Most cancer cells are believed to convert glucose avidly to lactate, even when oxygen is adequate for oxidative phosphorylation (OXPHOS). This phenomenon is known as the Warburg effect or aerobic glycolysis, which contrasts with anaerobic glycolysis, which is triggered in hypoxic normal cells or cancer cells adapting to hypoxia (6, 9, 11, 13, 20). In this issue, Fogal et al. report that knockdown of p32, a receptor for a tumor-specific homing peptide, in breast cancer cell lines enhanced glycolysis but, surprisingly, impaired tumorigenesis (7). Their observations unlink glycolysis from tumorigenic potential and question the paradigm stipulated by the Warburg effect.
In recent years, the interest in cancer cell metabolism has reemerged, with the Warburg effect as the flagship leading the current conceptual framework for a better understanding of the molecular basis of altered cancer cell metabolism. The central tenet posed by the Warburg effect is that cancer cells have defunct mitochondria and hence rely on glycolysis, which provides a hypothetical metabolic advantage to cancer cells that exist in a pervasively hypoxic microenvironment (3, 21). The tumor microenvironment is characterized by poor oxygen delivery from the host to cancer cells through the abnormal, often leaky and ineffective tumor neovasculature. Because hypoxia in vivo can induce hypoxia-inducible factor 1 (HIF-1), which promotes a transcriptional program for glycolysis with increased expression of glucose transporters and glycolytic enzyme genes, hypoxic cancer cells can undergo anaerobic glycolysis as an adaptive mechanism that is not the Warburg effect, per se (3).
Activation of oncogenes, such as the MYC, Ras, Akt, and phosphatidylinositol 3-kinase (PI3K) genes, or loss of tumor suppressors, such as p53 and VHL, triggers transcriptional or posttranscriptional alterations that rewire cells to acquire a cell-autonomous phenotype of enhanced glycolysis or the Warburg effect (11). In the case of MYC, p53, and VHL (which mediates its activity through HIF), deregulation of their genes results in transcriptional reprogramming, which increases glycolytic enzyme gene expression and expression of genes that attenuate mitochondrial respiration. Ras, Akt, and PI3K mediate the Warburg effect posttranscriptionally through phosphorylation of targets that enhance glycolysis or inhibit OXPHOS. The connections of oncogenes and tumor suppressors with metabolism are believed to provide the molecular basis for the Warburg effect.
Although the Warburg effect stipulates that mitochondrial respiration is decreased in cancer cells, continued mitochondrial activity is essential for cancer cell survival because, in addition to respiration, the mitochondrion also participates in many essential and fundamental metabolic pathways including the synthesis of pyrimidines through dihydroorotate dehydrogenase (DHODH). DHODH is a mitochondrial enzyme that is involved in the synthesis of uridine, and it requires direct coupling to the mitochondrial electron transport chain to catalyze the synthesis of orotic acid, a pyrimidine precursor. Furthermore, the mitochondrion is essential for providing the carbon backbone from glucose for lipid synthesis. The mitochondrion also serves as a hub for heme synthesis and fatty acid oxidation. It is not surprising, therefore, that this organelle also serves as a metabolic sensing station that can trigger cell death through the release of cytochrome c to initiate the intrinsic apoptotic pathway when intracellular growth signaling outstrips energy availability.
The role of the mitochondrion in cancer cell metabolism has been marginalized in the literature by the central tenet posed by the Warburg effect; however, its essential role has been documented by studies dating back to several decades ago. In addition to glucose, glutamine was documented to be essential for mammalian cell proliferation in vitro, particularly in a number of cancer cell lines. In fact, HeLa cells were shown to use glutamine as a major energy source, compared with glucose (17). In addition, studies of hepatomas documented that increased glutamine oxidation correlated with the degree of tumor growth in vivo, suggesting that glutamine oxidation and mitochondria play an essential role in liver cancer (12, 14, 15). These observations suggest that this nonessential amino acid is, in fact, facultatively essential depending on the tissue cell type; cells that do not express glutamine synthetase depend on extracellular glutamine (5).
Glutamine is transported by permeases into cells and is subsequently used as an amino acid for protein synthesis, as a precursor of glutathione, or as a nitrogen donor for nucleotide biosynthesis (5). In addition, glutamine is transported into the mitochondrion and converted by glutaminase to glutamate, which is a precursor of a pivotal tricarboxylic acid (TCA) cycle intermediate, α-ketoglutarate. In the presence of oxygen, α-ketoglutarate is catabolized in the TCA cycle to succinate, fumarate, and malate. Malate can exit the TCA cycle and the mitochondrion and is then converted to pyruvate by the malic enzyme. Pyruvate can then be converted to lactate or can reenter the TCA cycle as acetyl-coenzyme A (CoA). This pathway provides a route for the metabolism of glutamine to lactate, with TCA intermediates providing biosynthetic carbons for the production of macromolecules. As with glucose, the consumption of glutamine via the TCA cycle produces high-energy electrons that are processed through the electron transport chain for the production of ATP. Thus, glutamine is also a major cancer cell energy and anabolic substrate that requires functional mitochondria for its catabolism.
Fogal et al. report studies that further add to the positive role of the mitochondrion in tumorigenesis, such that knocking down p32 in breast cancer cell lines MDA-MB-435 and MDA-MB-231 resulted in diminished mitochondrial respiration and tumor xenograft growth (7). p32 was identified as a cell surface receptor for a tumor-homing peptide. This receptor, previously identified as HABP1, associates with the RNA splicing factor SF-2, which is also known to participate in tumorigenesis (10). p32 or HABP1 is identical to the complement C1q binding protein, and hence the official gene symbol is C1QBP. Although the exact molecular function(s) of p32 is not definitively delineated, Fogal et al. document that loss of p32 function was associated with diminished mitochondrial protein translation that affects a number of electron transport complexes which contain mitochondrially encoded components (7). Among those affected are mitochondrial complexes III, IV, and V, but not II. These alterations due to p32 knockdown were associated with decreased cellular oxygen consumption, increased glucose consumption, and lactate production. Furthermore, metabolic tracer studies using [13C]glucose indicate that diminished p32 levels caused enhanced glycolytic and pentose phosphate pathway fluxes and decreased TCA cycling. In essence, decreased p32 levels enhanced the Warburg effect in cancer cells.
If the Warburg effect is favored by cancer cells, then knockdown of p32 is expected to increase tumorigenesis, but this was in fact not the case. Knockdown of p32 in both MDA-MB-453 and MDA-MB-231 breast cancer cell lines diminished their xenograft tumor growth rates. Intriguingly, knockdown of p32 levels was associated with a persistently elevated pyruvate kinase M2 (PKM2) level, which has been associated with the Warburg effect and tumorigenicity (1, 2). In the case of p32 knockdown, which enhances the Warburg effect, persistent PKM2 expression is not sufficient to retain tumorigenicity of the breast cancer cell lines. With p32 knockdown, loss of mitochondrial function decreased both cellular respiration and tumorigenicity, suggesting that mitochondrial function is essential for tumor formation. Because mitochondrial function could not be rescued independent of p32, a conservative interpretation of the data presented by Fogal et al. is that p32 itself is necessary for full tumorigenic potential (7). Because p32 appears to have pleiotropic effects, it is possible that other functions associated with p32 are also essential for tumorigenesis, such as its association with and inhibition of the splicing factor SF-2 (SFRS1) (16). Hence, it remains possible that diminished mitochondrial respiration associated with decreased p32 may not responsible for decreased tumorigenicity. The evidence, however, suggests otherwise because of the prominent roles of glutamine, which requires mitochondrial function, and the mitochondria themselves in cancer cell metabolism. Furthermore, it is also notable that cancer cell heterogeneity within a tumor consists of glycolytic cells that produce lactate and oxidative cells that could recycle lactate as pyruvate for mitochondrial oxidation (18, 19).
Observations accumulated from a number of different laboratories now point to an interplay between the metabolism of glucose and glutamine and the recycling of lactate, which require oxidative phosphorylation (2, 18). It is intriguing that Myc could stimulate both glycolysis and glutamine metabolism, in part through the repression of miR-23a and miR-23b, and induce the expression of p32 (4, 7, 8, 22). These links suggest that p32 could play an integral role in the regulation of glutamine metabolism by Myc, and hence future studies to delineate p32's potential role in glutamine oxidation will be of great general interest (Fig. 1). The richer understanding of the complexity of the tumor tissue suggests, as do Fogal et al., that oxidative phosphorylation plays a pivotal role in tumorigenesis. Hence, is the Warburg effect a lot of hot air? Or is air available to cancer cells immediately surrounding blood vessels required to burn lactate and glutamine for the progression of tumors, which also contain cancer cells located away from blood vessels in hypoxic regions in which glycolysis dominates and where the Warburg effect provides a tumor cell survival advantage?
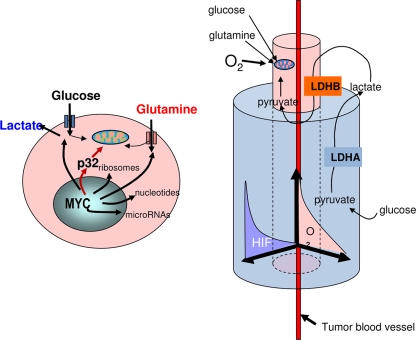
FIG. 1.
Schematic of a cancer cell with adequate oxygen (left) depicts the activation of p32 by Myc as well as Myc target genes involved in ribosomal biogenesis and glucose, glutamine, and nucleotide metabolism. p32 is shown to contribute to mitochondrial oxidative phosphorylation. Cancer cells undergoing oxidative phosphorylation around the tumor blood vessel (right) via the oxidation of glucose and glutamine proliferate and migrate away from the tumor blood vessel into hypoxic regions. Within the hypoxic tumor microenvironment, where hypoxia-inducible factors (HIF) are activated, the Warburg effect may permit survival because glycolysis proceeds independently of oxygen. Lactate produced by hypoxic cells is shown to be recycled for oxidation by cells that undergo OXPHOS.
Acknowledgments
This work is supported by the Leukemia & Lymphoma Society, NIH, NCI, and the AACR-SU2C initiative.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Christofk, H. R., M. G. Vander Heiden, M. H. Harris, A. Ramanathan, R. E. Gerszten, R. Wei, M. D. Fleming, S. L. Schreiber, and L. C. Cantley. 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452:230-233. [DOI] [PubMed] [Google Scholar]
- 2.Dang, C. V. 2009. PKM2 tyrosine phosphorylation and glutamine metabolism signal a different view of the Warburg effect. Sci. Signal. 2:pe75. [DOI] [PubMed] [Google Scholar]
- 3.Dang, C. V., J. W. Kim, P. Gao, and J. Yustein. 2008. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 8:51-56. [DOI] [PubMed] [Google Scholar]
- 4.Dang, C. V., A. Le, and P. Gao. 2009. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin. Cancer Res. 15:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deberardinis, R. J., and T. Cheng. 2 November 2009. Q.'s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene [Epub ahead of print.] doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed]
- 6.Deberardinis, R. J., N. Sayed, D. Ditsworth, and C. B. Thompson. 2008. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18:54-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogal, V., A. Richardson, P. P. Karmali, I. E. Scheffler, J. W. Smith, and E. Ruoslahti. 2010. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol. Cell. Biol. 30:1303-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, P., I. Tchernyshyov, T. C. Chang, Y. S. Lee, K. Kita, T. Ochi, K. I. Zeller, A. M. De Marzo, J. E. Van Eyk, J. T. Mendell, and C. V. Dang. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458:762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu, P. P., and D. M. Sabatini. 2008. Cancer cell metabolism: Warburg and beyond. Cell 134:703-707. [DOI] [PubMed] [Google Scholar]
- 10.Karni, R., E. de Stanchina, S. W. Lowe, R. Sinha, D. Mu, and A. R. Krainer. 2007. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J. W., and C. V. Dang. 2006. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 66:8927-8930. [DOI] [PubMed] [Google Scholar]
- 12.Kovacevic, Z., and H. P. Morris. 1972. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 32:326-333. [PubMed] [Google Scholar]
- 13.Kroemer, G., and J. Pouyssegur. 2008. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13:472-482. [DOI] [PubMed] [Google Scholar]
- 14.Linder-Horowitz, M., W. E. Knox, and H. P. Morris. 1969. Glutaminase activities and growth rates of rat hepatomas. Cancer Res. 29:1195-1199. [PubMed] [Google Scholar]
- 15.Matsuno, T., and I. Goto. 1992. Glutaminase and glutamine synthetase activities in human cirrhotic liver and hepatocellular carcinoma. Cancer Res. 52:1192-1194. [PubMed] [Google Scholar]
- 16.Petersen-Mahrt, S. K., C. Estmer, C. Ohrmalm, D. A. Matthews, W. C. Russell, and G. Akusjarvi. 1999. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reitzer, L. J., B. M. Wice, and D. Kennell. 1979. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 254:2669-2676. [PubMed] [Google Scholar]
- 18.Semenza, G. L. 2008. Tumor metabolism: cancer cells give and take lactate. J. Clin. Invest. 118:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonveaux, P., F. Vegran, T. Schroeder, M. C. Wergin, J. Verrax, Z. N. Rabbani, C. J. De Saedeleer, K. M. Kennedy, C. Diepart, B. F. Jordan, M. J. Kelley, B. Gallez, M. L. Wahl, O. Feron, and M. W. Dewhirst. 2008. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest. 118:3930-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden, M. G., L. C. Cantley, and C. B. Thompson. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warburg, O. 1930. The metabolism of tumours, English ed. Constable & Co. Ltd., London, United Kingdom.
- 22.Wise, D. R., R. J. DeBerardinis, A. Mancuso, N. Sayed, X. Y. Zhang, H. K. Pfeiffer, I. Nissim, E. Daikhin, M. Yudkoff, S. B. McMahon, and C. B. Thompson. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U. S. A. 105:18782-18787. [DOI] [PMC free article] [PubMed] [Google Scholar]