Abstract
In the purple sulfur bacterium Allochromatium vinosum, the reverse-acting dissimilatory sulfite reductase (DsrAB) is the key enzyme responsible for the oxidation of intracellular sulfur globules. The genes dsrAB are the first and the gene dsrR is the penultimate of the 15 genes of the dsr operon in A. vinosum. Genes homologous to dsrR occur in a number of other environmentally important sulfur-oxidizing bacteria utilizing Dsr proteins. DsrR exhibits sequence similarities to A-type scaffolds, like IscA, that partake in the maturation of protein-bound iron-sulfur clusters. We used nuclear magnetic resonance (NMR) spectroscopy to solve the solution structure of DsrR and to show that the protein is indeed structurally highly similar to A-type scaffolds. However, DsrR does not retain the Fe-S- or the iron-binding ability of these proteins, which is due to the lack of all three highly conserved cysteine residues of IscA-like scaffolds. Taken together, these findings suggest a common function for DsrR and IscA-like proteins different from direct participation in iron-sulfur cluster maturation. An A. vinosum ΔdsrR deletion strain showed a significantly reduced sulfur oxidation rate that was fully restored upon complementation with dsrR in trans. Immunoblot analyses revealed a reduced level of DsrE and DsrL in the ΔdsrR strain. These proteins are absolutely essential for sulfur oxidation. Transcriptional and translational gene fusion experiments suggested the participation of DsrR in the posttranscriptional control of the dsr operon, similar to the alternative function of cyanobacterial IscA as part of the sense and/or response cascade set into action upon iron limitation.
Reduced sulfur compounds, like sulfide or thiosulfate, are utilized as electron donors by quite a diverse group of prokaryotes. Among them are phototrophic purple and green sulfur bacteria, chemotrophic sulfur-oxidizing bacteria, like Thiobacillus denitrificans, and several symbiotic sulfur oxidizers, like “Candidatus Ruthia magnifica” strain Cm, the endosymbiont of the deep-sea hydrothermal-vent clam Calyptogena magnifica. In many sulfur-oxidizing prokaryotes and in all of the organisms mentioned above, sulfur globules are formed as intermediates during the oxidation of sulfide or thiosulfate (16, 51). The only gene region proven to be essential for the oxidation of sulfur stored in sulfur globules has been identified in the purple sulfur bacterium Allochromatium vinosum (51). This gene region, the dsr (dissimilatory sulfite reductase) operon, encompasses 15 genes (dsrABEFHCMKLJOPNRS). The first two, dsrAB, encode the key enzyme of this pathway, reverse sulfite reductase (15, 51). The enzyme is thought to catalyze the oxidation of activated sulfur to sulfite (13, 26, 58). dsrR is the second-to-last gene of the operon. Homologues are found in environmentally important free-living chemotrophic sulfur oxidizers, like T. denitrificans and Thioalkalivibrio sp. HL-EbGR7, as well as in the endosymbiotic sulfur oxidizers “Candidatus Ruthia magnifica” and “Candidatus Vesicomyosocius okutanii.” The gene dsrR is absent from the genome of the green sulfur bacterium Chlorobaculum tepidum (formerly Chlorobium tepidum) (32) and the purple sulfur bacterium Halorhodospira halophila (6, 15, 23). The dsrR gene encodes a soluble cytoplasmic protein of unknown function. It shows a certain similarity to A-type scaffolds, such as IscA and SufA, which partake in the maturation of protein-bound iron-sulfur clusters (3, 25, 34), but it lacks the highly conserved cysteine residues of these proteins (15).
In this report, we describe the nuclear magnetic resonance (NMR) structure of A. vinosum DsrR, examine the similarities between IscA and DsrR, and present the results gained by in-frame deletion mutagenesis of dsrR in A. vinosum. We also discuss the effects of the deletion of dsrR on the transcriptional and translational expression of other dsr genes and on the formation of various Dsr proteins in an attempt to describe the in vivo function of the DsrR protein.
MATERIALS AND METHODS
Overproduction, purification, and preparation of recombinant DsrR.
All general molecular-genetic techniques, media, and growth conditions were described previously (17). dsrR was amplified using primers that introduced NdeI and XhoI restriction sites. The NdeI/XhoI-digested PCR product was cloned into pET-15b, yielding pREX. E. coli BL21(DE3) cells containing pREX were cultured in 500 ml LB medium with 200 μM Fe(NH4)2(SO4)2 and 100 μg ampicillin ml−1 at 37°C and 180 rpm. At an optical density at 600 nm (OD600) of 0.5, IPTG (isopropyl-β-d-thiogalactopyranoside) (0.1 mM) was added, and the cells were harvested after 2 h. The cell pellet was resuspended in buffer A (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 7.5), and Complete protease inhibitor cocktail, EDTA free (Roche), and 1 mg ml−1 lysozyme were added. The cells were disrupted by sonication (2 min ml−1; Cell Disruptor B15; Branson) and centrifuged at 10,000 × g for 30 min at 4°C. The N-terminally His-tagged DsrR was purified using a nickel agarose column (Qiagen). After a second passage of the DsrR-containing fractions through a nickel agarose column, the fractions were dialyzed against 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl. The protein was concentrated to a final volume of no more than 2 ml via Centriprep-10 (Amicon). The purity was estimated to be >95% by electrophoretic analysis on 15% (wt/vol) SDS-PAGE. The state of oligomerization of the protein was investigated by gel filtration chromatography on Superdex-75 (GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 8.0), 500 mM NaCl. The His tag was removed by utilizing a thrombin cleavage capture kit (Novagen) according to the manufacturer's instructions. The success of the cleavage was verified by the failure of the protein to bind to nickel agarose and by SDS-PAGE analysis.
Protein techniques.
Immunoblot (Western) analysis and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described by Dahl et al. (15). DsrR was detected with an antiserum raised against the following oligopeptide: H2N-LNPRDPTYRPPSGG-CONH2, encompassing highly immunogenic epitopes deduced from the nucleotide sequence (Eurogentec, Seraing, Belgium). The antiserum was used at a dilution of 1:500.
Iron-binding assays.
Iron-binding assays were performed by anoxic incubation of 50 μM protein with no or 400 μM Fe(NH4)2(SO4)2 in the presence of 2.5 mM dithiothreitol (DTT) at 10°C for 16 h. The samples were separated from unbound iron by passing them through a HiTrap desalting column (GE Healthcare). The UV-visible absorption spectra of the samples were recorded in 1-ml quartz cuvettes using a Lambda 11 UV-Vis spectrometer (Perkin Elmer). The spectra were calibrated to an absorbance at 260 nm of 1.0. The ability to bind Fe-S clusters was tested by adding 400 μM Na2S to the above-mentioned preparation. The protein IscA was used for comparison. Overproduction and purification were performed as described for DsrR with the exception that 50 μg kanamycin ml−1 instead of ampicillin was added to the media. The plasmid pTISCA for overproduction of N-terminally His-tagged IscA (19) was kindly provided by H. Ding (Louisiana State University).
Preparations of samples for NMR.
NMR samples of U-13C, 15N-DsrR and 5% biosynthetically directed 13C, U-15N-DsrR were prepared as previously described (13) in 0.5-liter cultures. The cells were induced for 10.5 h at 30°C. The purified protein yield was approximately 10 mg liter−1. Following exchange into NMR buffer (50 mM Tris-HCl, pH 7.4, at 25°C, 500 mM NaCl) on a PD-10 column, DTT (5 mM), NaN3 (0.02% [wt/vol]), and D2O (5% [vol/vol]) were added. The final protein concentration in NMR samples was approximately 0.9 mM. The samples were free of precipitate.
NMR spectroscopy.
NMR experiments were conducted at the Environmental Molecular Sciences Laboratory of Pacific Northwest National Laboratory, Richland, WA, as described previously (13), on Varian Inova 600 and 750 instruments using standard triple-resonance pulse sequences (10). All experiments were from the Varian BioPack library except the four-dimensional (4D) 1H-13C heteronuclear multiple quantum coherence (HMQC)-nuclear Overhauser enhancement spectroscopy (NOESY)-HMQC (45). Data were collected at 20°C. NOESY mixing times were 80 ms. Chemical shifts were assigned manually.
Calculation and analysis of the structural ensemble.
The structural ensemble was generated as described previously (13) using AutoStructure (30) to generate a preliminary ensemble based on automated NOESY peak assignments, followed by manual refinement using Xplor-NIH and CNS. NOESY distance restraints had uniform lower bounds of 1.8 Å and upper bounds of 2.8, 3.5, 4.0, 5.0, or 5.5 Å. Amide 1H-15N-heteronuclear single quantum coherence (HSQC) cross peaks still present 45 min after dissolution of a lyophilized sample in D2O were used to derive hydrogen bond restraints with bounds of 1.7 to 2.3 Å for the HN-O distance and 2.7 to 3.3 Å for the N-O distance, provided preliminary structural ensembles indicated the correct acceptor atom. Dihedral restraints for Φ and Ψ were derived from TALOS (12). Φ restraints had bounds of −55° ± 30° for helical residues and −120° ± 50° for extended residues. Ψ restraints had values of −47° ± 30° for helical residues and 140° ± 50° for extended residues. Restraints derived from TALOS were included only for residues in helices and β-sheets, with consideration for the evident secondary-structure propensities in preliminary ensembles of structures.
The resulting distance restraints, H-bond restraints, and dihedral restraints were used to generate a set of 40 structures with Xplor-NIH (59) using restrained molecular dynamics simulated annealing. The routines sa.inp and dgsa.inp were used as provided, except that an initial temperature of 2,000 K was used with 20,000 high-temperature steps and 20,0000 cooling steps. Sum averaging was used for methyl groups and methylene proton pairs. A final refinement in explicit water was performed in CNS (39). Twenty (out of a total of 40) structures were selected to form the final ensemble on the basis of minimal restraint violations and energies. The structural ensemble was analyzed with PSVS (7). A structure validation report is available at http://www.nesg.org/. A table of structural statistics is included as Table S3 in the supplemental material.
Sequence and structure analyses.
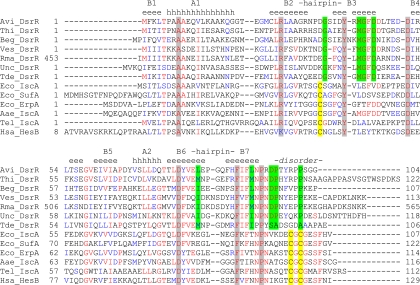
All amino acid sequences were obtained from GenBank. PSI-BLAST was used with default parameters to generate the protein sequence family (1). ClustalW was used to generate the multiple-sequence alignment (11). ConSurf was used to visualize the structural distribution of conserved residues (27). The multiple-sequence alignment of DsrR sequences used as input for ConSurf was generated using the seven DsrR sequences shown in Fig. 1, together with several more DsrR sequences from draft genome sequences. The alignment of IscA-like sequences was a precalculated alignment available from the ConSurf server and contained more than 100 sequences, including a few DsrR sequences. The surface electrostatic features of the protein were examined using ABPS (4) with PyMOL (18). Structure similarity searches using Dali (28) were conducted using the residues 1 to 94 of the first structure from the ensemble as a representative structure for similarity searching.
FIG. 1.
Multiple-sequence alignment of DsrR and IscA sequences. Highly conserved residues in both DsrR and IscA-like proteins are shaded gray. Residues conserved only in DsrR are shaded green. Cysteine residues characteristic for IscA-like proteins are shaded yellow. Avi, Allochromatium vinosum DSM 180; Thi, Thioalkalivibrio sp. HL-EbGR7; Beg, Beggiatoa sp. PS; Ves, “Candidatus Vesicomyosocius okutanii” HA; Rma, “Candidatus Ruthia magnifica” strain Cm; Unc, uncultured bacterium BAC13K9BAC; Tde, Thiobacillus denitrificans ATCC 25259; Eco, Escherichia coli K-12; Aae, Aquifex aeolicus VF5; Tel, Thermosynechococcus elongatus BP-1; Hsa, Homo sapiens.
Construction and characterization of dsrR in-frame deletion and complementation in A. vinosum.
In-frame deletion of dsrR was achieved by utilizing gene splicing by overlap extension PCR (29). An XbaI restriction site was introduced into the 5′ and 3′ ends of the final deletion-framing PCR fragment. The PCR amplicon was cloned into the XbaI site of the mobilizable suicide vector pK18mobsacB (57), resulting in the plasmid pK18mobsacBΔdsrR. The conjugative broad-host-range vector pBBR1-MCS-2 (36) was used to reintroduce the dsrR gene under the control of the dsrA promoter, dsrAp, into the ΔdsrR mutant. The dsrAp-containing region, as well as the dsr terminator region, was amplified in such a way that the resulting fragments overlapped each other and could serve as primers in a subsequent PCR. NheI and XmaJI restriction sites were introduced between the fused dsrA promoter and terminator, thus enabling the in-frame integration of any gene. Additionally, HindIII and XbaI restriction sites were introduced into the 5′ and 3′ ends of the PCR fragment. The promoter-terminator amplicon was ligated into pBBR1-MCS-2, yielding the plasmid pBBRdsrPT1. The dsrR gene was cloned into the NheI and XmaJI sites of pBBRdsrPT1. The resulting plasmid, pBBRdsrPT1-dsrR, was introduced into A. vinosum Rif50 as described by Lübbe et al. (41).
The photolithoautotrophic growth of A. vinosum wild-type and mutant strains was examined in batch culture under continuous illumination essentially as described by Prange et al. (52) in a medium containing sulfide as the sole sulfur compound. Two hundred fifty milliliters of a photoheterotrophically grown stationary-phase culture was harvested (5,900 × g; 10 min), and the cell material was used to inoculate 1 liter of modified Pfennig's medium (17) in a temperature-controlled fermentor. Kanamycin (10 μg ml−1) was added to the medium of the complementation mutant. Sulfur compounds were determined by high-performance liquid chromatography (HPLC) (Thermo Separation Products TSP) using the methods of Rethmeier et al. (53). Elemental sulfur content was determined by cyanolysis (35).
Expression studies by real-time RT-PCR.
Total RNAs of A. vinosum Rif50 and A. vinosum ΔdsrR grown under different growth conditions were isolated and measured as described by Prange et al. (52). RNA (250 ng) was used for real-time reverse transcription (RT)-PCR analysis utilizing the QuantiTect SYBR green RT-PCR kit (Qiagen) and the iCycler iQ real-time detection system (Bio-Rad) according to the manufacturers' instructions. “No-RT” control reactions were performed for each RNA sample. In case of DNA contamination, the RNA samples were digested with RNase-free DNase (Qiagen) and purified using an RNeasy Mini Kit (Qiagen). Fragments (approximately 200 bp) of dsrA (annealing temperature [Ta], 59°C), dsrE (Ta, 56°C), dsrC (Ta, 55°C), dsrL (Ta, 57°C), dsrR (Ta, 60°C), and dsrS (Ta, 58°C) were amplified with the respective primer pair, utilizing the indicated Tas. The RT-PCR conditions were as follows: 30 min at 50°C (reverse transcription); 15 min at 95°C (inactivation of the reverse transcriptase and activation of the polymerase); 40 cycles of 15 s at 94°C, 30 s at Ta, and 30 s at 72°C, followed by melting curve analysis in which the temperature was increased every 30 s by 0.5°C from 40°C to 100°C. RNA standards were generated as described by Fey et al. (24). The samples were automatically quantified by the iCycler iQ software (Bio-Rad) based on the RNA standards.
Construction of a transcriptional and translational gene fusion plasmid and β-galactosidase assay.
A 908-bp DNA fragment encompassing the dsrA promoter (dsrAp) region, not including the ribosome binding site of dsrA, was PCR amplified using A. vinosum Rif50 chromosomal DNA as a template. The EcoRI/PstI-digested fragment was ligated into the similarly prepared vector pK18mobsacB to yield pKdsrProm. The lacZ gene was amplified from E. coli K-12 genomic DNA. The resulting amplicon was digested with PstI and HindIII and ligated to the 6,560-bp PstI/HindIII fragment of pKdsrProm, yielding the transcriptional fusion plasmid pTS.
The promoterless lacZ gene was excised with SalI and EcoRI from the translational lacZ fusion vector pPHU235 (31). The SalI site was filled with the Klenow fragment of DNA polymerase, and the fragment was inserted into the 5,670-bp EcoRI/HindIII fragment of pK18mobsacB, yielding the plasmid pK235 (B. Franz, personal communication). The dsr promoter region including the first 12 bp of dsrA was amplified by PCR, introducing a PstI and a HindIII restriction site. The amplicon was inserted into the PstI/HindIII restriction sites of pK235, thus forming the dsrA′-lacZ translational fusion plasmid pTL.
The gene fusion plasmids were transferred into A. vinosum Rif50 and A. vinosum ΔdsrR by conjugation as described by Pattaragulwanit and Dahl (50). The plasmid-carrying strains were grown on modified Pfennig's medium (containing 10 μg ml−1 kanamycin) with 2 mM sulfide, thiosulfate, sulfite, and/or malate for 24 h before β-galactosidase activity was tested. One milliliter of culture was pelleted (13,000 × g; 3 min; 4°C) and resuspended in 1 ml Z buffer (43). After the addition of 100 μl chloroform and 50 μl 0.1% sodium dodecyl sulfate (SDS), the sample was vortexed for 10 s at maximum speed and incubated at 30°C for 10 min before 200 μl preheated o-nitrophenyl-β-d-galactopyranoside (ONPG) (4 mg ml−1) was added. As soon as a yellow color could be discerned, the reaction was stopped by adding 500 μl 1 M Na2CO3. After centrifugation (5 min; 15,000 × g), the absorption of the supernatant was measured in a 1-ml quartz cuvette at 460 nm against a chemical blank. The specific β-galactosidase activity was calculated by using the following formula: [nmol o-nitrophenol/(min·mg)] = {(OD420/k) × Vassay [ml]/tassay [min] × protein concentration [mg ml−1] × Vculture [ml]}. Under the above conditions, 1 μM o-nitrophenol had an optical density (420 nm) of 0.0044 (k). The protein concentration for each sample was determined by Bradford assay (9).
Accession numbers.
The structure was deposited in the Protein Data Bank (PDB) under accession no. (PDB ID) 2K4Z. The chemical shifts were deposited in BioMagResBank (Madison, WI) with accession no. BMRB-15816.
RESULTS
Monomeric DsrR does not have the iron- or Fe-S-binding properties of IscA.
DsrR resembles the protein IscA, which is involved in iron-sulfur cluster maturation. IscA functions either as an iron chaperone, delivering iron ions to nascent iron-sulfur clusters built on the IscU scaffold (21, 67), or as an alternative scaffold for [2Fe-2S] or [4Fe-4S] clusters (37, 49). Sequence alignments of several DsrR homologues of sulfur-oxidizing bacteria with IscA homologues of non-sulfur-oxidizing bacteria illustrate the sequence similarity between these proteins but also highlight the differences between them (Fig. 1). The overall sequence identity between A. vinosum DsrR and IscA is around 30%, while identity with the alternative A-type scaffolds SufA and ErpA is somewhat lower. Sequence identity among DsrR homologues varies but can be as low as 30%. All DsrR homologues lack the three invariant cysteine residues of the IscA protein family that are involved in iron-sulfur cluster coordination or iron binding (14, 20, 64). Sequence comparison also revealed a number of conserved acidic amino acid residues in DsrR-like proteins (A. vinosum DsrR Asp35, Asp45, Glu49, and Asp95). As acidic amino acids may in principle be able to bind iron, the possibility that DsrR retained a reduced iron-binding capability could not be entirely discounted.
In order to examine a possible IscA-like function for DsrR, we overproduced both DsrR and IscA in E. coli grown on Fe2+-containing media, evaluated the oligomerization states of the recombinant proteins via gel filtration chromatography on Superdex-75, and compared the iron- and Fe-S-binding capabilities of DsrR and IscA in vitro.
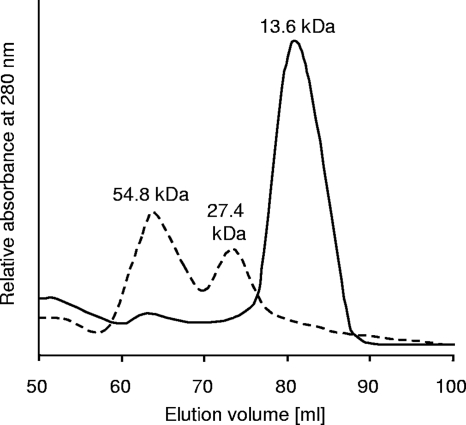
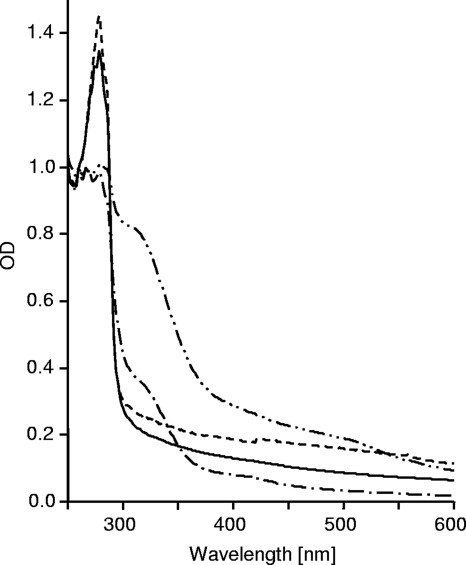
Unlike IscA, which occurred as a homodimer and a homotetramer, N-terminally His-tagged DsrR eluted in one clear peak corresponding to the monomer (Fig. 2). The NMR spectra of 1 mM DsrR samples were also not consistent with a dimeric species (data not shown). The isolated IscA contained a small amount of bound iron, visible as an absorption peak at 315 nm (Fig. 3), due to the presence of Fe(NH4)2(SO4)2 in the E. coli overproduction medium. The amplitude of the absorption peak at 315 nm dramatically increased after the protein was incubated with Fe(NH4)2(SO4)2 and dithiothreitol under anoxic conditions. The absorption spectra of freshly purified recombinant DsrR did not indicate any bound iron ions. Attempts to achieve in vitro reconstitution of DsrR with Fe2+ or Fe3+ ions under aerobic conditions in the absence of DTT failed. We therefore studied iron binding in the absence of oxygen, as has been described repeatedly for IscA (21, 37, 49, 67). While these experiments were clearly successful for IscA, reconstitution of DsrR with iron ions led to only a small increase of the amplitude at 315 nm (data not shown). This increase was due to the His tag, as DsrR without a His tag did not show the increase (Fig. 3). This observation is further corroborated by NMR spectroscopy, as the NMR spectra of DsrR did not reveal any characteristics of a paramagnetic species, arguing against the presence of bound iron(III). The NMR sample had no UV-visible absorption that was not attributable to the protein alone. In order to also exclude a potential Fe-S cluster binding function of DsrR, we added Na2S to the above-mentioned preparation but could not detect any change in the spectra. DsrR did not display the characteristic UV-visible absorption spectrum reported for IscA with a bound iron-sulfur cluster (references 37 and 67 and data not shown).
FIG. 2.
Gel filtration chromatogram of IscA (monomer, 13.7 kDa including His tag; dashed line) and DsrR (monomer, 13.6 kDa including His tag; solid line). The proteins were loaded onto a Superdex-75 column (GE Healthcare) preequilibrated with 50 mM Tris-HCl (pH 8.0), 500 mM NaCl. The flow rate was 0.5 ml min−1. The elution was monitored by absorption at 280 nm. The column had been calibrated with several proteins of known molecular masses.
FIG. 3.
Iron-binding assay of DsrR in comparison to IscA. Shown are UV-visible absorption spectra of DsrR (solid and dashed lines) and IscA (--·-- and --··--) after incubation with 0 or 400 μM Fe(NH4)2(SO4)2 in the presence of 2.5 mM DTT at 10°C for 16 h. The proteins were repurified by passing them through a HiTrap desalting column. The spectra were calibrated to an absorbance at 260 nm of 1.0.
The structural fold of DsrR is highly similar to that of IscA-like proteins.
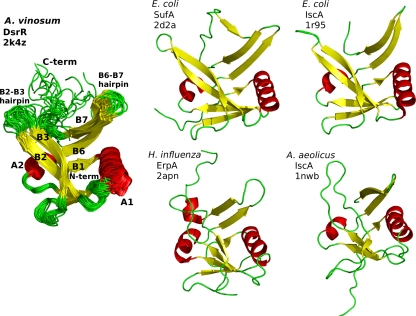
The structure of DsrR in aqueous solution determined with solution state NMR methods clearly shows that the protein has the same IscA-like fold (Fig. 4) seen in several NMR and X-ray structures of IscA (8, 14, 66), SufA (63), and ErpA (J. Orban, personal communication), all containing the conserved Cys-(Xn)-Cys-Gly-Cys motif (PDB IDs 1r95, 1s98, 1x0g, 2d2a, 2apn, and 1nwb). The IscA-like fold, also known as the HesB-like domain fold (46), appears to be a novel fold unique to this family (8, 66) in which two β-hairpins opposed in 2-fold symmetry form a deep cleft. Formation of an Fe-S cluster binding site by the conserved Cys residues occurs upon dimer and/or tetramer formation (8, 14, 44, 63). DsrR can be superimposed over the structured portion of IscA-like proteins to between 2- and 3-Å root mean square deviation (RMSD) for the backbone atoms, with the exception of 1nwb, the NMR structure of IscA-like protein aq_1857 from Aquifex aeolicus (3.6-Å backbone RMSD to DsrR). DsrR exhibits disorder in the C-terminal region beginning around residue Pro93, with no long-range NOESY peaks observed from this region to other parts of the protein, while the N terminus is ordered from the beginning of the DsrR sequence. IscA-like proteins without bound iron typically exhibit a similar lack of structure in the C terminus beginning at the equivalent proline residue, which is conserved throughout all DsrR and IscA-like proteins. This disordered region corresponds to the region containing the conserved Cys-Gly-Cys residues that form the Fe-S cluster binding site in IscA-like proteins. Several residues at the tips of the β2-β3 and β6-β7 hairpins of DsrR exhibit decreased convergence in the NMR ensemble, probably reflecting greater mobility in these regions, a characteristic also seen in the corresponding regions of IscA-like proteins.
FIG. 4.
Structural features of DsrR and comparison to IscA-like proteins. (Left) Structures of DsrR (overlaid ribbons for the ensemble). (Right) Ribbon structures for E. coli IscA and SufA, Haemophilus influenzae ErpA, and A. aeolicus aq_1857, the only IscA-like protein found in that organism. C-term, C terminus; N-term, N terminus.
DsrR also exhibits lower structural similarity to two uncharacterized proteins from a small family of proteins from lactobacilli, Lactobacillus salivarius UCC118 LSL_1730 (PBD ID 2p2e) and Lactobacillus acidophilus NCFM LBA0486 (2qgo). These proteins were identified by structure comparison; sequence similarity to DsrR and IscA is lower and was not identified by a PSI-BLAST search with DsrR as the query sequence. These proteins lack the two cysteine residues near the C terminus but have the first conserved cysteine of IscA-like proteins. Structural similarity to IscA-like proteins is lower than that found for DsrR, and the C terminus is structured, forming an additional β-strand along β7 and β3. Several other insertions of 1 to 5 residues make these sequences longer than the central folded portion of DsrR or IscA-like proteins. This feature, together with a more compact structure overall, creates a closed β-barrel that is unlike other β-barrel folds.
Some surface features on IscA-like proteins not involved with Fe-S or iron binding are conserved in DsrR, and other surface features are not conserved.
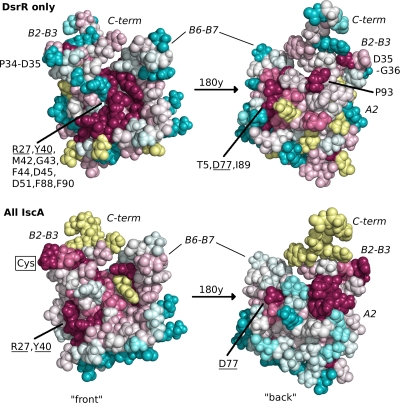
Conserved residues in the multiple-sequence alignment (Fig. 1) were mapped onto the surface of the DsrR structure according to whether they are conserved in DsrR only or in all IscA-like proteins (Fig. 5). The most striking differences are at the three conserved cysteine residues in IscA-like proteins, which are not present in any DsrR sequence, and the residues surrounding these cysteines. Several residues that are highly conserved in DsrR but not in IscA cluster along the β2-β3 and β6-β7 hairpins and in the cleft between them (Fig. 5). Among all IscA-like proteins, conserved surface residues mainly cluster on the hairpins. However, the regions surrounding these sites in DsrR and IscA are structurally similar, despite the sequence differences. Other residues on the surface are conserved in both IscA and DsrR, including three charged residues and two partially buried aromatic residues (Asp27, Tyr40, Asp51, Asp77, and Phe88 in DsrR). Though some of these occupy the cleft formed by the twin β-hairpins in the IscA fold, this cleft is also the site of several residues conserved only in DsrR (Fig. 5).
FIG. 5.
Space-filling structures of DsrR, colored according to the conservation of residues in multiple-sequence alignments of DsrR sequences only or all IscA-like sequences using ConSurf (27). Darker magenta colors indicate greater conservation, darker blue colors indicate lack of conservation, and lighter colors represent intermediate levels of residue conservation, while yellow indicates insufficient information, primarily in the C terminus, where the sequence alignment is uncertain. Residues that are particularly conserved in DsrR sequences are indicated, and those also conserved in IscA are underlined. The position on the DsrR structure where the conserved Cys residue in IscA on the β2-β3 hairpin was substituted is indicated. The other conserved IscA Cys residues are not indicated, as they are aligned with the disordered portion of the C terminus on DsrR.
Deletion of dsrR leads to a diminished sulfur oxidation rate and influences the production of several Dsr proteins.
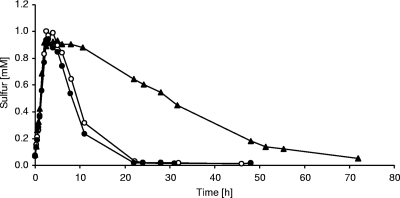
In an effort to assess the importance of DsrR for sulfur oxidation, we created and characterized an A. vinosum mutant carrying an in-frame deletion of dsrR. Neither sulfide oxidation nor formation of sulfur globules was affected in the ΔdsrR mutant (Table 1), as has been reported for other A. vinosum dsr mutants (15, 17, 41, 51, 56). On the other hand, the oxidation of intracellularly stored sulfur was severely reduced (Fig. 6). Compared to the wild type, the dsrR deletion mutant exhibited a specific sulfur oxidation rate that was reduced by ∼88%. Complementation of the ΔdsrR mutant by reintroducing dsrR in trans restored the oxidation rate to the wild-type level (Table 1), thereby confirming that the observed phenotype was exclusively caused by the lack of dsrR. The growth yields of all three strains were in the same range (Table 1). We surmise that DsrR is not absolutely essential for the oxidation of intermediary stored sulfur but that it clearly plays an important role in ensuring an undisturbed process.
TABLE 1.
Characteristics of the A. vinosum ΔdsrR deletion mutant compared to the wild type and the complementation mutant
| Characteristic | Value for A. vinosum straina: |
||
|---|---|---|---|
| Wild type | ΔdsrR | ΔdsrR+dsrR | |
| Sulfide oxidation rateb | 199.0 ± 18.2 | 205.3 ± 0.4 | 200.0 ± 16.8 |
| Sulfur globule formation rateb | 90.7 ± 0.6 | 90.7 ± 5.0 | 89.5 ± 2.1 |
| Sulfur oxidation rateb | 24.1 ± 0.3 | 2.9 ± 0.5 | 23.9 ± 0.65 |
| Growth yieldc | 8.8 ± 0.9 | 9.0 ± 0.9 | 8.9 ± 0.5 |
The results represent the means and standard deviations of three independent growth experiments. Initial sulfide concentration, 2 mM.
Oxidation and formation rates are given as nmol min−1 mg protein−1.
The growth yield is given as g protein mol sulfide−1.
FIG. 6.
Sulfur accumulation and oxidation by Allochromatium vinosum wild type (○), A. vinosum ΔdsrR (▴), and A. vinosum ΔdsrR+dsrR (•). The cultures were grown photolithoautotrophically in batch culture with 2 mM sulfide. The protein contents of the cultures were 49.7 μg ml−1 (wild type), 91.0 μg ml−1 (ΔdsrR), and 62.7 μg ml−1 (ΔdsrR+dsrR) at the start of the experiment. Representatives of three independent growth experiments for each strain are shown.
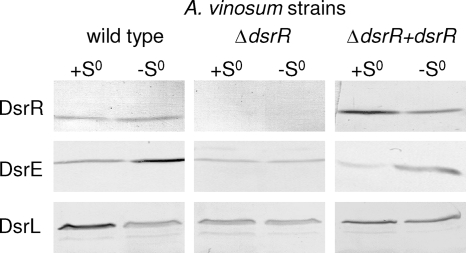
The presence of several Dsr proteins was examined in the A. vinosum wild type in comparison to ΔdsrR and the complemented mutant strain using protein-specific antisera against DsrE, DsrC, DsrK, DsrL, and DsrR. A. vinosum DsrR was predicted to be an 11.4-kDa cytoplasmic soluble protein (15). Appropriately, it was detected in the soluble fraction of the wild type and the complemented mutant but was absent in the dsrR deletion mutant (Fig. 7). The formation of DsrC and the membrane-associated cytoplasmic protein DsrK (15) was not influenced by the deletion of dsrR (data not shown). Considering, that DsrC is constitutively expressed (51), it is not surprising that it is regulated differently and as such is not affected by the deletion. The unchanged formation of DsrK, which is part of the transmembrane redox complex DsrMKJOP, on the other hand, is quite puzzling, and a closer look at the regulation of the dsr operon is necessary to elucidate this result. Surprisingly, the formation of DsrE, a subunit of the soluble cytoplasmic heterohexameric protein DsrEFH (17), was slightly reduced in the ΔdsrR mutant, especially after stored sulfur had been completely metabolized. It should be noted that DsrE appears to be present in larger amounts after the completion of sulfur oxidation than in the phase of active degradation of sulfur globules. Furthermore, the formation of DsrL (41), a soluble cytoplasmic iron-sulfur flavoprotein with NADH:acceptor oxidoreductase activity, was somewhat reduced in A. vinosum ΔdsrR. The diminished formation of DsrE and DsrL, both proteins essential to the sulfur oxidation pathway (17, 41), in the dsrR-deficient mutant could be restored approximately to wild-type level by complementation (Fig. 7).
FIG. 7.
Western blot analyses with antisera against DsrR (11.4 kDa), DsrE (14.6 kDa), and DsrL (71.4 kDa) were performed with soluble fractions of A. vinosum (96 μg protein), A. vinosum ΔdsrR (96 μg protein), and A. vinosum ΔdsrR+dsrR (68 μg) grown in batch culture on 2 mM sulfide and harvested either at the maximum content of intracellularly stored sulfur (+S0) or after the sulfur had been completely metabolized (−S0). The antisera were raised against oligopeptides comprising a highly immunogenic epitope deduced from the nucleotide sequence, and their specific reactions with the respective Dsr protein overproduced in E. coli were proven (15).
DsrR affects the translation of the dsr operon.
Balasubramanian et al. (5) recently proposed an alternative function for IscA in the regulation of iron homeostasis and the sensing of redox stress in cyanobacteria. In an attempt to test for an analogous function for DsrR, we examined the expression levels of dsrA, dsrE, dsrC, dsrL, dsrR, and dsrS in the wild type and the ΔdsrR mutant under photo-organoheterotrophic and photolithoautotrophic growth conditions via real-time RT-PCR. Major differences in expression could not be discerned for any of these genes (data not shown).
The effects of the deletion of dsrR on the translational level were analyzed by introducing a translational dsrA′-lacZ fusion into the A. vinosum wild type and the ΔdsrR mutant. The β-galactosidase activities were compared to those achieved with a transcriptional dsrAp-lacZ fusion. The reporter strains were grown for 24 h on modified Pfennig's medium with 2 mM malate or sulfide before the lacZ-mediated β-galactosidase activity was determined (Table 2). As was expected in light of the RT-PCR results, no major differences were detected between the β-galactosidase activities in the wild type and those in the ΔdsrR mutant. The 2-fold-higher LacZ activity in the malate-grown culture of ΔdsrR cannot be explained at the moment. Surprisingly, the translation of dsrA′-lacZ in the ΔdsrR mutant was reduced by 80.6% under both growth conditions. This implicates DsrR as a factor in posttranscriptional control, as apparently less DsrA is formed when DsrR is lacking than when it is present.
TABLE 2.
Specific β-galactosidase activities in A. vinosum wild type and A. vinosum ΔdsrR carrying transcriptional or translational gene fusionsa
| A. vinosum strainb | β-Galactosidase sp actc |
|
|---|---|---|
| Malate | Sulfide | |
| Wild type | ||
| dsrAp-lacZ | 2.9 ± 0.7 | 9.1 ± 0.9 |
| dsrA′-lacZ | 42.6 ± 1.7 | 96.2 ± 27.1 |
| ΔdsrR | ||
| dsrAp-lacZ | 6.7 ± 0.8 | 11.3 ± 1.7 |
| dsrA′-lacZ | 6.4 ± 0.3 | 18.7 ± 1.0 |
Cells were grown either photo-organoheterotrophically (malate) or photolithoautotrophically (sulfide).
Photo-organoheterotrophically grown cultures containing the transcriptional gene fusion (dsrAp-lacZ) or the translational gene fusion (dsrA′-lacZ), were used to inoculate 12 ml of modified Pfennig's medium with 2 mM malate or sulfide. The β-galactosidase activity was measured 24 h after inoculation.
The specific β-galactosidase activity is given as nmol o-nitrophenol min−1 mg protein−1. Under the given conditions, 1 nmol ml−1 o-nitrophenol had an optical density at 420 nm of 0.0044. The protein content of each sample was determined utilizing the Bradford method. The average protein content amounted to 500 μg ml−1. The results represent the means and standard deviations of three independent measurements.
DISCUSSION
Based on the sequence similarity to IscA, a protein involved in iron-sulfur cluster maturation (21, 37, 62), it is tempting to assume DsrR to have a function similar to that of IscA. When we consider that several iron-sulfur cluster-containing proteins are encoded within the dsr operon (DsrAB, DsrK, DsrO, and DsrL), it even appears appropriate to provide them with a protein-specific iron-sulfur cluster scaffold, as is assumed for NifIscA and the IscA homologue ErpA. In Azotobacter vinelandii, NifIscA supposedly serves as an alternative scaffold to NifU in nitrogen fixation-specific iron-sulfur cluster assembly (37). In Escherichia coli, ErpA, like DsrR in A. vinosum, is not encoded near any other Fe-S cluster biogenesis operon. ErpA is essential for isopentenyl diphosphate (IPP) biosynthesis, as it is probably involved in the maturation of the iron-sulfur cluster-containing key enzymes IspG and IspH (40). It may be interesting in this respect that dsrR is encoded immediately upstream of a gene annotated as ispH in the uncultured bacterium BAC13K9BAC (accession no. DQ068067).
A closer look at the amino acid sequence of DsrR, however, reveals an important difference between A-type scaffold proteins and DsrR (Fig. 1). Despite the high structural similarity, the DsrR protein family lacks all three of the invariant cysteine residues that are involved in the coordination of iron or iron-sulfur clusters (8, 14, 20, 33). The presence of conserved acidic amino acid residues instead of cysteines in A. vinosum DsrR, though at different positions in the sequence (Asp35, Asp45, Glu49, and Asp95), could indicate a reduced iron-binding capability of DsrR. However, the UV-visible spectra of DsrR after purification and after reconstitution with iron or with iron and sulfide showed no indication of bound iron or bound iron-sulfur clusters. Moreover, gel filtration analysis showed DsrR to be a monomer, unlike IscA, which occurs as a mixture of oligomeric forms with dimers and tetramers predominating (49, 65). IscA monomers associate to form a dimer of dimers with a central channel within which the conserved cysteine residues are presumed to form a “cysteine pocket” and where mononuclear iron or iron-sulfur clusters can be coordinated in a subunit-bridging manner (8, 14, 37). The loss of the cysteine residues, as well as the lack of a higher oligomerization state, strongly argue against the notion that DsrR serves as a Dsr protein-specific iron-sulfur cluster scaffold or as an iron chaperone delivering iron ions to nascent iron-sulfur clusters.
In the genome of “Candidatus Ruthia magnifica” (47), the sulfur-oxidizing symbiont of the giant hydrothermal-vent clam Calyptogena magnifica, dsrR is fused to dsrN, which is located immediately upstream. A separate ribosome binding site is not apparent for the dsrR gene, indicating cotranslation of the two genes in “Candidatus R. magnifica.” This observation provoked the idea that DsrN and DsrR may act together or even be subunits of a hetero-oligomeric protein (26, 55). DsrN is a siroheme amidase and is repsonsible for the amidation of siroheme to siroamide, the prosthetic group of sulfite reductase in A. vinosum (41). In light of the comparable phenotypes observed for the respective deletion mutants, i.e., severe reduction but not cessation of sulfur oxidation (41), this assumption at first seemed quite plausible. However, when we consider (1) that dsrR and dsrN do not always occur together (2); that dsrR is present neither in the genome sequences of green sulfur bacteria, e.g., C. tepidum, nor in that of the purple sulfur bacterium H. halophila; and (3) that dsrN is part of the core set of dsr genes present in all prokaryotes utilizing Dsr proteins in dissimilatory sulfur metabolism (56), this conclusion now appears premature. In fact, in the course of the work presented here, we accumulated evidence that DsrR performs a function completely different from that of DsrN.
The DsrR family is a subgroup of proteins belonging to the IscA superfamily occurring only in sulfur-oxidizing bacteria that use reverse-acting sulfite reductase to oxidize intracellularly stored sulfur. As outlined above, this subgroup is characterized by the striking absence of the three invariant cysteine residues present in hundreds of IscA superfamily sequences. DsrR must have acquired a specialized role in sulfur oxidation following gene duplication of an ancestral iscA gene. This role must not require binding of iron or Fe-S clusters directly, and loss of the cysteine residues may have been driven by negative selection for Fe-S- or iron-binding activity. A. vinosum and other organisms with dsrR still contain iscA or erpA genes. The structural similarity between DsrR and IscA-like proteins and the conservation of several surface residues suggest that DsrR retains functional characteristics other than coordination of Fe-S clusters or iron binding in common with its IscA-like paralogues. One possibility is that DsrR assists in the recruitment of iron or Fe-S cluster donors and the facilitation of metal transfer to an Fe-S cluster-containing Dsr protein, such as DsrAB, DsrK, DsrO, or DsrL, without actually interacting with the metal itself. In this sense, DsrR would not be unlike other IscA paralogues, such as NifIscA and ErpA, that are specialized in the maturation of specific Fe-S cluster-containing proteins, except that DsrR would not interact directly with iron or the Fe-S cluster.
There is some evidence suggesting that cysteineless scaffold proteins are mediators of protein-protein interaction. The iron-sulfur cluster scaffold protein NfuA of E. coli consists of two domains. NfuA is essential for E. coli to grow under oxidative stress and iron-starvation conditions (2). The C terminus resembles the Nfu domain of NifU, and the N-terminal domain shows similarity to A-type scaffold proteins (IscA, SufA, and ErpA) but, like DsrR, has lost the three invariant cysteine residues of this protein family. The NfuA N-terminal domain is not particularly similar to that of DsrR, however, so this seems to be a second instance of loss of cysteine residues from an ancestral IscA-like protein rather than the occurrence of a DsrR-like protein in an organism that does not have other dsr genes and does not oxidize intracellularly stored sulfur. Complementation studies showed that both domains are essential for the function of NfuA in vivo (2). Interestingly, several transcriptomic analyses reported that nfuA (also known as yhgI and gntY) is upregulated in response to translational stress, either due to misfolded protein (38), kanamycin (60), or heat shock (48) and, of course, under iron limitation (42). These facts led Angelini et al. (2) to the conclusion that NfuA possesses functions as a chaperone and in repairing damaged iron-sulfur proteins.
A similar case involving the loss of cysteine residues from one domain of a protein with duplicate domains involved in Fe-S cluster assembly is found in OsCnfU-1A of Oryza sativa (54). The two domains of OsCnfU-1A resemble the Nfu domain of NifU, the IscU-like iron-sulfur cluster scaffold for maturation of the nitrogenase iron-sulfur cluster in Azotobacter vinelandii (22, 61). The second domain does not have the invariant cysteine residues typical of Nfu domains. It is responsible for the mediation of the interaction with the apoprotein ferredoxin through an extensive basic surface, which makes it ideal for the interaction with the predominantly negatively charged ferredoxin (54). DsrR could in principle be another example of a cysteineless scaffold protein mediating protein-protein interactions.
Further evidence of IscA-like proteins lacking Fe-S- or Fe-binding activity exists in the structures of two similar proteins of unknown function from Lactobacillus species deposited in the structure database recently (L. salivarius LSL_1730 [PDB ID 2p2e] and L. acidophilus LBA0486 [2qgo]). They adopt the IscA fold; however, they and their homologues all have only one of the three conserved cysteines (the Cys in the first hairpin loop, but not the two near the C terminus) of IscA/SufA/HesB/ErpA. The sequence similarity between these proteins and DsrR or IscA-like proteins is low, but together with the structural similarity, it may suggest a common IscA-like ancestor with a role in Fe-S cluster biosynthesis.
The results obtained in the course of this study, however, point in another direction, i.e., they indicate the participation of DsrR in posttranscriptional regulation. Comparison of the effects of the deletion of dsrR from A. vinosum on transcriptional versus translational reporter gene fusions showed that transcription was not affected but that translation was diminished by 80.6%. Reporter gene activities in the ΔdsrR strain for the translational gene fusion decreased to approximately the same extent as the sulfur oxidation rate (∼88%). The decreased sulfur oxidation rate and the diminished presence of DsrE and DsrL, both of which are absolutely essential for sulfur oxidation (17, 41), can be explained by a reduced rate of posttranscriptional processes in the ΔdsrR mutant. Taken together, these findings suggest DsrR as a player in posttranscriptional regulation. The protein could either be involved in stabilizing ribosome-mRNA interaction, thus enhancing translation, or play a role during translational attenuation, i.e., inducing a conformational change in the mRNA and thereby permitting translation initiation. Interestingly, the entire Shine-Dalgarno sequence of dsrA is part of a predicted stem-loop possibly preventing ribosomal access (free energy of formation, −38 kJ mol−1 according to the DINAMelt Server [http://dinamelt.bioinfo.rpi.edu/quikfold.php]). It is also interesting in this respect that Balasubramanian et al. (5) suggested a regulatory role for IscA, in addition to its function as an Fe-S cluster assembly scaffold. These authors described IscA as part of the sense and/or response cascade set into action upon iron limitation in cyanobacteria.
We have clearly shown that the region encompassing the ribosome binding site is required for the downregulation of the accumulation of DsrA′-LacZ protein in the absence of DsrR. As the NMR structure of DsrR did not reveal any RNA-binding motifs, it seems improbable that DsrR itself binds directly to mRNA. The first coprecipitation experiments with His-tagged DsrR and soluble crude extract of A. vinosum or pure recombinant Dsr proteins, like DsrC, DsrEFH, and DsrL, have not yet indicated specific binding partners for DsrR. However, more detailed experiments need to be performed to elucidate the likely indirect contribution of DsrR to posttranscriptional regulation as part of a signal-transducing reporter chain cascade. Similar functions should now be considered for IscA-like proteins, as well.
Supplementary Material
Acknowledgments
The NMR spectra were acquired in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy Office of Biological and Environmental Research) located at Pacific Northwest National Laboratory. We thank Jacob Bitoun for the guidelines to iron-binding assays, Bernd Masepohl for his helpful tips concerning gene fusion, Fabian Grein for introduction into and upkeep of the anaerobic chamber, and Bettina Franz for insightful discussions.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) grants Da 351/3-4 and Da 351/3-5 to C.D. J.R.C. acknowledges support from the NIH Protein Structure Initiative (grant P50-GM62413-02). DsrR is community-selected target OP5 of the Northeast Structural Genomics Consortium.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelini, S., C. Gerez, S. Ollagnier-de-Choudens, Y. Sanakis, M. Fontecave, F. Barras, and B. Py. 2008. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283:14084-14091. [DOI] [PubMed] [Google Scholar]
- 3.Ayala-Castro, C., A. Saini, and F. W. Outten. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72:110-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian, R., G. Shen, D. A. Bryant, and J. H. Golbeck. 2006. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 188:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beller, H. R., P. S. G. Chain, T. E. Letain, A. Chakicherla, F. W. Larimer, P. M. Richardson, M. A. Coleman, A. P. Wood, and D. P. Kelly. 2006. The genome sequence of the obligately chemolithoautotrophic, facultatively anaerobic bacterium Thiobacillus denitrificans. J. Bacteriol. 188:1473-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya, A., R. Tejero, and G. T. Montelione. 2007. Evaluating protein structures determined by structural genomics consortia. Proteins 66:778-795. [DOI] [PubMed] [Google Scholar]
- 8.Bilder, P. W., H. Ding, and M. E. Newcomer. 2004. Crystal structure of the ancient, Fe-S scaffold IscA reveals a novel protein fold. Biochemistry 43:133-139. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh, J., W. J. Fairbrother, A. G. I. Palmer, and N. J. Skelton. 1996. Protein NMR spectroscopy, principle and practice. Academic Press, San Diego, CA.
- 11.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13:289-302. [DOI] [PubMed] [Google Scholar]
- 13.Cort, J. R., U. Selan, A. Schulte, F. Grimm, M. A. Kennedy, and C. Dahl. 2008. Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 382:692-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cupp-Vickery, J. R., J. J. Silberg, D. T. Ta, and L. E. Vickery. 2004. Crystal structure of IscA, an iron-sulfur cluster assembly protein from Escherichia coli. J. Mol. Biol. 338:127-137. [DOI] [PubMed] [Google Scholar]
- 15.Dahl, C., S. Engels, A. S. Pott-Sperling, A. Schulte, J. Sander, Y. Lübbe, O. Deuster, and D. C. Brune. 2005. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187:1392-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl, C., and A. Prange. 2006. Bacterial sulfur globules: occurrence, structure and metabolism, p. 21-51. In J. M. Shively (ed.), Inclusions in prokaryotes. Springer-Verlag, Heidelberg, Germany.
- 17.Dahl, C., A. Schulte, Y. Stockdreher, C. Hong, F. Grimm, J. Sander, R. Kim, S.-H. Kim, and D. H. Shin. 2008. Structural and molecular genetic insight into a widespread sulfur oxidation pathway. J. Mol. Biol. 384:1287-1300. [DOI] [PubMed] [Google Scholar]
- 18.Delano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 19.Ding, H., and R. J. Clark. 2004. Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem. J. 379:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, H., R. J. Clark, and B. Ding. 2004. IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J. Biol. Chem. 279:37499-37504. [DOI] [PubMed] [Google Scholar]
- 21.Ding, H., K. Harrison, and J. Lu. 2005. Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. J. Biol. Chem. 280:30432-30437. [DOI] [PubMed] [Google Scholar]
- 22.Dos Santos, P. C., A. D. Smith, J. Frazzon, V. L. Cash, M. K. Johnson, and D. R. Dean. 2004. Iron-sulfur cluster assembly. NifU-directed activation of the nitrogenase Fe protein. J. Biol. Chem. 279:19705-19711. [DOI] [PubMed] [Google Scholar]
- 23.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. U. S. A. 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fey, A., S. Eichler, S. Flavier, R. Christen, M. G. Höfle, and C. A. Guzmán. 2004. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70:3618-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fontecave, M., and S. Ollagnier-de-Choudens. 2008. Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474:226-237. [DOI] [PubMed] [Google Scholar]
- 26.Frigaard, N.-U., and C. Dahl. 2009. Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54:103-200. [DOI] [PubMed] [Google Scholar]
- 27.Glaser, F., T. Pupko, I. Paz, R. E. Bell, D. Bechor-Shental, E. Martz, and N. Ben-Tal. 2003. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19:163-164. [DOI] [PubMed] [Google Scholar]
- 28.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis: SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Y. J., H. N. B. Moseley, M. C. Baran, C. Arrowsmith, R. Powers, R. Tejero, T. Szyperski, and G. T. Montelione. 2005. An integrated platform for automated analysis of protein NMR structures. Methods Enzymol. 394:111-141. [DOI] [PubMed] [Google Scholar]
- 31.Hübner, P., J. C. Willison, P. Vignais, and T. A. Bickle. 1991. Expression of regulatory nif genes in Rhodobacter capsulatus. J. Bacteriol. 173:2993-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imhoff, J. F. 2003. Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int. J. Syst. Evol. Microbiol. 53:941-951. [DOI] [PubMed] [Google Scholar]
- 33.Jensen, L. T., and C. Culotta. 2000. Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol. 20:3918-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 35.Kelly, D. P., L. A. Chambers, and P. A. Trudinger. 1969. Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41:898-901. [Google Scholar]
- 36.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 37.Krebs, C., J. N. Agar, A. D. Smith, J. Frazzon, D. R. Dean, B. H. Huynh, and M. K. Johnson. 2001. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry 40:14069-14080. [DOI] [PubMed] [Google Scholar]
- 38.Lesley, S. A., J. Graziano, C. Y. Cho, M. W. Knuth, and H. E. Klock. 2002. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 15:153-160. [DOI] [PubMed] [Google Scholar]
- 39.Linge, J. P., and M. Nilges. 1999. Influence of non-bonded parameters on the quality of NMR structures: a new force field for NMR structure calculation. J. Biomol. NMR 13:51-59. [DOI] [PubMed] [Google Scholar]
- 40.Loiseau, L., C. Gerez, M. Bekker, S. Ollagnier-de-Choudens, B. Py, Y. Sanakis, J. Teixeira de Mattos, M. Fontecave, and F. Barras. 2007. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:13626-13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lübbe, Y., H.-S. Youn, R. Timkovich, and C. Dahl. 2006. Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c-diamide synthase, for sulphur oxidation. FEMS Microbiol. Lett. 261:194-202. [DOI] [PubMed] [Google Scholar]
- 42.McHugh, J. P., F. Rodríguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1972. Assay of β-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Morimoto, K., E. Yamashita, Y. Kondou, S. J. Lee, F. Arisaka, T. Tsukihara, and M. Nakai. 2006. The asymmetric IscA homodimer with an exposed [2Fe-2S] cluster suggests the structural basis of the Fe-S cluster biosynthetic scaffold. J. Mol. Biol. 360:117-132. [DOI] [PubMed] [Google Scholar]
- 45.Muhandiram, D. R., G. Y. Xu, and L. E. Kay. 1993. An enhanced-sensitivity pure absorption gradient 4D 15N, 13C-edited NOESY experiment. J. Biomol. NMR 3:463-470. [Google Scholar]
- 46.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 47.Newton, I. L. G., T. Woyke, T. A. Auchtung, G. F. Dilly, R. J. Dutton, M. C. Fisher, K. M. Fontanez, E. Lau, F. J. Stewart, P. M. Richardson, K. W. Barry, E. Saunders, J. C. Detter, D. Wu, J. A. Eisen, and C. M. Cavanaugh. 2007. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315:998-1000. [DOI] [PubMed] [Google Scholar]
- 48.Nonaka, G., M. Blankschien, C. Herman, and C. A. Gross. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20:1776-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ollagnier-de-Choudens, S., T. Mattioli, Y. Takahashi, and M. Fontecave. 2001. Iron-sulfur cluster assembly. Characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 276:22604-22607. [DOI] [PubMed] [Google Scholar]
- 50.Pattaragulwanit, K., and C. Dahl. 1995. Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 164:217-222. [Google Scholar]
- 51.Pott, A. S., and C. Dahl. 1998. Sirohaem sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144:1881-1894. [DOI] [PubMed] [Google Scholar]
- 52.Prange, A., H. Engelhardt, H. G. Trüper, and C. Dahl. 2004. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT PCR. Arch. Microbiol. 182:165-174. [DOI] [PubMed] [Google Scholar]
- 53.Rethmeier, J., A. Rabenstein, M. Langer, and U. Fischer. 1997. Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high-performance liquid chromatography methods. J. Chromatogr. A 760:295-302. [Google Scholar]
- 54.Saio, T., H. Kumeta, K. Ogura, M. Yokochi, M. Asayama, E. Katoh, K. Teshima, and F. Inagaki. 2007. The cooperative role of OsCnfU-1A domain I and domain II in the iron-sulphur cluster transfer process as revealed by NMR. J. Biochem. 142:113-121. [DOI] [PubMed] [Google Scholar]
- 55.Sander, J., and C. Dahl. 2009. Metabolism of inorganic sulfur compounds in purple bacteria, p. 595-622. In C. N. Hunter, F. Daldal, M. C. Thurnauer, and J. T. Beatty (ed.), Purple bacteria. Springer, Dordrecht, The Netherlands.
- 56.Sander, J., S. Engels-Schwarzlose, and C. Dahl. 2006. Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 186:357-366. [DOI] [PubMed] [Google Scholar]
- 57.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 58.Schedel, M., M. Vanselow, and H. G. Trüper. 1979. Siroheme sulfite reductase isolated from Chromatium vinosum. Purification and investigation of some of its molecular and catalytic properties. Arch. Microbiol. 121:29-36. [Google Scholar]
- 59.Schwieters, C. D., J. J. Kuszewski, N. Tjandra, and G. M. Clore. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65-73. [DOI] [PubMed] [Google Scholar]
- 60.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 61.Smith, A. D., G. N. L. Jameson, P. C. Dos Santos, J. N. Agar, S. Naik, C. Krebs, J. Frazzon, D. R. Dean, B. H. Huynh, and M. K. Johnson. 2005. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44:12955-12969. [DOI] [PubMed] [Google Scholar]
- 62.Tokumoto, U., and Y. Takahashi. 2001. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J. Biochem. 130:63-71. [DOI] [PubMed] [Google Scholar]
- 63.Wada, K., Y. Hasegawa, Z. Gong, Y. Minami, K. Fukuyama, and Y. Takahashi. 2005. Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: implications for a functional dimer. FEBS Lett. 579:6543-6548. [DOI] [PubMed] [Google Scholar]
- 64.Wollenberg, M., C. Berndt, E. Bill, J. D. Schwenn, and A. Seidler. 2003. A dimer of the FeS cluster biosynthesis protein IscA from cyanobacteria binds a [2Fe2S] cluster between two protomers and transfers it to [2Fe2S] and [4Fe4S] apo proteins. Eur. J. Biochem. 270:1662-1671. [DOI] [PubMed] [Google Scholar]
- 65.Wu, G., S. S. Mansy, C. Hemann, R. Hille, K. K. Surerus, and J. A. Cowan. 2002. Iron-sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J. Biol. Inorg. Chem. 7:526-532. [DOI] [PubMed] [Google Scholar]
- 66.Xu, D., G. Liu, R. Xiao, T. Acton, S. Goldsmith-Fischman, B. Honig, G. T. Montelione, and T. Szyperski. 2004. NMR structure of the hypothetical protein AQ-1857 encoded by the Y157 gene from Aquifex aeolicus reveals a novel protein fold. Proteins 54:794-796. [DOI] [PubMed] [Google Scholar]
- 67.Yang, J., J. P. Bitoun, and H. Ding. 2006. Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J. Biol. Chem. 281:27956-27963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.