Abstract
Xanthomonas campestris pv. campestris, the causal agent of black rot disease of brassicas, is known for its ability to catabolize a wide range of plant compounds. This ability is correlated with the presence of specific carbohydrate utilization loci containing TonB-dependent transporters (CUT loci) devoted to scavenging specific carbohydrates. In this study, we demonstrate that there is an X. campestris pv. campestris CUT system involved in the import and catabolism of N-acetylglucosamine (GlcNAc). Expression of genes belonging to this GlcNAc CUT system is under the control of GlcNAc via the LacI family NagR and GntR family NagQ regulators. Analysis of the NagR and NagQ regulons confirmed that GlcNAc utilization involves NagA and NagB-II enzymes responsible for the conversion of GlcNAc-6-phosphate to fructose-6-phosphate. Mutants with mutations in the corresponding genes are sensitive to GlcNAc, as previously reported for Escherichia coli. This GlcNAc sensitivity and analysis of the NagQ and NagR regulons were used to dissect the X. campestris pv. campestris GlcNAc utilization pathway. This analysis revealed specific features, including the fact that uptake of GlcNAc through the inner membrane occurs via a major facilitator superfamily transporter and the fact that this amino sugar is phosphorylated by two proteins belonging to the glucokinase family, NagK-IIA and NagK-IIB. However, NagK-IIA seems to play a more important role in GlcNAc utilization than NagK-IIB under our experimental conditions. The X. campestris pv. campestris GlcNAc NagR regulon includes four genes encoding TonB-dependent active transporters (TBDTs). However, the results of transport experiments suggest that GlcNAc passively diffuses through the bacterial envelope, an observation that calls into question whether GlcNAc is a natural substrate for these TBDTs and consequently is the source of GlcNAc for this nonchitinolytic plant-associated bacterium.
Xanthomonas campestris pv. campestris, the causal agent of black rot disease of brassicas, produces extracellular plant cell wall-degrading enzymes which contribute to its pathogenicity by facilitating its spread through plant tissues and give the bacterium access to a ready source of nutrients via the carbohydrate utilization loci containing TonB-dependent transporters (CUT loci) (7, 16, 35). The CUT loci are characterized by the presence of genes encoding regulators, degradative enzymes, inner membrane transporters, and outer membrane TonB-dependent transporters (TBDTs), which have been identified as active carbohydrate transporters (7, 33, 44). However, recently, an example of passive diffusion through a TBDT in Caulobacter crescentus was described (17). X. campestris pv. campestris has 72 TBDTs and belongs to a class of bacteria in which TBDTs are overrepresented (7). Our previous study suggested that there are several CUT loci or systems in this bacterium (7).
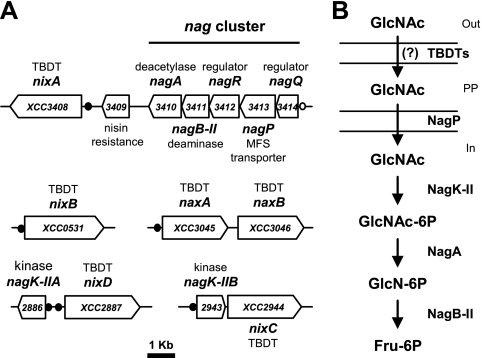
N-Acetylglucosamine (GlcNAc) is an amino sugar that is used for the synthesis of cell surface structures in bacteria and plays an important role in supplying carbon and energy by entering the glycolytic pathway after it is converted into fructose-6-phosphate (fructose-6P) (1, 9). In a recent comparative study of bacterial GlcNAc utilization pathways and regulatory networks, Yang and coworkers identified conserved and distinct features of the GlcNAc utilization pathway in proteobacteria (48). The expression of X. campestris pv. campestris GlcNAc-specific genes was proposed to be controlled by NagR and NagQ regulators belonging to the LacI and GntR families, respectively. In X. campestris pv. campestris strain ATCC 33913, one predicted binding motif specific for NagQ (designated the NagQ box) consists of two imperfect repeats of the TGGTATT sequence separated by 4 bp and is located upstream of the nagQ gene (XCC3414) (Fig. 1A) (48). This gene is part of the nag cluster and is followed by genes encoding the major facilitator superfamily (MFS) inner membrane transporter NagP (XCC3413), the regulator NagR (XCC3412), the GlcN-6P deaminase NagB-II (XCC3411), and the GlcNAc-6P deacetylase NagA (XCC3410) (Fig. 1A). NagR boxes contain the palindromic sequence AATGACARCGYTGTCATT (bold type indicates less highly conserved nucleotides) and are upstream of genes encoding two glucokinase-like NagK-II proteins (XCC2886 [nagK-IIA] and XCC2943 [nagK-IIB]), as well as 5 genes encoding TBDTs (XCC0531, XCC2887, XCC3045, XCC3408, and XCC2944 located downstream of XCC2943) (Fig. 1A). All of the X. campestris pv. campestris genes located downstream of NagR or NagQ boxes were proposed to belong to a GlcNAc utilization pathway involved in uptake of GlcNAc through the bacterial envelope and subsequent phosphorylation, deacetylation, and deamination, which finally leads to the common metabolic intermediate fructose-6-phosphate (Fig. 1B) (48). It was recently demonstrated that in C. crescentus the TBDT CC0446 gene, which is clustered with other nag genes, is responsible for the uptake of GlcNAc (17). The presence of TBDTs in the GlcNAc regulon, which has been observed in Alteromonadales and Xanthomonadales (48), suggests that genes belonging to the GlcNAc utilization pathway define a new CUT system.
FIG. 1.
X. campestris pv. campestris N-acetylglucosamine (GlcNAc) utilization pathway. (A) Organization of genes in the proposed GlcNAc utilization pathway. NagR boxes are indicated by filled circles, and the NagQ box is indicated by an open circle. (B) GlcNAc is proposed to be transported through the outer membrane by TBDTs and then transported across the inner membrane by the MFS transporter NagP. GlcNAc would then be phosphorylated by nagK-II-encoded enzymes. Subsequent metabolism via the nagA-encoded (GlcNAc-6P deacetylase) and nagB-II-encoded (GlcN-6P deaminase) enzymes results in fructose 6-phosphate (Fru-6P) (48). MFS, major facilitator superfamily; PP, periplasm; TBDT, TonB-dependent transporter.
Here we describe characterization of the X. campestris pv. campestris GlcNAc utilization pathway and regulatory network, which involves at least the repressors NagR and NagQ. TBDTs are associated with this pathway, confirming the presence of a GlcNAc CUT system in X. campestris pv. campestris. In this bacterium, GlcNAc entry and catabolism imply that novel families containing a GlcNAc inner membrane transporter and GlcNAc kinases are involved.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The X. campestris pv. campestris strains and plasmids used in this study are listed in Table 1. X. campestris pv. campestris cells were grown at 30°C in MOKA (7) or KADO (4) rich medium or in minimal medium (MME) (3). Sodium-free minimal medium contained 10.5 g/liter K2HPO4 and 4.5 g/liter KH2PO4. Escherichia coli cells were grown on Luria-Bertani medium at 37°C. For solid media, agar was added at a final concentration of 1.5% (wt/vol).
TABLE 1.
Plasmids and X. campestris pv. campestris strains used or generated in this study
| Strain or plasmid | Characteristicsa | Locationb | Designation | Reference |
|---|---|---|---|---|
| Xanthomonas strains | ||||
| Wild type | Wild-type strain; rifampin-resistant derivative of X. campestris pv. campestris LMG568 (= ATCC 33913) | 31 | ||
| nixB::pVO | XCC0531::pVO155; Rifr Kmr | 736 | XP010 | 7 |
| nixC::pVO | XCC2944::pVO155; Rifr Kmr | 334 | XP041 | 7 |
| nixD::pVO | XCC2887::pVO155; Rifr Kmr | 1558 | XP040 | 7 |
| naxA::pVO | XCC3045::pVO155; Rifr Kmr | 1572 | XP044 | 7 |
| naxB::pVO | XCC3046::pVO155; Rifr Kmr | 702 | XP045 | 7 |
| nixA::pVO | XCC3408::pVO155; Rifr Kmr | 1897 | XP059 | 7 |
| nagQ::pVO | XCC3414::pVO155; Rifr Kmr | 659 | XP108 | This study |
| nagR::pVO | XCC3412::pVO155; Rifr Kmr | 519 | XP109 | This study |
| nagA::pVO | XCC3410::pVO155; Rifr Kmr | 236 | XP110 | This study |
| nagK-IIA::pVO | XCC2886::pVO155; Rifr Kmr | 303 | XP111 | This study |
| ΔnagQ | ΔXCC3414; Rifr | From 203 to stop | XP112 | This study |
| ΔnagR | ΔXCC3412; Rifr | From start to stop | XP113 | This study |
| ΔnagP | ΔXCC3413; Rifr | From start to stop | XP114 | This study |
| ΔnagA | ΔXCC3410; Rifr | From start to stop | XP115 | This study |
| ΔnagB-II | ΔXCC3411; Rifr | From start to stop | XP116 | This study |
| ΔnagK-IIA | ΔXCC2886; Rifr | From start to stop | XP117 | This study |
| ΔnagK-IIB | ΔXCC2943; Rifr | From start to stop | XP118 | This study |
| ΔnagK-IIAB | ΔXCC2886 ΔXCC2943; Rifr | From start to stop | XP119 | This study |
| ΔnagA pC-nagA | ΔXCC3410 pC-XCC3410; Rifr Tetr | XP120 | This study | |
| ΔnagB-II pC-nagB-II | ΔXCC3411 pC-XCC3411; Rifr Tetr | XP121 | This study | |
| ΔnagB-II pC-nagB-II-nagA | ΔXCC3411 pC-XCC3411-XCC3410; Rifr Tetr | XP122 | This study | |
| ΔnagK-IIA pC-nagK-IIA | ΔXCC2886 pC-XCC2886; Rifr Tetr | XP123 | This study | |
| ΔnagK-IIB pC-nagK-IIB | ΔXCC2943 pC-XCC2943; Rifr Tetr | XP124 | This study | |
| ΔnagK-IIAB pC-nagK-IIA | ΔXCC2886 ΔXCC2943 pC-XCC2886; Rifr Tetr | XP125 | This study | |
| ΔnagK-IIAB pC-nagK-IIB | ΔXCC2886 ΔXCC2943 pC-XCC2943; Rifr Tetr | XP126 | This study | |
| ΔnagR nagK-IIA::pVO | ΔXCC3412 XCC2886::pVO155; Rifr Kmr | XP127 | This study | |
| ΔnagQ nagK-IIA::pVO | ΔXCC3414 XCC2886::pVO155; Rifr Kmr | XP128 | This study | |
| ΔnagP nagA::pVO | ΔXCC3413 XCC3410::pVO155; Rifr Kmr | XP129 | This study | |
| ΔnagK-IIAB nagA::pVO | ΔXCC2886 ΔXCC2943 XCC3410::pVO155; Rifr Kmr | XP130 | This study | |
| ΔnagP ΔnagA | ΔXCC3413 ΔXCC3410; Rifr | XP131 | This study | |
| ΔnagK-IIAB ΔnagA | ΔXCC2886 ΔXCC2943 ΔXCC3410; Rifr | XP132 | This study | |
| Plasmids | ||||
| pVO155 | pUC119 derivative containing the promoterless gus (uidA) reporter gene encoding β-glucuronidase, used for insertion mutagenesis; Kmr Ampr | 34 | ||
| pFAJ1700 | pTR102-derived expression vector containing a multiple-cloning site and transcriptional terminators in both orientations; Tetr Ampr | 15 | ||
| pSC150 | pET-26b(+) derivative vector with a Ptac promoter sequence; Kmr | 13 | ||
| pCZ917 | pFAJ1700 derivative containing 2,094 bp of pSC150 with lacI, Ptac promoter, and T7 terminator; Tetr Ampr | This study | ||
| pC-nagA | pCZ917-XCC3410; Tetr Kmr | From −20 to stop | This study | |
| pC-nagB-II | pCZ917-XCC3411; Tetr Kmr | From −19 to stop | This study | |
| pC-nagB-II-nagA | pCZ917-XCC3411-XCC3410; Tetr Kmr | From −19 of XCC3411 to stop of XCC3410 | This study | |
| pC-nagK-IIA | pCZ917-XCC2886; Tetr Kmr | From −21 to stop | This study | |
| pC-nagK-IIB | pCZ917-XCC2943; Tetr Kmr | From −19 to stop | This study |
Rif: rifampin; Km: kanamycin; Tet: tetracycline.
Position of insertion, deletion, or X. campestris pv. campestris sequence cloned relative to the putative start codon.
Antibiotics were used at the following concentrations: for X. campestris pv. campestris, 50 μg/ml rifampin, 50 μg/ml kanamycin, and 5 μg/ml tetracycline; for E. coli, 50 μg/ml ampicillin, 50 μg/ml kanamycin, and 10 μg/ml tetracycline.
Mutagenesis of X. campestris pv. campestris.
X. campestris pv. campestris insertion mutants were constructed using the suicide plasmid pVO155 (34) with a 300- to 500-bp PCR amplicon internal to each open reading frame (ORF) (Table 1). Deletion mutants were constructed by using the cre-lox system adapted by Angot et al. (2) from the system of Marx and colleagues (30) or by using the sacB system (43). Deleted regions are indicated in Table 1. Oligonucleotide primers used for PCR amplification will be provided upon request.
Plasmids were introduced into E. coli by electroporation and into Xanthomonas strains by triparental conjugation, as described by Turner et al. (45).
Plasmid constructs.
DNA manipulations were performed as described previously (42). For complementation studies, PCR amplicons (oligonucleotide primers used for PCR amplification will be provided upon request) were cloned into pCZ917, a derivative of pFAJ1700 (15) containing a 2,094-bp fragment of pSC150 (13) with the lacI gene, Ptac promoter, and T7 terminator.
Expression studies.
Bacterial cultures grown in the appropriate medium were harvested after 6 h of incubation for β-glucuronidase assays (25).
The methods used for quantitative reverse transcription-PCR (qRT-PCR) experiments were adapted from the methods of Blanvillain et al. (7). A 2-μg sample of RNA was treated with RNase-free DNase I (Amersham) for 30 min at 37°C. After DNase inactivation (10 min at 75°C), RNAs were reverse transcribed with Superscript II (Invitrogen) using random hexamers (Biolabs) for 10 min at room temperature and then for 1 h at 42°C. Oligonucleotide primers used for quantitative PCR amplification will be provided upon request. 16S rRNA was used as a control for real-time PCR (7, 32).
Growth curves.
Growth curves were generated using a FLUOStar Omega apparatus (BMG Labtech, Offenburg, Germany) with four replicates. Growth was measured using 96-well flat-bottom microtiter plates with 200-μl preparations inoculated at an optical density at 600 nm (OD600) of 0.1 from 4 independent washed overnight precultures. The microplates were shaken continuously at 700 rpm using the double-orbital-shaking mode.
[14C]GlcNAc transport experiments.
Transport experiments with radiolabeled GlcNAc (specific activity, 2.04 GBq/mmol; PerkinElmer) were performed as previously described (7). For competition experiments, unlabeled sugars were added to [14C]GlcNAc at final concentrations of 50 and 500 μM, and cells were incubated for 1 h before collection. The initial concentration-dependent GlcNAc transport was determined using the rapid dilution method as previously described (7, 33).
GlcNAc phosphorylation assays.
GlcNAc kinase activity assays were performed using an enzyme-linked assay based on the NAD+/NADH ratio (19). Fifty milliliters of an overnight culture in minimal medium supplemented with 10 mM GlcNAc was centrifuged and resuspended in 2 ml of resuspension buffer (0.05 mM Tris HCl [pH 8], 13.3 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol). Cells were disrupted with a French press and centrifuged, and 100 μl of supernatant was added to 900 μl of reaction buffer (0.1 M Tris HCl [pH 7.5], 10 mM MgCl2, 1 mM phosphoenolpyruvate, 4 mM ATP, 0.2 mM NADH, 10 mM GlcNAc, 4 U lactate dehydrogenase [Sigma], 4 U pyruvate kinase [Sigma]) prewarmed for 5 min at 37°C. The OD340 was determined every 10 s for 5 min at 37°C. A decrease in the OD340 corresponded to production of NAD+ from NADH and was enzymatically coupled to GlcNAc phosphorylation to form GlcNAc-6P. Protein concentrations of cell lysates were determined using the Bradford assay (Bio-Rad).
RESULTS
GlcNAc and chitobiose, but not chitin, are carbon and nitrogen sources for X. campestris pv. campestris.
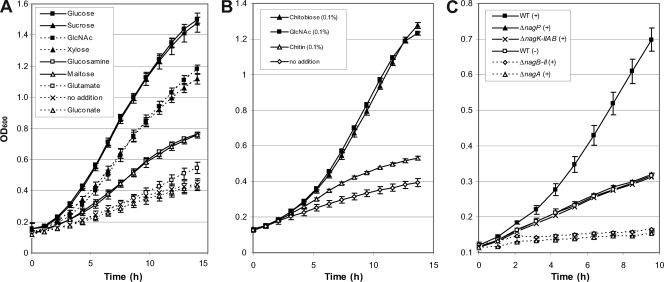
The presence in X. campestris pv. campestris of genes proposed to belong to a GlcNAc utilization pathway suggests that GlcNAc can be metabolized by X. campestris pv. campestris. Therefore, the growth rates of X. campestris pv. campestris cultures in MME supplemented with GlcNAc and with other carbon sources were compared. After sucrose and glucose, GlcNAc and the GlcNAc dimer chitobiose were among the best carbon sources for X. campestris pv. campestris (Fig. 2A and B). In the presence of the GlcNAc homopolymer chitin, slight growth was reproducibly observed (Fig. 2B), probably due to the presence of small amounts of free GlcNAc or chitobiose molecules. This result suggests that X. campestris pv. campestris is not able to efficiently degrade chitin, a suggestion corroborated by the absence of any obvious chitinase-encoding gene in the genome of X. campestris pv. campestris strain ATCC 33913 (14, 48).
FIG. 2.
Growth of X. campestris pv. campestris wild-type and mutant strains in the presence of N-acetylglucosamine (GlcNAc) or other carbon sources. (A) Growth of the wild-type strain in the presence of carbon sources and in the presence of carbon and nitrogen sources. (B) Growth of the wild-type strain in the presence of GlcNAc, chitobiose, or chitin. (C) Growth of the wild type (WT) and mutant strains in the presence (+) or in the absence (−) of GlcNAc. After overnight growth in complete medium, cells were harvested, washed, and resuspended in minimal medium. Carbohydrates were added at a final concentration of 10 mM (A and C) or 0.1% (B). The error bars indicate the standard deviations obtained from 4 independent experiments.
GlcNAc is also a nitrogen source for X. campestris pv. campestris, since this bacterium grows in nitrogen-depleted MME (MME without Casamino Acids and NH4SO4 [see Materials and Methods]) in the presence of GlcNAc, whereas no growth was observed in the presence of glucose (data not shown).
GlcNAc pathway genes are induced by GlcNAc.
The expression of genes located downstream of putative NagR or NagQ boxes was measured to assess the relationship of these genes to utilization of GlcNAc. The TBDT gene XCC3046 located downstream of the TBDT gene XCC3045 might belong to the same operon (Fig. 1B) and was therefore included in this analysis. None of the primer pairs designed for qRT-PCR experiments yielded reliable results for the XCC2886 and XCC3046 genes. Therefore, transcriptional fusions with the promoterless uidA gene were constructed by pVO155 insertion mutagenesis (34). The resulting mutants were used to measure the expression of these genes with a β-glucuronidase assay. All of the ORFs in the proposed GlcNAc CUT system, including the nagQ and nagR putative regulatory genes, were clearly induced in the presence of GlcNAc (Table 2). However, XCC3045 and XCC3046 were repressed. Based on these results, the TBDT genes XCC3408, XCC0531, XCC2944, and XCC2887 were designated nixA, nixB, nixC, and nixD, respectively (N-acetylglucosamine-induced genes in Xanthomonas), while the TBDT genes XCC3045 and XCC3046 were designated naxA and naxB, respectively (N-acetylglucosamine-associated genes in Xanthomonas).
TABLE 2.
Relative expression ratios for genes in the N-acetylglucosamine utilization pathway
| Gene | Designation | Function | Expression ratios (SD) |
||
|---|---|---|---|---|---|
| Wild type with GlcNAc/wild type in MME | ΔnagQ mutant in MME/wild type in MME | ΔnagR mutant in MME/wild type in MME | |||
| XCC3414a | nagQ | GntR repressor | 7.11 (0.70)d | NDe | 0.47 (0.04) |
| XCC3413a | nagP | MFS transporter | 6.43 (0.62)d | 2.28 (0.90)d | 0.83 (0.10) |
| XCC3412a | nagR | LacI repressor | 6.99 (0.73)d | 7.91 (3.34)d | ND |
| XCC3411a | nagB-II | Deaminase | 6.72 (0.46)d | 3.17 (0.16)d | 0.88 (0.06) |
| XCC3410a | nagA | Deacetylase | 3.16 (0.16)d | 8.33 (2.76)d | 0.41 (0.28) |
| XCC2886b | nagK-IIA | GlcNAc kinase | 2.70 (0.12)d | 0.88 (0.02) | 4.88 (1.06)d |
| XCC2943a | nagK-IIB | GlcNAc kinase | 3.00 (0.81)d | 1.70 (0.73)c | 27.11 (7.00)c,d |
| XCC3408a | nixA | TBDT | 6.32 (0.84)d | 0.31 (0.08) | 7.77 (1.17)d |
| XCC0531a | nixB | TBDT | 3.97 (0.49)d | 0.63 (0.24) | 1.08 (0.47) |
| XCC2944a | nixC | TBDT | 9.21 (1.17)d | 0.42 (0.15) | 40.14 (14.35)d |
| XCC2887a | nixD | TBDT | 106.70 (12.56)d | 0.86 (0.55) | 47.82 (15.16)d |
| XCC3045a | naxA | TBDT | 0.42 (0.12)d | 0.61 (0.34) | 0.26 (0.08) |
| XCC3046b | naxB | TBDT | 0.25 (0.06)d | ND | ND |
Data from real-time quantitative reverse transcriptase PCR performed in at least three independent experiments. Calculation of the relative expression included normalization with the 16S rRNA data.
Data from β-glucuronidase assays performed in at least three independent experiments using pVO155 insertion mutations leading to transcriptional fusions with the promotorless uidA gene. Insertions were made in the wild-type strain and, for XCC2886, in the ΔnagR and ΔnagQ strains.
qRT-PCR expression values were obtained from nagR and nagQ pVO155 insertion mutants instead of deletion mutants.
The levels of expression in the conditions compared were significantly different (P < 0.05) as determined using a Student t test.
ND, not determined.
To determine whether expression of the other TBDT genes of X. campestris pv. campestris is affected by GlcNAc, β-glucuronidase assays were performed using pVO155 insertion mutants with mutations in each of the 72 TBDT genes (7). The nixA, nixB, nixC, and nixD genes were the only TBDT genes induced by GlcNAc (data not shown). It is worth noting that the expression of the nix TBDT genes was not as strongly induced in the pVO155 insertion mutant (the induction levels ranged from 1.8-fold for nixB to 36-fold for nixD [data not shown]) as was expected based on the results of qRT-PCR for a wild-type background (for which the induction levels ranged from 3.97-fold for nixB to 106.7-fold for nixD [Table 2]).
The expression of GlcNAc-induced genes was then measured after growth in MME supplemented with a range of GlcNAc concentrations. Representative results obtained with the nixD::pVO mutant are reported here because this mutant displayed one of the highest levels of induction in the presence of GlcNAc and because its growth was not impaired in MME supplemented with GlcNAc (see below). The reporter gene was induced with 5 μM to 20 mM GlcNAc. The maximal induction (around 30-fold) was observed with 50 μM GlcNAc (data not shown). Induction was also observed with high concentrations of glucosamine (GlcN), but the maximal induction was only 3-fold (data not shown).
NagQ and NagR are GlcNAc pathway-specific regulators.
The involvement of two presumptive regulators, NagQ and NagR, was evaluated by comparing nix gene expression in the wild-type strain and nix gene expression in nagQ and nagR mutants in MME without added GlcNAc. Mutants with insertions and deletions of these two regulatory genes were constructed, but deletion mutants were chosen to avoid possible polar effects, since both regulatory genes may be expressed as part of an operon (Fig. 1A).
The levels of expression of the nagP, nagR, nagB-II, and nagA genes were clearly higher in the ΔnagQ deletion mutant than in the wild-type strain (Table 2). These genes are downstream of the nagQ gene, which is itself downstream of the unique putative NagQ box detected in the X. campestris pv. campestris genome (Fig. 1A). This result suggests that NagQ regulates its own expression and that the genes from nagQ to nagA form an operon. The expression of the other GlcNAc pathway genes was not significantly affected by deletion of nagQ.
The expression of NagQ-regulated genes was not affected by deletion of nagR. The expression of nixA, nixC, nixD, nagK-IIA, and nagK-IIB was derepressed in the ΔnagR deletion mutant compared to the expression in the wild-type strain (Table 2). This is in agreement with the presence of putative NagR boxes in the promoter regions of these genes or operons, as determined by Yang and coworkers (48). Surprisingly, the GlcNAc-induced TBDT nixB gene located downstream of a NagR box seemed not to be regulated by NagR under our conditions (Table 2), suggesting that four of the five putative NagR boxes are functional. This result prompted us to generate a position weight matrix with the PREDetector program (23) using the four functional NagR boxes for screening the X. campestris pv. campestris genome. Of the 61 predicted targets, 16 are located in intergenic regions (see Table S1 in the supplemental material). Sequences upstream of nixA, nixB, nixD, nagK-IIA, and nagK-IIB each had strong predicted NagR-binding sites. However, the score obtained for the nixB promoter site was close to the scores for weak sites (see Table S1 in the supplemental material). This low score might explain the poor NagR regulation of nixB, a gene which is nevertheless induced by GlcNAc.
A sequence logo was generated by WebLogo (http://weblogo.berkley.edu/; 11) from the alignment of the four putative functional NagR boxes, which resulted in discovery of a new NagR box (GTTGACARCGYTGTCANC). This NagR box differed at positions 1, 2, and 18 from the previously proposed NagR box (AATGACARCGYTGTCATT) (48).
Together, these results show that NagR and NagQ are functional repressors of genes belonging to the GlcNAc CUT system. Proteins encoded by NagR- and NagQ-regulated genes can be classified into two main categories: transport and metabolism of GlcNAc.
Transport of GlcNAc in X. campestris pv. campestris. (i) Free GlcNAc passively diffuses through the envelope.
GlcNAc uptake rates in the X. campestris pv. campestris wild-type strain were compared after overnight preculture in the presence of GlcNAc (induced) and after overnight preculture in the presence of xylose (uninduced), a substrate that results in a growth rate similar to that obtained with GlcNAc (Fig. 2A) but does not affect the expression of GlcNAc-induced TBDT genes (data not shown). Before transport experiments were performed with [14C]GlcNAc, cells were washed to remove nonradiolabeled GlcNAc from the medium. The GlcNAc uptake rates under the two conditions were not significantly different (data not shown), suggesting that GlcNAc import is limited by a GlcNAc-independent transport step.
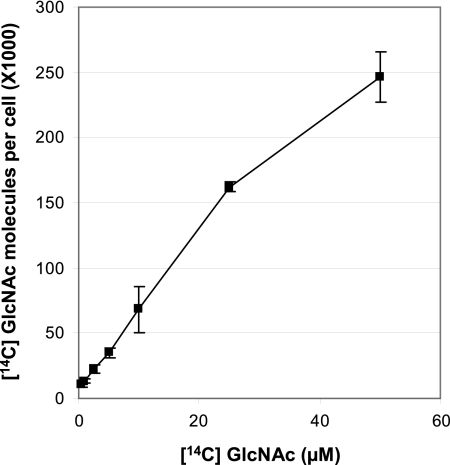
The initial concentration-dependent [14C]GlcNAc transport, reflecting the dissociation constant (Kd) for GlcNAc uptake, was determined using the previously described rapid dilution method (7, 33). The kinetic values revealed that the uptake rate was low and monophasic (Fig. 3), suggesting either that the outer and inner membrane transporters have similar affinities for GlcNAc or that transport through the outer membrane is limiting and masks transport through the inner membrane. The deduced Kd (138.9 μM) is more than 100-fold higher than the Kd estimated for passive uptake of GlcNAc through the CC0446 TBDT in C. crescentus (17) and is in a range similar to the range for Kd values obtained for passive diffusion through porins (18). Therefore, free GlcNAc uptake through the X. campestris pv. campestris envelope seems to occur via passive diffusion rather than by active uptake, although the X. campestris pv. campestris GlcNAc regulon contains at least four TBDT genes encoding active outer membrane transporters.
FIG. 3.
Concentration-dependent transport of 14C-labeled N-acetylglucosamine (GlcNAc) into X. campestris pv. campestris. Cells were grown in minimal medium without GlcNAc, and transport was measured for 15 s at the [14C]GlcNAc concentrations indicated.
A 100-fold excess of unlabeled glucose, galactose, sucrose, mannose, xylose, fructose, or GlcNAc-6P had no effect on radiolabeled GlcNAc uptake (Table 3). With unlabeled GlcNAc, the concentration for inhibition of the transport rate to one-half of the control rate was estimated to be 193.7 μM, which is in accordance with the Kd deduced from the results of the initial concentration-dependent GlcNAc transport assays. Glucosamine and chitobiose both inhibit radioactive GlcNAc uptake as much as unlabeled GlcNAc (Table 3). Inhibition of GlcNAc uptake by chitobiose could be due either to the chitobiose molecule itself or to degradation of this molecule to GlcNAc. These competition experiments suggest that GlcNAc, glucosamine, and probably chitobiose are transported across the envelope via the same transporters.
TABLE 3.
Inhibition of uptake of 0.5 μM [14C]GlcNAc by various carbohydrates in X. campestris pv. campestris wild-type strain after 1 h of incubation
| Carbohydrate | % of control uptake (SD) at carbohydrate concn ofa: |
|
|---|---|---|
| 50 μM | 500 μM | |
| N-Acetylglucosamine | 65 (1) | 18 (2) |
| N-Acetylglucosamine-6P | 101 (4) | 105 (4) |
| Glucosamine | 66 (3) | 41 (0) |
| Chitobiose | 58 (1) | 34 (4) |
| Glucose | 98 (3) | 84 (5) |
| Galactose | 105 (5) | 103 (5) |
| Mannose | 95 (1) | 94 (5) |
| Fructose | 100 (3) | 101 (4) |
| Xylose | 99 (5) | 102 (11) |
| Sucrose | 100 (5) | 101 (7) |
Standard deviations were calculated from the results of three independent experiments.
(ii) None of the GlcNAc-induced TBDTs seems to play a major role in utilization of free GlcNAc.
The rates of [14C]GlcNAc uptake in the X. campestris pv. campestris wild-type strain and in GlcNAc regulon TBDT insertion mutants were compared, and none of the TBDT mutants exhibited a significant effect in GlcNAc uptake (Table 4). Furthermore, the growth rates of strains with mutations in the nix and nax TBDT genes in the presence of 10 mM GlcNAc were similar to the growth rate of the wild-type strain (data not shown). The absence of a phenotype for mutants with single mutations in TBDT genes could be due to the redundant functions of the transporters.
TABLE 4.
Rates of 14C-labeled N-acetylglucosamine transport compared to the rate in X. campestris pv. campestris wild-type straina
| Strain | Transporter family | Mean % transport (SD)b |
|---|---|---|
| Wild type | 100 (9.9) | |
| nixA::pVO | TBDT | 96.6 (8.8) |
| nixB::pVO | TBDT | 115.2 (6.4) |
| nixC::pVO | TBDT | 111.2 (12.5) |
| nixD::pVO | TBDT | 106.8 (11.8) |
| naxA::pVO | TBDT | 114 (11.3) |
| naxB::pVO | TBDT | 115 (15.7) |
| ΔnagP | MFS | 1.2 (0.4) |
| ΔnagP pC-nagP | 98 (4) |
Transport rates were measured 60 min after addition of 14C-labeled N-acetylglucosamine.
Standard deviations were calculated from three independent experiments.
(iii) NagP is the major GlcNAc inner membrane transporter in X. campestris pv. campestris.
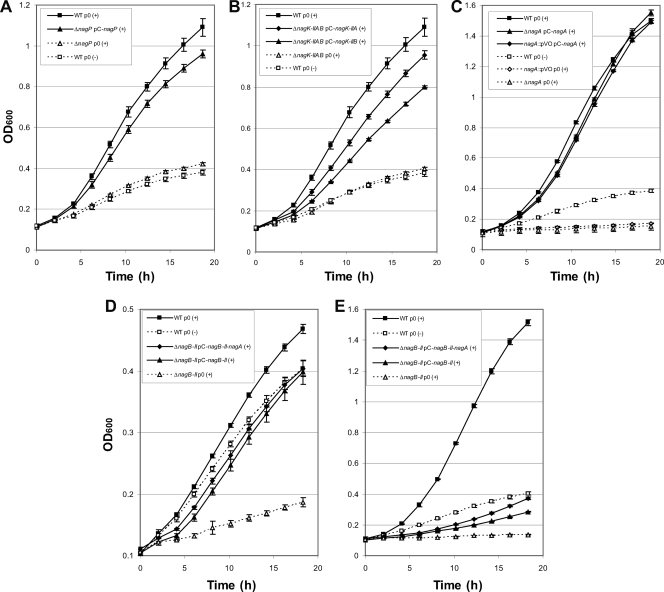
The nagP gene was deleted to test the putative role of NagP, which belongs to the major facilitator superfamily (MFS), in the transport of GlcNAc. Growth of the ΔnagP strain was impaired on MME containing GlcNAc as the sole carbon source (Fig. 2C), suggesting that this transporter could be involved in the uptake of GlcNAc through the inner membrane. The rate of uptake of radiolabeled GlcNAc obtained for the ΔnagP strain was only 1.2% of the rate obtained for the wild-type strain (Table 4). In the nagP-complemented strain, GlcNAc transport capacity (Table 4) and growth on GlcNAc-containing MME (Fig. 4A) were restored, confirming that NagP is the major transporter of GlcNAc across the inner membrane.
FIG. 4.
Complementation of mutants with mutations in genes encoding proteins in the X. campestris pv. campestris N-acetylglucosamine (GlcNAc) utilization pathway. After overnight growth in complete medium, cells were harvested, washed, and resuspended in minimal medium containing 200 μM isopropyl-β-thiogalactoside (IPTG), 5 μg/ml of tetracycline, and 10 mM (A, B, C, and E) or 100 μM (D) GlcNAc. The error bars indicate the standard deviations obtained from 4 independent experiments. WT, wild-type strain.
Catabolism of GlcNAc in X. campestris pv. campestris. (i) NagK-IIA and NagK-IIB phosphorylate GlcNAc.
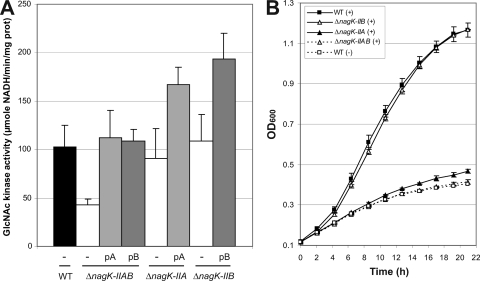
In the cytoplasm, the first step in the X. campestris pv. campestris GlcNAc utilization pathway is phosphorylation of GlcNAc (Fig. 1B). Two genes, nagK-IIA and nagK-IIB, coding for proteins belonging to the glucokinase family, belong to the GlcNAc regulon (Table 2), and their products have been proposed to act as putative GlcNAc kinases in Xanthomonas (48). To test the function of these proteins in the phosphorylation of GlcNAc, ΔnagK-IIA and ΔnagK-IIB single mutants, as well as a ΔnagK-IIAB double mutant, were constructed. The GlcNAc kinase activity of the ΔnagK-IIAB double mutant was about 41% of the wild-type activity (Fig. 5A). The GlcNAc kinase activity of the double mutant could have been due to the presence of residual ADP or pyruvate in the crude extracts used in the experiments. Wild-type GlcNAc kinase activity was restored when either nagK-IIA or nagK-IIB was supplied in trans on an expression plasmid, suggesting that both proteins phosphorylate GlcNAc. However, the activities obtained for each single mutant did not differ significantly from the wild-type activity (Fig. 5A), suggesting that these two proteins are functionally redundant.
FIG. 5.
Phosphorylation of N-acetylglucosamine by X. campestris pv. campestris NagK-II enzymes. (A) In vitro GlcNAc kinase assay based on the NAD+/NADH ratio of the wild-type strain (WT), strains with single deletions (ΔnagK-IIA and ΔnagK-IIB), or a strain with a double deletion (ΔnagK-IIAB) containing plasmid pC-nagK-IIA (pA), plasmid pC-nagK-IIB (pB), or no plasmid (−). The activity observed in ATP-depleted medium was subtracted to normalize the assay results. Strains were cultured in minimal medium supplemented with 10 mM N-acetylglucosamine (GlcNAc) to induce expression of the nagK-II genes. The error bars indicate the standard deviations obtained from 4 independent experiments. (B) Growth curves for mutant strains cultivated in minimal medium supplemented (+) or not supplemented (−) with GlcNAc. After overnight growth in complete medium, cells were harvested, washed, and resuspended in minimal medium containing 10 mM N-acetylglucosamine. The error bars indicate the standard deviations obtained from 4 independent experiments.
The growth of the ΔnagK-IIAB double mutant was clearly impaired in GlcNAc-containing minimal medium (Fig. 2C and Fig. 5B). The growth of the ΔnagK-IIA single mutant was also affected, but to a lesser extent, whereas the ΔnagK-IIB mutant grew like the wild-type strain (Fig. 5B). Growth of the ΔnagK-IIAB double mutant in GlcNAc minimal medium was partially restored when nagK-IIA or nagK-IIB was overexpressed in trans on an expression plasmid; however, better complementation was observed with nagK-IIA (Fig. 4B). These results suggest that although both GlcNAc kinases are enzymatically functional, NagK-IIA plays a major role in GlcNAc utilization, whereas NagK-IIB, which is apparently not essential, can also function in this capacity.
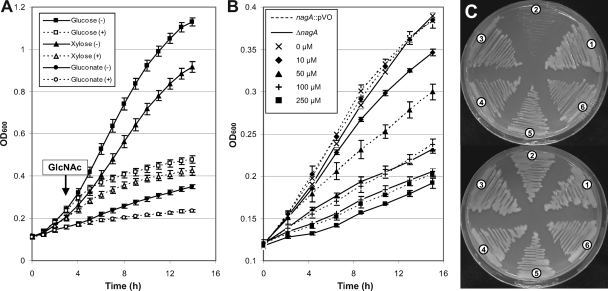
(ii) GlcNAc is toxic for nagA and nagB-II mutants.
NagA deacetylase (XCC3410) catalyzes the conversion of GlcNAc-6P to GlcN-6P, and the NagB-II deaminase (XCC3411) deaminates and isomerizes GlcN-6P to fructose-6P (Fig. 1B). Because the NagA and NagB-II proteins of X. campestris pv. campestris are very similar to those of Shewanella oneidensis strain MR-1 (48), their enzymatic activities were not studied in vitro. However, their biological importance for the X. campestris pv. campestris GlcNAc utilization pathway was studied genetically. Addition of GlcNAc to the medium resulted in rapid inhibition of growth of the ΔnagA mutant (Fig. 2C and 6A). Complete inhibition was observed with a GlcNAc concentration of 1 mM (data not shown). This “amino sugar sensitivity” phenomenon, previously observed in E. coli (6, 46), has been proposed to be due to accumulation of GlcNAc-6P, leading to pentose starvation (6). In X. campestris pv. campestris, GlcNAc sensitivity was still observed when gluconate or glucose was added to the medium (Fig. 6A). Intriguingly, a nagA::pVO insertion mutant seemed to be less sensitive to GlcNAc than the ΔnagA deletion mutant, since significant differences in inhibition of growth between these two strains were observed for GlcNAc concentrations ranging from 10 μM to 250 μM (Fig. 6B). This difference is likely not due to any polar effect on the expression of a downstream gene because GlcNAc sensitivity was abolished when nagA was supplied in trans in both mutants (Fig. 4C).
FIG. 6.
X. campestris pv. campestris nagA mutants are sensitive to N-acetylglucosamine. (A) Effect of addition of N-acetylglucosamine (GlcNAc) on growth of the ΔnagA mutant cultivated in minimal medium containing various carbon sources at a concentration of 20 mM. After 3 h of growth, media were supplemented (+) or not supplemented (−) with N-acetylglucosamine at a concentration of 10 mM. The error bars indicate the standard deviations obtained from 4 independent experiments. (B) Effects of different concentrations of N-acetylglucosamine (GlcNAc) on growth of nagA::pVO (dashed lines) and ΔnagA (solid lines) mutants. Cells were grown in minimal medium containing the concentrations of GlcNAc indicated. The error bars indicate the standard deviations obtained from 4 independent experiments. (C) Effects of secondary mutations on the GlcNAc sensitivity of the ΔnagA mutant. Strains were streaked on complete medium plates containing 10 mM N-acetylglucosamine (upper plate) or 10 mM xylose (lower plate). Region 1, wild-type strain; region 2, ΔnagA mutant; region 3, ΔnagP mutant; region 4, ΔnagP ΔnagA mutant; region 5, ΔnagK-IIAB mutant; region 6, ΔnagK-IIAB ΔnagA mutant. The photograph was taken after 96 h of growth. Identical results were obtained on minimal medium plates.
Growth of the ΔnagB-II mutant was also inhibited in the presence of GlcNAc (Fig. 2C), but the sensitivity of this mutant was less pronounced than that of the ΔnagA strain since no inhibition was observed at GlcNAc concentrations below 10 μM with the ΔnagB-II mutant (data not shown). A difference in sensitivity of nagA and nagB-II mutants has also been observed for E. coli and could be due to the gradual assimilation of GlcN-6P in the nagB-II mutant, whereas GlcNAc-6P that accumulates in the nagA mutant cannot be metabolized further (6). When nagB-II or nagB-II and nagA were supplied in trans, GlcNAc sensitivity was abolished in the presence of 100 μM GlcNAc (Fig. 4D), but the growth rate of each complemented strain was lower than that of the wild-type strain. Surprisingly, very faint complementation was observed in the presence of 10 mM GlcNAc, even with isopropyl-β-d-thiogalactopyranoside (IPTG) induction of complementing genes (Fig. 4E).
(iii) NagP, NagK-II, and NagA enzymes are in the same metabolic pathway.
The GlcNAc sensitivity of nagA mutants with a second mutation in either nagP or the nagK-IIA and nagK-IIB genes was tested to confirm that the NagP transporter and NagK-IIA and NagK-IIB GlcNAc kinases are located upstream of NagA deacetylase in the GlcNAc utilization pathway. The absence of GlcNAc entry or phosphorylation should prevent the formation of GlcNAc-6P and therefore reduce or eliminate its accumulation in nagA mutants. A ΔnagP ΔnagA double mutant and a ΔnagK-IIAB ΔnagA triple mutant were constructed, and their sensitivities to GlcNAc were tested. Growth inhibition of nagA mutants by GlcNAc was abolished when the inner membrane transporter was absent (ΔnagP ΔnagA mutant) or when the two GlcNAc kinases were mutated (ΔnagK-IIAB ΔnagA mutant) (Fig. 6C). Identical results were obtained for nagA::pVO mutants (data not shown). This epistatic effect confirms that the NagP, NagK-II, and NagA enzymes are all part of the same metabolic pathway.
DISCUSSION
Efficient bacterial exploitation (sensing, uptake, and catabolism) of nutrients requires close regulation of specific genetic programs. In X. campestris pv. campestris, carbohydrate utilization loci containing TonB-dependent transporters (CUT loci) have been proposed to play a major role in carbohydrate scavenging (7). In this paper, we characterize the X. campestris pv. campestris GlcNAc utilization pathway and propose that this pathway comprises a CUT system since the TBDTs were coregulated with the other genes in this pathway.
The MFS transporters comprise a novel family of GlcNAc transporters in bacteria.
The Major facilitator superfamily (MFS) inner membrane transporters comprise the largest family of secondary active transporters and have a diverse range of substrates, including ions, sugars, drugs, nucleosides, and peptides, but individual members of this family show stringent specificity. NagP is the major GlcNAc transporter in X. campestris pv. campestris and is required for bacterial growth on GlcNAc as the sole carbon source. To our knowledge, this is the first report of bacterial GlcNAc uptake through an MFS transporter. Previously, two important families of inner membrane transporters for the uptake of GlcNAc in bacteria were identified: the phosphotransferase transporter systems (PTS), analogous to NagE in E. coli (28), and the ATP-binding cassette transporters (ABC), such as NgcEFG in Streptomyces olivaceoviridis (47). Our results indicate that the MFS is a third inner membrane transporter family involved in the uptake of GlcNAc. The gene encoding NagP is present in all Xanthomonadales strains sequenced so far, suggesting that it has a conserved role in GlcNAc uptake in these bacteria.
The MFS NagP transporter is a nonphosphorylating transport system. Therefore, in contrast to E. coli, in which GlcNAc uptake through the PTS is coupled with phosphorylation (28), the first step in the X. campestris pv. campestris GlcNAc utilization pathway in the cytoplasm is phosphorylation of GlcNAc.
GlcNAc: a new substrate for glucokinase enzymes.
NagK-IIA and NagK-IIB, which are very similar (54% identity), can both phosphorylate GlcNAc. These glucokinase family proteins (PF02685 in the Pfam database [5]) possess a ROK domain (PF0480) and might be members of group B of the hexokinase superfamily (27). Although the physiological functions of most members of this group are unknown, several enzymes belonging to this family have been shown to phosphorylate glucose and other hexoses (8, 21). Our work provides the first evidence of a role in phosphorylation of GlcNAc for this protein family. Therefore, these glucokinase family proteins represent a new subfamily of GlcNAc kinases, as proposed by Yang and coworkers (48). X. campestris and Xanthomonas axonopodis strains possess both nagK-II genes. However, in the other sequenced Xanthomonadaceae (Stenotrophomonas, Xylella, and Xanthomonas oryzae) an ortholog of nagK-IIB is not present or not functional (data not shown), raising a question about the role of NagK-IIB in strains that also possess NagK-IIA. Although NagK-IIB is not required for X. campestris pv. campestris growth on GlcNAc-containing media, this protein can substitute for NagK-IIA in GlcNAc phosphorylation. The glucokinase family protein GlcK from Bacillus sphaericus has been reported to have substrate ambiguity, phosphorylating not only glucose but also fructose and mannose (21). It is thus possible that GlcNAc is an alternative substrate for NagK-IIB. It was recently shown that in X. campestris pv. campestris strain 8004 NagK-IIB was not involved in the phosphorylation of glucose, arabinose, xylose, sorbitol, mannose, mannitol, sorbose, fructose, galactose, rhamnose, sucrose, or maltose (29). Since this enzyme is under the control of NagR, its substrate is probably related to GlcNAc metabolism. Therefore, it could be interesting to determine the role of NagK-IIB in the phosphorylation of other hexosamines, such as glucosamine or even chitobiose.
The X. campestris pv. campestris nag cluster encodes two functional repressors, NagR and NagQ.
At least three nonorthologous types of transcriptional regulators were proposed previously to control the expression of GlcNAc utilization genes in proteobacteria: the ROK family protein NagC, the LacI family protein NagR, and the GntR-type protein NagQ (48). Interestingly, bacteria in the genus Xanthomonas are the only bacteria known to have two genes coding for these GlcNAc-specific regulators (i.e., nagR and nagQ) (48), both of which are located in the nag cluster. The work performed here showed that both of the regulators are functional. Repression by NagQ seems to be restricted to the nag operon. Therefore, NagQ can be considered a local transcription factor. On the other hand, NagR likely acts at multiple sites, since several NagR boxes were identified and are scattered throughout the X. campestris pv. campestris genome. The NagR regulon could be broader, and it would be interesting to identify additional targets of this repressor in global transcriptomic analyses.
In X. campestris pv. campestris, NagQ represses nagR. The importance of this regulatory loop in GlcNAc catabolism is a matter of conjecture. Furthermore, the GlcNAc regulatory network could be much more complicated, and additional regulatory elements could participate in this network. Indeed, nixA, naxA, and nagK-IIA belong to the Clp regulon, a conserved global regulator shown to play a central role in the regulation of virulence factors in X. campestris pv. campestris (22).
The nagQ gene is specific for Xanthomonas strains. Indeed, in the sequenced plant-pathogenic Xylella strains, which have reduced genomes (37), nagQ is partially deleted, suggesting that this gene has become vestigial and can therefore be lost during evolution. This gene is also not present in Stenotrophomonas maltophilia, a non-plant-pathogenic species belonging to the Xanthomonadaceae family that includes free-living as well as endophytic isolates and opportunistic human pathogens (12, 41). Interestingly, in both bacteria, the NagQ box located upstream of the nag cluster is replaced by a candidate NagR box (48).
GlcNAc passively diffuses through the outer membrane.
Although four X. campestris pv. campestris active outer membrane TBDT genes are induced by GlcNAc, three of which are under the control of NagR, GlcNAc passively diffuses through the outer membrane. Furthermore, none of the GlcNAc-induced TBDTs was individually required for growth on GlcNAc and for uptake of this molecule. This implies either that several of these TBDTs allow diffusion of GlcNAc or, alternatively, that GlcNAc can diffuse through the outer membrane via other transporters, such as porins.
TBDTs, which are well known for their role in iron and vitamin B12 uptake (36), were shown previously to be involved in the active uptake of carbohydrates, such as maltodextrins in C. crescentus (33) or sucrose in X. campestris pv. campestris (7). In C. crescentus, TBDT CC0446 is essential for growth on the chito-oligosaccharides (GlcNAc)3 and (GlcNAc)5 in a TonB-dependent manner, but transport of GlcNAc apparently occurs by passive diffusion through this transporter, a process which is TonB independent (17). Therefore, we propose that X. campestris pv. campestris TBDTs belonging to the GlcNAc regulon are involved in the active transport of complex molecules containing GlcNAc, as observed for C. crescentus (17). Consequently, this implies that the source of GlcNAc could be more complex than monomeric GlcNAc.
What is the source of GlcNAc for X. campestris pv. campestris in the environment?
There are two principal sources of GlcNAc in nature: chitin and bacterial cell walls. In E. coli, about 50% of cell wall peptidoglycan is broken down each generation (20, 24). Recent bioinformatic analyses identified the gene coding for the d-Ala-d-Ala aminopeptidase that is responsible for catabolism of the cell wall precursor d-Ala-d-Ala as a part of the Streptomyces coelicolor DasR regulon (39). This pleiotropic regulator belonging to the GntR family is essential for development and is involved in the regulation of genes encoding both the GlcNAc PTS and the GlcN-6P deaminase NagB (38). Therefore, there is a direct relationship between peptidoglycan recycling and GlcNAc utilization. Although this link was not studied in X. campestris pv. campestris, a role for an X. campestris pv. campestris GlcNAc-induced TBDT(s) in the active uptake of peptidoglycan degradation products can be readily envisaged.
Alternatively, X. campestris pv. campestris could have developed a system to exploit chito-oligosaccharides derived from fungal and insect chitin degraded by plant chitinases (26) or by chitinolytic bacteria. Occupation of niches in the plant phyllosphere by epiphytic, saprophytic, and pathogenic fungi and bacteria and the interactions of these organisms are important for the Xanthomonas life cycle (40). In this environment, the growth of X. campestris pv. campestris on chitin or its polymeric subunits relies on other organisms for chitin degradation, as has recently been proposed for the nonchitinolytic aquatic bacterium C. crescentus (17). To exploit chito-oligosaccharides, X. campestris pv. campestris must produce enzymes involved in degradation of these compounds. This bacterium has one of the largest glycobiomes, as determined using the CAZy database (http://www.cazy.org/) (10). Interestingly, downstream of the nixD TBDT gene (XCC2887), we identified several putative enzyme-encoding genes which could be involved in the degradation of oligosaccharides that contain GlcNAc. Among these, XCC2889 codes for a protein belonging to the GH-18 family, which includes endo-beta-N-acetylglucosaminidase (EC 3.2.1.96), and XCC2890 codes for a protein belonging to the glycoside hydrolase GH-20 family, which includes the β-hexosaminidases (EC 3.2.1.52). Work is now under way to further characterize these genes, to determine whether they are part of the GlcNAc CUT system, and to determine which molecules are targeted by these enzymes.
Supplementary Material
Acknowledgments
We are grateful to Scott Soby for critical reading of the manuscript. We also acknowledge Laurent Noël and Servane Baufumé for valuable discussions and critical reading of the manuscript.
A. Boulanger and G. Déjean were supported by a grant from the French Ministère de la Recherche et de l'Enseignement Supérieur. We gratefully acknowledge financial support from the Département Santé des Plantes et Environnement de l'Institut National de la Recherche Agronomique (grant 2007_0441_02) and from the French Agence Nationale de la Recherche (grant ANR-08-BLAN-0193-01).
Footnotes
Published ahead of print on 14 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez-Anorve, L. I., M. L. Calcagno, and J. Plumbridge. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 187:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angot, A., N. Peeters, E. Lechner, F. Vailleau, C. Baud, L. Gentzbittel, E. Sartorel, P. Genschik, C. Boucher, and S. Genin. 2006. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U. S. A. 103:14620-14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlat, M., C. L. Gough, C. E. Barber, C. Boucher, and M. J. Daniels. 1991. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 4:593-601. [DOI] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernheim, N. J., and W. J. Dobrogosz. 1970. Amino sugar sensitivity in Escherichia coli mutants unable to grow on N-acetylglucosamine. J. Bacteriol. 101:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through tonb-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigham, C. J., and M. H. Malamy. 2005. Characterization of the RokA and HexA broad-substrate-specificity hexokinases from Bacteroides fragilis and their role in hexose and N-acetylglucosamine utilization. J. Bacteriol. 187:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calcagno, M., P. J. Campos, G. Mulliert, and J. Suastegui. 1984. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from Escherichia coli. Biochim. Biophys. Acta 787:165-173. [DOI] [PubMed] [Google Scholar]
- 10.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crossman, L. C., V. C. Gould, J. M. Dow, G. S. Vernikos, A. Okazaki, M. Sebaihia, D. Saunders, C. Arrowsmith, T. Carver, N. Peters, E. Adlem, A. Kerhornou, A. Lord, L. Murphy, K. Seeger, R. Squares, S. Rutter, M. A. Quail, M. A. Rajandream, D. Harris, C. Churcher, S. D. Bentley, J. Parkhill, N. R. Thomson, and M. B. Avison. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunnac, S. 2004. Identification à l'échelle génomique des effecteurs dépendant du système de sécrétion de type III de la bactérie phytopathogène Ralstonia solanacearum. Ph.D. thesis. Université Toulouse III Paul Sabatier, Toulouse, France.
- 14.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. V. Sluys, N. F. Almeida, L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. A. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. N. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 15.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 16.Dow, J. M., and M. J. Daniels. 1994. Pathogenicity determinants and global regulation of pathogenicity of Xanthomonas campestris pv. campestris. Curr. Top. Microbiol. Immunol. 192:29-41. [DOI] [PubMed] [Google Scholar]
- 17.Eisenbeis, S., S. Lohmiller, M. Valdebenito, S. Leicht, and V. Braun. 2008. NagA-dependent uptake of N-acetyl-glucosamine and N-acetyl-chitin oligosaccharides across the outer membrane of Caulobacter crescentus. J. Bacteriol. 190:5230-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freundlieb, S., U. Ehmann, and W. Boos. 1988. Facilitated diffusion of p-nitrophenyl-alpha-d-maltohexaoside through the outer membrane of Escherichia coli. Characterization of LamB as a specific and saturable channel for maltooligosaccharides. J. Biol. Chem. 263:314-320. [PubMed] [Google Scholar]
- 19.Gonzali, S., L. Pistelli, L. De Bellis, and A. Alpi. 2001. Characterization of two Arabidopsis thaliana fructokinases. Plant Sci. 160:1107-1114. [DOI] [PubMed] [Google Scholar]
- 20.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, B., H. Liu, X. Hu, Y. Cai, D. Zheng, and Z. Yuan. 2007. Molecular characterization of a glucokinase with broad hexose specificity from Bacillus sphaericus strain C3-41. Appl. Environ. Microbiol. 73:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Y. W., A. Y. Ng, M. Xu, K. Lin, L. H. Wang, Y. H. Dong, and L. H. Zhang. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281-292. [DOI] [PubMed] [Google Scholar]
- 23.Hiard, S., R. Maree, S. Colson, P. A. Hoskisson, F. Titgemeyer, G. P. van Wezel, B. Joris, L. Wehenkel, and S. Rigali. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357:861-864. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferson, R. A., T. A. Kavanagh, and M. W. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasprzewska, A. 2003. Plant chitinases—regulation and function. Cell. Mol. Biol. Lett. 8:809-824. [PubMed] [Google Scholar]
- 27.Kawai, S., T. Mukai, S. Mori, B. Mikami, and K. Murata. 2005. Hypothesis: structures, evolution, and ancestor of glucose kinases in the hexokinase family. J. Biosci. Bioeng. 99:320-330. [DOI] [PubMed] [Google Scholar]
- 28.Lengeler, J. W., K. Jahreis, and U. F. Wehmeier. 1994. Enzymes II of the phosphoenol pyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim. Biophys. Acta 1188:1-28. [DOI] [PubMed] [Google Scholar]
- 29.Lu, G. T., Z. J. Yang, F. Y. Peng, Y. N. Tan, Y. Q. Tang, J. X. Feng, D. J. Tang, Y. Q. He, and J. L. Tang. 2007. The role of glucose kinase in carbohydrate utilization and extracellular polysaccharide production in Xanthomonas campestris pathovar campestris. Microbiology 153:4284-4294. [DOI] [PubMed] [Google Scholar]
- 30.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, D., E. Lauber, D. Roby, M. Arlat, and T. Kroj. 2005. Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 6:327-333. [DOI] [PubMed] [Google Scholar]
- 32.Morales, C. Q., J. Posada, E. Macneale, D. Franklin, I. Rivas, M. Bravo, J. Minsavage, R. E. Stall, and M. C. Whalen. 2005. Functional analysis of the early chlorosis factor gene. Mol. Plant-Microbe Interact. 18:477-486. [DOI] [PubMed] [Google Scholar]
- 33.Neugebauer, H., C. Herrmann, W. Kammer, G. Schwarz, A. Nordheim, and V. Braun. 2005. ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187:8300-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 35.Onsando, J. 1992. Black rot of crucifers. Dis. Veg. Oil Seed Crops 2:243-252. [Google Scholar]
- 36.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 37.Puhler, A., M. Arlat, A. Becker, M. Gottfert, J. P. Morrissey, and F. O'Gara. 2004. What can bacterial genome research teach us about bacteria-plant interactions? Curr. Opin. Plant Biol. 7:137-147. [DOI] [PubMed] [Google Scholar]
- 38.Rigali, S., H. Nothaft, E. E. Noens, M. Schlicht, S. Colson, M. Muller, B. Joris, H. K. Koerten, D. A. Hopwood, F. Titgemeyer, and G. P. van Wezel. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237-1251. [DOI] [PubMed] [Google Scholar]
- 39.Rigali, S., F. Titgemeyer, S. Barends, S. Mulder, A. W. Thomae, D. A. Hopwood, and G. P. van Wezel. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudolph, K. 1993. Infection of the plant by Xanthomonas, p. 193-264. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman and Hall, London, United Kingdom.
- 41.Ryan, R. P., S. Monchy, M. Cardinale, S. Taghavi, L. Crossman, M. B. Avison, G. Berg, D. van der Lelie, and J. M. Dow. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7:514-525. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 44.Schauer, K., D. A. Rodionov, and H. de Reuse. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33:330-338. [DOI] [PubMed] [Google Scholar]
- 45.Turner, P., C. E. Barber, and M. J. Daniels. 1985. Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 199:338-343. [Google Scholar]
- 46.White, R. J. 1968. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, X., F. Wang, A. Saito, J. Majka, A. Schlosser, and H. Schrempf. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol. Genet. Genomics 267:429-439. [DOI] [PubMed] [Google Scholar]
- 48.Yang, C., D. A. Rodionov, X. Li, O. N. Laikova, M. S. Gelfand, O. P. Zagnitko, M. F. Romine, A. Y. Obraztsova, K. H. Nealson, and A. L. Osterman. 2006. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 281:29872-29885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.