles during proliferation (48). More recent studies, using a chemical genetic approach, demonstrate that these Jnk1−/− and Jnk2−/− phenotypes reflect adaptation and gain-of-function of the remaining JNK isoform (25). Thus, Jnk2−/− MEFs have increased JNK1 protein kinase activity compared to wild-type MEFs (25). These adaptive changes may account for the effects of JNK1 or JNK2 deficiency on ES cells. For example, Jnk1−/− ES cells formed colonies more slowly and Jnk2−/− ES cells formed colonies more rapidly than wild-type ES cells (Fig. 1D). Similarly, the increased luminal space within JNK1- or JNK2-deficient EBs may reflect adaptation by the remaining JNK isoforms. Together, these considerations illustrate the need for studies of compound mutants that completely lack JNK expression.
It has been established that compound mutant Jnk1−/− Jnk2−/− MEFs proliferate slowly (54). Subsequent studies using conditional gene ablation and chemical genetic analysis demonstrated that loss of JNK function resulted in rapid p53-dependent senescence of MEFs (12). These data suggest that JNK plays a critical role in cell proliferation. However, our analysis of JNK-deficient ES cells indicates that the role of JNK may be more complex that previously anticipated. We show that compound mutant Jnk1−/− Jnk2−/− ES cells proliferate more rapidly than wild-type ES cells when cultured as undifferentiated colonies on feeder layers of inactivated MEF (Fig. 1D). However, these JNK-deficient ES cells exhibit a marked proliferation defect when cultured as EBs (Fig. 2 and 3). This analysis demonstrates that there is no obligate role of JNK in the cell cycle, but JNK does influence cell cycle progression in a context-specific manner.
We speculate that these different roles of JNK in cell cycle regulation reflect the interaction of JNK with the p53 pathway. JNK deficiency does not cause growth inhibition in ES cells that exhibit rapid cell cycles that lack fully formed gap (G1/G2) phases (57). Moreover, it is established that the p53 pathway fails to mediate growth arrest in ES cells (2, 23), in part, because of cytoplasmic retention of p53 (52). In contrast, ES cells grown in EBs initiate a differentiation program that changes the cell cycle to include fully formed gap (G1/G2) phases (57). Similarly, the cell cycle of MEFs includes fully formed gap phases (57). The effect of JNK deficiency to cause p53-dependent growth inhibition therefore correlates with the presence of gap (G1/G2) phases in the cell cycle.
JNK and developmental apoptosis.
Cavity formation in EBs is a critical process that is required for differentiation and maturation (3, 40). The mechanism of cavity formation is mediated by programmed cell death of the inner ectodermal cells and survival of the outer ectodermal cells that form a columnar epithelium mediated by contact with the basement membrane (11). This mechanism of dual signaling to selectively target ectodermal cells for survival and death is an essential step in development. Previous studies have established that compound mutant embryos lacking Jnk1 and Jnk2 exhibited severe dysregulation of cell survival and apoptosis (32). It was therefore possible that JNK may contribute to cavitation. Indeed, Jnk1−/− Jnk2−/− ES cells formed EBs without cavitation (Fig. 2). This loss of cavitation was associated with reduced apoptosis and also with reduced proliferation (Fig. 3 and 4). These data demonstrate that JNK is required for a very early morphogenetic process that involves developmentally regulated apoptosis. It is likely that the loss of cavitation has profound consequences for differentiation. This is consistent with the finding that Jnk1−/− Jnk2−/− ES cells exhibit a reduced capacity to differentiate (Fig. 5 to 8).
Stem cell self-renewal and differentiation.
The observation that Jnk1−/− Jnk2−/− ES cells exhibit a profound defect in differentiation (Fig. 5 to 8) could reflect either a requirement of JNK for the maintenance of stem cell totipotency or a requirement of JNK for differentiation. We used genetic complementation assays to distinguish between these possibilities. We found that the ectopic expression of JNK in Jnk1−/− Jnk2−/− ES cells was fully capable of restoring the differentiation of these cells to a mesodermal lineage (Fig. 8). This finding demonstrates that the self-renewal potential of ES cells does not require JNK. However, JNK is required for specific differentiation programs. This conclusion is consistent with the finding that radiation chimeras prepared by transplantation of Jnk1−/− Jnk2−/− bone marrow into lethally irradiated recipient mice results in the reconstitution of these animals by Jnk1−/− Jnk2−/− hematopoietic stem cells (13).
Conclusions.
We report that JNK is not required for stem cell self-renewal. However, JNK does play a major role in the suppression of senescence, proliferation, apoptosis, and differentiation of ES cells that have committed to a specific cell lineage.
Supplementary Material
TABLE 2.
Quantitation of teratoma cell typea
| Tissue type | Mean % area |
|||||
|---|---|---|---|---|---|---|
| ++++ | +++ | ++ | + | +/- | − | |
| Cuboidal ciliated epithelium and goblet cells | >15 | >10 | >5 | >1 | >0.1 | 0 |
| Serous glandlike cells | >0.4 | >0.3 | >0.2 | >0.1 | >0.01 | 0 |
| Stratified columnar ciliated epithelium | >4 | >3 | >2 | >1 | >0.1 | 0 |
| Muscle | >20 | >15 | >10 | >5 | >0.1 | 0 |
| Cartilage | >0.5 | >0.1 | >0.05 | >0.01 | >0.005 | 0 |
| Fat | >0.5 | >0.1 | >0.05 | >0.01 | >0.005 | 0 |
| Keratinized stratified squamous epithelium | >0.5 | >0.1 | >0.05 | >0.01 | >0.005 | 0 |
| Neuroepithelium | >30 | >20 | >10 | >1 | >0.1 | 0 |
| Skinlike structures | >1 | >0.5 | >0.1 | >0.05 | >0.01 | 0 |
Sections of teratomas stained with H&E were examined to identify differentiated cell types (Fig. 6). The teratoma area for each differentiated cell type was quantitated using ImageJ software (n = 20). The mean percent area for each cell type was calculated and is represented on a scale from highest (“++++”) to lowest (“−”) as discussed in the text.
Acknowledgments
We thank Steve Jones for providing feeder cells; Jian-Hua Liu, Judith Reilly, and Tamera Barrett for expert technical assistance; and Kathy Gemme for administrative assistance.
These studies were supported by grants from the National Institutes of Health (CA065861 and NS054948). Core facilities used by these studies were supported by the NIDDK Diabetes and Endocrinology Center (P30 DK32520) at the University of Massachusetts. R.J.D. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 11 January 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akala, O. O., I. K. Park, D. Qian, M. Pihalja, M. W. Becker, and M. F. Clarke. 2008. Long-term hematopoietic reconstitution by Trp53−/− p16Ink4a−/− p19Arf−/− multipotent progenitors. Nature 453:228-232. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., B. T. Spike, L. W. Rodewald, T. J. Hope, M. Klemm, R. Jaenisch, and G. M. Wahl. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8:145-155. [DOI] [PubMed] [Google Scholar]
- 3.Andollo, N., M. D. Boyano, R. Andrade, M. M. Zalduendo, C. Eguizabal, A. Asumendi, J. Arlucea, and J. Arechaga. 2005. Structural and functional preservation of specific sequences of DNA and mRNA in apoptotic bodies from ES cells. Apoptosis 10:417-428. [DOI] [PubMed] [Google Scholar]
- 4.Beddington, R. S., and E. J. Robertson. 1999. Axis development and early asymmetry in mammals. Cell 96:195-209. [DOI] [PubMed] [Google Scholar]
- 5.Blumenfeld, M., M. Maury, T. Chouard, M. Yaniv, and H. Condamine. 1991. Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development 113:589-599. [DOI] [PubMed] [Google Scholar]
- 6.Boheler, K. R., J. Czyz, D. Tweedie, H. T. Yang, S. V. Anisimov, and A. M. Wobus. 2002. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 91:189-201. [DOI] [PubMed] [Google Scholar]
- 7.Cassia, R., L. Besnard, L. Fiette, A. Espinosa de los Monteros, P. Ave, M. C. Py, M. Huerre, J. de Vellis, M. M. Zakin, and F. Guillou. 1997. Transferrin is an early marker of hepatic differentiation, and its expression correlates with the postnatal development of oligodendrocytes in mice. J. Neurosci. Res. 50:421-432. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, S. M., C. A. Shaw, C. Gatza, C. J. Fisk, L. A. Donehower, and M. A. Goodell. 2007. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 5:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., S. W. Kubalak, S. Minamisawa, R. L. Price, K. D. Becker, R. Hickey, J. Ross, Jr., and K. R. Chien. 1998. Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem. 273:1252-1256. [DOI] [PubMed] [Google Scholar]
- 10.Cipelletti, B., G. Avanzini, L. Vitellaro-Zuccarello, S. Franceschetti, G. Sancini, T. Lavazza, D. Acampora, A. Simeone, R. Spreafico, and C. Frassoni. 2002. Morphological organization of somatosensory cortex in Otx1−/− mice. Neuroscience 115:657-667. [DOI] [PubMed] [Google Scholar]
- 11.Coucouvanis, E., and G. R. Martin. 1995. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83:279-287. [DOI] [PubMed] [Google Scholar]
- 12.Das, M., F. Jiang, H. K. Sluss, C. Zhang, K. M. Shokat, R. A. Flavell, and R. J. Davis. 2007. Suppression of p53-dependent senescence by the JNK signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 104:15759-15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, M., G. Sabio, F. Jiang, M. Rincon, R. A. Flavell, and R. J. Davis. 2009. Induction of hepatitis by JNK-mediated expression of TNF-alpha. Cell 136:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 15.Dong, C., D. D. Yang, C. Tournier, A. J. Whitmarsh, J. Xu, R. J. Davis, and R. A. Flavell. 2000. JNK is required for effector T-cell function but not for T-cell activation. Nature 405:91-94. [DOI] [PubMed] [Google Scholar]
- 16.Dong, C., D. D. Yang, M. Wysk, A. J. Whitmarsh, R. J. Davis, and R. A. Flavell. 1998. Defective T-cell differentiation in the absence of Jnk1. Science 282:2092-2095. [DOI] [PubMed] [Google Scholar]
- 17.Dufort, D., L. Schwartz, K. Harpal, and J. Rossant. 1998. The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development 125:3015-3025. [DOI] [PubMed] [Google Scholar]
- 18.Dumble, M., L. Moore, S. M. Chambers, H. Geiger, G. Van Zant, M. A. Goodell, and L. A. Donehower. 2007. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 109:1736-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edmondson, D. G., G. E. Lyons, J. F. Martin, and E. N. Olson. 1994. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120:1251-1263. [DOI] [PubMed] [Google Scholar]
- 20.Gangwani, L., R. A. Flavell, and R. J. Davis. 2005. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol. Cell. Biol. 25:2744-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Cao, I., M. Garcia-Cao, J. Martin-Caballero, L. M. Criado, P. Klatt, J. M. Flores, J. C. Weill, M. A. Blasco, and M. Serrano. 2002. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21:6225-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrmann, B. G., and A. Kispert. 1994. The T genes in embryogenesis. Trends Genet. 10:280-286. [DOI] [PubMed] [Google Scholar]
- 23.Hong, Y., and P. J. Stambrook. 2004. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 101:14443-14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iida, M., T. Heike, M. Yoshimoto, S. Baba, H. Doi, and T. Nakahata. 2005. Identification of cardiac stem cells with FLK1, CD31, and VE-cadherin expression during embryonic stem cell differentiation. FASEB J. 19:371-378. [DOI] [PubMed] [Google Scholar]
- 25.Jaeschke, A., M. Karasarides, J. J. Ventura, A. Ehrhardt, C. Zhang, R. A. Flavell, K. M. Shokat, and R. J. Davis. 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 23:899-911. [DOI] [PubMed] [Google Scholar]
- 26.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 27.Kanai-Azuma, M., Y. Kanai, J. M. Gad, Y. Tajima, C. Taya, M. Kurohmaru, Y. Sanai, H. Yonekawa, K. Yazaki, P. P. Tam, and Y. Hayashi. 2002. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129:2367-2379. [DOI] [PubMed] [Google Scholar]
- 28.Kawase, E., Y. Yamazaki, T. Yagi, R. Yanagimachi, and R. A. Pedersen. 2000. Mouse embryonic stem (ES) cell lines established from neuronal cell-derived cloned blastocysts. Genesis 28:156-163. [PubMed] [Google Scholar]
- 29.Kelley, C., H. Blumberg, L. I. Zon, and T. Evans. 1993. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development 118:817-827. [DOI] [PubMed] [Google Scholar]
- 30.Kemler, R., P. Brulet, M. T. Schnebelen, J. Gaillard, and F. Jacob. 1981. Reactivity of monoclonal antibodies against intermediate filament proteins during embryonic development. J. Embryol. Exp. Morphol. 64:45-60. [PubMed] [Google Scholar]
- 31.Kennedy, N. J., H. K. Sluss, S. N. Jones, D. Bar-Sagi, R. A. Flavell, and R. J. Davis. 2003. Suppression of Ras-stimulated transformation by the JNK signal transduction pathway. Genes Dev. 17:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuan, C. Y., D. D. Yang, D. R. Samanta Roy, R. J. Davis, P. Rakic, and R. A. Flavell. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22:667-676. [DOI] [PubMed] [Google Scholar]
- 33.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, J. A., J. J. Ventura, P. Hess, R. A. Flavell, and R. J. Davis. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 11:1479-1489. [DOI] [PubMed] [Google Scholar]
- 35.Lints, T. J., L. M. Parsons, L. Hartley, I. Lyons, and R. P. Harvey. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:419-431. [DOI] [PubMed] [Google Scholar]
- 36.Maier, B., W. Gluba, B. Bernier, T. Turner, K. Mohammad, T. Guise, A. Sutherland, M. Thorner, and H. Scrable. 2004. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 18:306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallamaci, A., S. Mercurio, L. Muzio, C. Cecchi, C. L. Pardini, P. Gruss, and E. Boncinelli. 2000. The lack of Emx2 causes impairment of Reelin signaling and defects of neuronal migration in the developing cerebral cortex. J. Neurosci. 20:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez, S. 2001. The isthmic organizer and brain regionalization. Int. J. Dev. Biol. 45:367-371. [PubMed] [Google Scholar]
- 39.Matheu, A., A. Maraver, P. Klatt, I. Flores, I. Garcia-Cao, C. Borras, J. M. Flores, J. Vina, M. A. Blasco, and M. Serrano. 2007. Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448:375-379. [DOI] [PubMed] [Google Scholar]
- 40.Maye, P., S. Becker, E. Kasameyer, N. Byrd, and L. Grabel. 2000. Indian hedgehog signaling in extraembryonic endoderm and ectoderm differentiation in ES embryoid bodies. Mech. Dev. 94:117-132. [DOI] [PubMed] [Google Scholar]
- 41.McGowan, K. M., and P. A. Coulombe. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143:469-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meletis, K., V. Wirta, S. M. Hede, M. Nister, J. Lundeberg, and J. Frisen. 2006. p53 suppresses the self-renewal of adult neural stem cells. Development 133:363-369. [DOI] [PubMed] [Google Scholar]
- 43.Mendrysa, S. M., K. A. O'Leary, M. K. McElwee, J. Michalowski, R. N. Eisenman, D. A. Powell, and M. E. Perry. 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 20:16-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzger, J. M., W. I. Lin, R. A. Johnston, M. V. Westfall, and L. C. Samuelson. 1995. Myosin heavy chain expression in contracting myocytes isolated during embryonic stem cell cardiogenesis. Circ. Res. 76:710-719. [DOI] [PubMed] [Google Scholar]
- 45.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 46.Mroz, K., L. Carrel, and P. A. Hunt. 1999. Germ cell development in the XXY mouse: evidence that X chromosome reactivation is independent of sexual differentiation. Dev. Biol. 207:229-238. [DOI] [PubMed] [Google Scholar]
- 47.Nagy, A. 2003. Manipulating the mouse embryo: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 48.Sabapathy, K., K. Hochedlinger, S. Y. Nam, A. Bauer, M. Karin, and E. F. Wagner. 2004. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15:713-725. [DOI] [PubMed] [Google Scholar]
- 49.Schreiber, M., A. Kolbus, F. Piu, A. Szabowski, U. Mohle-Steinlein, J. Tian, M. Karin, P. Angel, and E. F. Wagner. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharpless, N. E., and R. A. DePinho. 2007. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell. Biol. 8:703-713. [DOI] [PubMed] [Google Scholar]
- 51.Small, E. M., and P. A. Krieg. 2003. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev. Biol. 261:116-131. [DOI] [PubMed] [Google Scholar]
- 52.Solozobova, V., A. Rolletschek, and C. Blattner. 2009. Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol. 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tailleux, A., P. Duriez, J. C. Fruchart, and V. Clavey. 2002. Apolipoprotein A-II, HDL metabolism, and atherosclerosis. Atherosclerosis 164:1-13. [DOI] [PubMed] [Google Scholar]
- 54.Tournier, C., P. Hess, D. D. Yang, J. Xu, T. K. Turner, A. Nimnual, D. Bar-Sagi, S. N. Jones, R. A. Flavell, and R. J. Davis. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288:870-874. [DOI] [PubMed] [Google Scholar]
- 55.Tyner, S. D., S. Venkatachalam, J. Choi, S. Jones, N. Ghebranious, H. Igelmann, X. Lu, G. Soron, B. Cooper, C. Brayton, S. Hee Park, T. Thompson, G. Karsenty, A. Bradley, and L. A. Donehower. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415:45-53. [DOI] [PubMed] [Google Scholar]
- 56.Ventura, J. J., N. J. Kennedy, J. A. Lamb, R. A. Flavell, and R. J. Davis. 2003. c-Jun NH2-terminal kinase is essential for the regulation of AP-1 by tumor necrosis factor. Mol. Cell. Biol. 23:2871-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White, J., and S. Dalton. 2005. Cell cycle control of embryonic stem cells. Stem Cell Rev. 1:131-138. [DOI] [PubMed] [Google Scholar]
- 58.Xu, P., S. Hashimoto, H. Miyazaki, K. Asabe, S. Shiraishi, and K. Sueishi. 1998. Morphometric analysis of the immunohistochemical expression of Clara cell 10-kDa protein and surfactant apoproteins A and B in the developing bronchi and bronchioles of human fetuses and neonates. Virchows Arch. 432:17-25. [DOI] [PubMed] [Google Scholar]
- 59.Xu, P., K. Yoshioka, D. Yoshimura, Y. Tominaga, T. Nishioka, M. Ito, and Y. Nakabeppu. 2003. In vitro development of mouse embryonic stem cells lacking JNK/stress-activated protein kinase-associated protein 1 (JSAP1) scaffold protein revealed its requirement during early embryonic neurogenesis. J. Biol. Chem. 278:48422-48433. [DOI] [PubMed] [Google Scholar]
- 60.Yang, D. D., D. Conze, A. J. Whitmarsh, T. Barrett, R. J. Davis, M. Rincon, and R. A. Flavell. 1998. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity 9:575-585. [DOI] [PubMed] [Google Scholar]
- 61.Yuasa, S., Y. Itabashi, U. Koshimizu, T. Tanaka, K. Sugimura, M. Kinoshita, F. Hattori, S. Fukami, T. Shimazaki, S. Ogawa, H. Okano, and K. Fukuda. 2005. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat. Biotechnol. 23:607-611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.