Abstract
Genes encoding the tail proteins of the temperate phage 16-3 of the symbiotic nitrogen-fixing bacterium Sinorhizobium meliloti 41 have been identified. First, a new host range gene, designated hII, was localized by using missense mutations. The corresponding protein was shown to be identical to the 85-kDa tail protein by determining its N-terminal sequence. Electron microscopic analysis showed that phage 16-3 possesses an icosahedral head and a long, noncontractile tail characteristic of the Siphoviridae. By using a lysogenic S. meliloti 41 strain, mutants with insertions in the putative tail region of the genome were constructed and virion morphology was examined after induction of the lytic cycle. Insertions in ORF017, ORF018a, ORF020, ORF021, the previously described h gene, and hII resulted in uninfectious head particles lacking tail structures, suggesting that the majority of the genes in this region are essential for tail formation. By using different bacterial mutants, it was also shown that not only the RkpM and RkpY proteins but also the RkpZ protein of the host takes part in the formation of the phage receptor. Results for the host range phage mutants and the receptor mutant bacteria suggest that the HII tail protein interacts with the capsular polysaccharide of the host and that the tail protein encoded by the original h gene recognizes a proteinaceous receptor.
The Sinorhizobium meliloti-Medicago symbiosis is an important model for endosymbiotic nitrogen fixation. The genome sequence of S. meliloti (strain 1021) has been established (14), and the Medicago truncatula genome is under intensive investigation (3). Phage 16-3 is a temperate, double-stranded DNA phage of S. meliloti strain 41. It is by far the best-studied rhizobiophage and serves as a tool in analyses of rhizobium genetics, in the isolation of some symbiotic mutants, and in the construction of special vectors. Genetic determinants and molecular mechanisms of many aspects of the 16-3 life cycle, such as phage integration and excision (8, 26, 38), regulation of the lytic/lysogenic switch (5, 6, 9, 24, 28), immunity to superinfection (4), phage DNA packaging (15), and the role of gene h in the host range (32), have been examined in detail. Moreover, the complete 60-kb phage genome sequence (accession no. DQ500118) has been determined recently (P. P. Papp et al., unpublished results). However, little is known about the genes and structural elements involved in the interaction between the phage and its host, and furthermore, only one study of the 16-3 virion proteins has been reported (11).
The initial interaction between a tailed phage and its bacterial host cell is mediated by the distal part of the phage tail, which specifically binds to the phage receptor located on the host surface. Earlier results demonstrated that phage 16-3 adsorption is connected to the strain-specific capsular polysaccharide of S. meliloti 41, the KR5 antigen. So far, three bacterial gene clusters involved in KR5 antigen production, including the rkp-1, rkp-2, and rkp-3 regions, have been described. rkp mutants are defective in the invasion of the host plant for symbiosis. In addition, they cannot adsorb phage 16-3, suggesting that the KR5 antigen is required for both functions (19, 20, 30).
In order to elucidate the molecular mechanism of phage 16-3 and S. meliloti 41 recognition, bacterial mutants carrying an altered phage receptor and host range phage mutants able to overcome the adsorption block have been characterized previously (32). It was shown that the RkpM protein, together with other yet uncharacterized elements, is a component of the phage receptor. With the use of rkpM mutants, host range mutations in phage gene h, which probably encodes the tail fiber protein, were identified. Interestingly, some mutations influencing phage-host recognition could not be localized in the rkpM and h genes, indicating that on both sides, additional components are important for bacteriophage-host recognition.
The aim of this study was to identify additional genetic determinants involved in S. meliloti 41 and phage 16-3 recognition by characterizing new host range and receptor mutants. Furthermore, by using insertional mutagenesis, we examined a region of the phage chromosome supposed to be responsible for tail formation and identified six new genes essential for phage assembly.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, phage techniques, and growth conditions.
Escherichia coli strains XL1-Blue (2), DH5α (16), and WA321 (10) were used for cloning procedures. S. meliloti strain 41 (isolate Rm41) and its exoB derivative AK631 (33) were used for bacteriophage propagation and genetic experiments. Phage strains 16-3ΔNC (9) and 16-3cti3 (24) were used as background strains for the isolation of host range and insertional mutants, respectively. Host range mutant h109 was isolated earlier on strain GH4180 (32). New host range mutants described in this study were isolated on different bacterial mutants by the same procedure: h182 was isolated on strain AT313 (AK631 rkpZ::Tn5) (20), while h842 and h843 were isolated on strain PP4073 (25).
Plasmids pBluescript II SK(+) (Stratagene, La Jolla, CA), pBBR1MCS-5 (21), and pPAG160 (15) were used for cloning. pCU999 was used as the source of the kanamycin resistance cassette (27). Cosmid clones pDH1 and pDH79 (23) and pDH114 (7) of the 16-3cti3 chromosome and pMW23, harboring the rkpZ gene of S. meliloti 41 (41), have been described elsewhere. In triparental matings, helper plasmid pRK2013 (12) was used for the transfer of pBBR1MCS-5 and pLAFR1 cosmid clones, and pCU101 (39) was used for the transfer of pPAG160 derivatives. Media, antibiotic concentrations, and culture conditions for E. coli, S. meliloti, and phage 16-3 have been described previously (34, 35).
DNA procedures, sequence determination, and bioinformatics.
Basic DNA manipulation procedures including DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation of E. coli were performed according to standard techniques (37) or as recommended by the material suppliers.
To determine the nucleotide sequences encompassing the mutations in h109, h182, and h843, the 1.5-kb-long EcoRI-L fragments were amplified using primers L1U (ACTGGATGTTCGCTGGTTTG) and L1L (GGTAGGCACTGTCCGTCTCG). Sequences of both DNA strands were determined directly by using these PCR primers and four additional primers: L2U (CTCTCCAATAAAACGACAGC), L3U (GATAAAGGTCAAAGGTCTCG), L2L (GATACCATACTCTTCGTTAGC), and L3L (AGTTGTAGATCACCACCGAG). Primers corresponding to the previously described h gene (32) were used to examine the mutation in phage h842. DNA sequence determination was done with the BigDye Terminator kit and an Applied Biosystems 373A sequencer (Perkin Elmer, Wellesley, MA).
For bioinformatic analyses of the predicted proteins, the following tools were used: the BLAST server (blast.ncbi.nlm.nih.gov), the HHpred server (toolkit.tuebingen.mpg.de/hhpred), and BetaWrap prediction (groups.csail.mit.edu/cb/betawrap/).
Marker rescue analysis.
In order to localize the temperature-sensitive (ts) mutations in phages h109 and the h843, cosmid clones pDH1, pDH79, and pDH114 (7, 23) and pBBR1MCS-5 derivatives harboring the EcoRI-C, EcoRI-D, EcoRI-L, and EcoRI-H fragments of the 16-3cti3 phage chromosome were used (Fig. 1). To exclude the effect of immX, which provides immunity against superinfection with homoimmune phages (4), mutant derivatives of pDH1 and pDH79 cosmid clones were created by inserting a kanamycin cassette into the gene immXI via homologous recombination. The clones described above were introduced into S. meliloti 41 (Rm41) by triparental conjugation. Mutant phages h109 and h843 were propagated by one-step growth on these transconjugants. Phage progeny were plated onto an Rm41 lawn and incubated at a restrictive temperature (37°C) in order to count wild-type recombinants (and revertants). The frequency of reversion was detected by propagating the mutant phages on Rm41 harboring empty vectors at 37°C. Representatives of the ts revertants were scored and tested for host range. As expected from the ts nature of the alleles in h109 and h843, all ts revertants showed a wild-type host range.
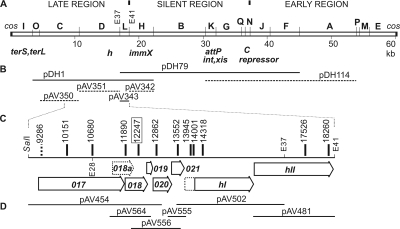
FIG. 1.
Mutational analysis of the 16-3 phage tail region. (A) EcoRI restriction map of the 16-3 phage genome. The functions of known gene products are indicated below the map. EcoRI restriction site numbers (E37 and E41) and alphabetic designations of fragments refer to physical map positions on the 16-3 chromosome (9). (B) Fragments used for marker rescue in h109 and h843 host range phage mutants. Solid lines represent fragments where marker rescue occurred, and broken lines represent fragments where marker rescue was not detected. (C) Tail region of the genome with ORF017 to the hII gene. Vertical bars represent positions of the insertions isolated in this study. The different markings reflect the distinct phenotypes corresponding to the insertions (see the text). Numbers show the exact coordinates of the insertions according to the 16-3 phage genome sequence. (D) Fragments used for genetic complementation of the insertional mutations.
Temperature shift experiments.
Cultures and lysates were warmed up to the restrictive temperature (37°C), and approximately 105 PFU of h109 phage particles was added to 2 × 108 CFU of Rm41 bacteria. Cultures were shaken at the restrictive temperature and were cooled down to the permissive temperature (25°C) at different time points. Phages were propagated for 180 min altogether to complete the life cycle. The number of phage progeny was compared to the number of phages in the control lysate, which was grown for 180 min at 25°C.
Construction of insertional mutants of phage 16-3.
EcoRI-C, EcoRI-D, and EcoRI-L DNA fragments of the 16-3 phage genome (Fig. 1) were cloned into the pBluescript II SK(+) vector and used for insertional mutagenesis. All but one insertion was constructed in vitro by using MuA transposase with the kanamycin resistance transposon Entranceposon F779 from the Template Generation System II (TGS II) kit (Finnzymes, Espoo, Finland). The approximate position of each insertion was determined by PCR using the MuEnd primer from the kit and the M13 reverse primer specific to pBluescript II SK(+). The correct positions of the selected insertions were determined by DNA sequencing with SeqE or SeqW primers from the TGS II kit. The mutation at position 10680 was constructed by inserting a 1.2-kb-long kanamycin cassette, derived from pCU999 (27), directly into the EcoRI(28) site.
DNA fragments carrying the insertions were recloned into pPAG160, harboring a spectinomycin resistance marker. The plasmid derivatives were introduced by triparental mating into a lysogenic Rm41 strain harboring a temperature-inducible prophage (16-3cti3). Since pPAG160 is unable to replicate in S. meliloti, kanamycin-resistant but spectinomycin-sensitive colonies should have acquired the mutations by homologous recombination. The absence of the vector sequences was tested by PCRs with primer pairs GCTTGCGAGGGTGCTACTTA/CGGTTACGAGATCCATTTGCT and GCGCGATTTTGCCGGTTACT/GATGTTACGCAGCAGGGCAGTC, specific for the pSC101 region and the omega fragment of pPAG160, respectively. Integration of the mutations was verified by PCR using primers specific for the Entranceposon and for the given open reading frame (ORF) (data not shown). Excision of prophage mutants was induced at 37°C for 30 min, and phage maturation was completed by incubation of the cultures for an additional 180 min at 28°C.
Plasmid construction for genetic complementation experiments.
Derivatives of pBBR1MCS-5 containing different fragments of the 16-3cti3 chromosome were as follows: pAV454, carrying the 4,315-bp-long SalI fragment (including ORF017, ORF018a, ORF018, and ORF019); pAV564, carrying the 1,284-bp-long ClaI-PvuII fragment (including ORF018a and ORF018); pAV556, carrying the 1,565-bp-long NotI-SphI fragment (including ORF019 and ORF020); pAV555, carrying the 895-bp-long SalI fragment (including ORF021); pAV502, carrying the previously identified h gene on the 3,199-bp-long fragment from the SphI site to EcoRI(37) site; pAV481, carrying the 2,580-bp-long fragment from primer hterU1 (GGTACCCCACCCGCCGACAGC) to EcoRI(41); and pAV482, carrying the 2,538-bp-long fragment from primer hterU2 (GGTACCGCGCGGGCTTTTTCTTTA) to EcoRI(41) (both fragments in plasmids pAV481 and pAV482 cover the gene herein designated hII). Underlining in sequences of primers hterU1 and hterU2 shows an extra KpnI site for cloning (Fig. 1D).
Purification of phage particles and protein techniques.
Purification of bacteriophages by cesium chloride step gradient centrifugation was performed by conventional methods (37). Purified phage samples were dialyzed against SM buffer (100 mM NaCl, 20 mM Tris-Cl, 5 mM MgSO4, 1 mM CaCl2, pH 7.5). DNA contents of the purified phage particles were assayed by agarose gel electrophoresis.
Protein separation was carried out with a NuPage gel electrophoresis system according to the protocols of Invitrogen (Carlsbad, CA). Purified, dialyzed phage samples were mixed with sample buffer and reducing agent, boiled for 10 min, and loaded into NuPage Novex bis-Tris-4 to 12% polyacrylamide gels. Electrophoresis was performed at a constant voltage of 200 V with either MES (morpholineethanesulfonic acid)- or MOPS (morpholinepropanesulfonic acid)-sodium dodecyl sulfate (SDS) running buffer. Protein bands were visualized by SimpleBlue staining. Western blotting was performed with phage proteins separated by electrophoresis and subsequently electroblotted onto polyvinylidene difluoride membranes by using an XCell II blot module and NuPage transfer buffer. N-terminal amino acid sequencing was performed with an Applied Biosystems (model 471) protein sequencer using Edman degradation chemistry.
Electron microscopy methods.
Formvar-coated copper grids (300/400 mesh) were floated on drops of CsCl-purified, dialyzed phage samples for 5 min, excess fluid was carefully removed with the edge of a filter paper, and the preparations were negatively stained with 3% (wt/vol) phosphotungstic acid (PTA) solution, pH 6.6, for 40 s. Excess PTA was removed, and the remainder was left to air dry. The preparations were examined on a JEOL 1200 EXII transmission electron microscope with an accelerating voltage of 80 kV.
RESULTS
The hII gene represents a new host range locus.
In a previous study, we have shown that in addition to the h gene, there is at least one other gene influencing host recognition since one host range mutation (that in mutant h109) could not be localized within the h gene (32). To delimit the location of the h109 mutation on the physical map of the phage genome, marker rescue experiments were carried out. We have observed that the h109 mutant is temperature sensitive, producing minute, hardly visible plaques at 37°C but forming normal, wild-type plaques at 28°C. Therefore, the emergence of recombinant wild-type phages at the restrictive temperature can be easily detected. In marker rescue experiments, mutant phages were propagated by single-step growth on S. meliloti 41 (Rm41) transconjugants that harbored different fragments of the 16-3 chromosome (Fig. 1B). When wild-type phages appeared at least two orders of magnitude more frequently than those in the control population (recombinants versus revertants), the cloned fragment was considered to carry the wild-type sequence of the mutated region. The shortest DNA fragment that resulted in wild-type recombinants was the 1.5-kb-long EcoRI-L fragment of the phage genome (pAV343 in Fig. 1B). This region covers the majority of the 2,412-bp-long coding region designated ORF023. After PCR amplification, the nucleotide sequence of the h109 allele was determined. One missense mutation resulting in the substitution D783N was detected in the coding frame for the putative protein (Fig. 1C and Table 1). This new host range locus was designated hII. The original h locus is hereinafter referred to as hI.
TABLE 1.
Characteristics of phage 16-3 host range mutantsa
| Phage strain | Phage genotype or mutated gene (amino acid change) | Propagation efficiencyd on: |
||||
|---|---|---|---|---|---|---|
| Rm41 (wild type) | GH4046 [rkpM (L252F)] | PP4073 [rkpY (L552P)] | GH4180 (rkpZ) | AT313 (rkpZ::Tn5) | ||
| 16-3 | Wild type | ++ | − | − | + | + |
| h5 | hIb (G588D) | ++ | +* | + | ++ | ++ |
| h105 | hIb (G588D) | ++ | + | + | ++* | ++ |
| h842 (ts) | hIb (G588V) | ++ | ++ | ++* | ++ | ++ |
| h182 | hIIc (N666K) | ++ | − | + | ++ | ++* |
| h109 (ts) | hIIc (D783N) | ++ | − | + | ++* | ++ |
| h843 (ts) | hIIc (D783G) | ++ | − | +* | ++ | ++ |
References for and additional information on bacterial and phage strains are given in the text.
hI (ORF022) is 2,112 bp long; mutations affected the GGC codon from bp 1762 to 1764. Mutants have a GAC or GTC triplet.
hII (ORF023) is 2,412 bp long; mutations affected either the AAC codon from bp 1996 to 1998, with mutant h182 having an AAA triplet, or the codon GAC from bp 2347 to 2349, with mutants h109 and h843 having AAC and GGC codons, respectively.
*, the host range mutant was isolated on bacteria of the indicated strain; ++, the mutant formed large, sharp-contoured plaques, with spots of confluent lysis; +, the mutant formed small plaques with pale spots; −, no plaque formation occurred.
hII host range mutants can also be isolated on rkpZ::Tn5 bacteria.
Host range phage mutant h109 was isolated earlier on GH4180, a receptor mutant of strain Rm41 (32). Genetic complementation experiments showed that the mutation in GH4180 is located in the rkp-3 region, harboring genes for capsular polysaccharide biosynthesis. According to the deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE) analysis, the capsular polysaccharide pattern of strain GH4180 was similar to that of the rkpZ mutants. In further complementation experiments, we have shown that the wild-type allele of rkpZ on plasmid pMW23 was able to convert the phage resistance of GH4180 to phage sensitivity, suggesting that the mutation in this strain affects the rkpZ gene. This finding implies that the rkpZ gene product has a role in 16-3 phage adsorption.
Earlier studies indicated that most mutants with Tn5 transposon insertions in the rkp genes are unable to bind phage 16-3 and are not suitable for the isolation of host range phage mutants (19, 32). Interestingly, the rkpZ::Tn5 insertional mutant showed a distinct phenotype. The wild-type phage was able to infect this strain very weakly, but host range mutants produced large plaques. To examine whether the new host range mutations are located in the hII gene, the nucleotide sequence of the EcoRI-L fragment of a representative mutant (h182) was determined. Similar to mutant h109, mutant h182 had a missense mutation localized in hII but at a different position from that in h109. The codon AAC was changed to AAA at bp 1998, resulting in the N666K substitution (Table 1).
Additional mutations affecting phage-host recognition.
A missense mutation in host gene rkpY (yielding strain PP4073) that also allows the isolation of host range phage mutants was identified recently (25). Several phage mutants were isolated on this strain and tested with bacterial receptor mutants. These phages showed two different phenotypes: (i) some of them, represented by h842, were able to infect all the bacterial receptor mutants; (ii) another group, represented by h843, were not able to propagate on strain GH4046 and were temperature sensitive (Table 1). Thus, with respect to the host range pattern, mutant h842 resembled the previously described hI gene mutant h5 (32) and mutant h843 resembled hII gene mutant h109 (characterized above) (Table 1; Fig. 1). Therefore, the nucleotide sequences of gene hI from mutant h842 and gene hII from mutant h843 were determined. The new mutations in the hI and hII genes (in mutants h842 and h843) affected the same codons as the mutations in the prototype mutants h5 and h109, respectively. However, in both cases, these mutations resulted in different base substitutions and hence led to different codons and amino acid substitutions (Table 1).
The hII gene encodes a structural protein.
The hII locus was identified as a functional gene by using host range mutations as described above. The putative 803-amino-acid-long HII protein showed no significant homology to any other proteins with known functions when analyzed using the BLAST server. However, by using the HHpred tool, a well-defined part of the HII protein (from amino acid residue 300 to the end) was found to show strong homology to sugar interaction proteins (40 entries had probabilities of over 90%). All of these proteins display a parallel-beta-helix structure. The beta-helical fold seems to be a feature of HII as well, indicating that it may interact with the host surface capsular polysaccharide.
The predicted molecular mass of HII is about 85 kDa. A protein with a similar molecular mass in purified samples was identified earlier as a tail component of phage 16-3 (11). To establish whether the detected protein corresponds to HII, phage proteins were separated by SDS-PAGE and the N-terminal sequence of the appropriate band was determined. As expected, the resulting amino acid sequence (AITAAEAFRDY) was the same as that encoded after the putative start codon.
To get more information about the role of protein HII, temperature shift experiments were carried out using host range mutant phage h109. We aimed to investigate when the restrictive temperature (37°C) affects the propagation of mutants. Since h109 is a host range mutant, the first question raised was whether the adsorption event is blocked at 37°C. Interestingly, in temperature shift experiments, we found that this mutant was not sensitive to the restrictive temperature for the first 120 min of the phage life cycle, which includes the adsorption of the phage particles to the host cells. The phage yield was dramatically decreased when the restrictive temperature was applied for more than 120 min. When the heat treatment was extended over 180 min, the yield of the mutant phages was 1% of that of the mutants cultured at 25°C (for one entire life cycle at 25°C, a minimum of 135 min is required). When maturated h109 phage particles were incubated at 37°C, no effect of the elevated temperature was detected, suggesting that the assembled tail structure of the mutant phage is no longer temperature sensitive. These results indicate a role for HII protein in a late event in virus morphogenesis.
Identification of functional genes in the tail region.
The complete sequence of the phage 16-3 genome was deposited recently in the nucleotide databases under accession number DQ500118 (Papp et al., unpublished). Upstream of genes hI and hII, five additional genes (ORF017 to ORF021) (Fig. 1C) that were suspected to encode the proteins of the tail structure were predicted. This part of the genome is referred to as the tail region. In order to identify tail structural and tail assembly protein genes, this region was selected for directed insertional mutagenesis.
Phage 16-3 mutants were obtained by introducing insertions with a kanamycin resistance marker into each of the selected ORFs by homologous recombination. For this purpose, a lysogenic Rm41 strain harboring a thermoinducible prophage, 16-3cti3, was used. Into this background, derivatives of the pPAG160 vector carrying insertions in the putative genes were introduced. Since pPAG160 is unable to replicate in S. meliloti, kanamycin-resistant exconjugants represent double-crossover products (i.e., products of allelic replacement by the Km-tagged allele) or cointegrants (i.e., products of single crossovers by Campbell recombination) (see details in Materials and Methods). The positions of the integrated mutations and the absence of plasmid sequences were verified by different PCR experiments, and the mutants which carried the Km-tagged allelic replacements (Fig. 1C) were kept for further investigations. No insertion in the 159-bp ORF019 was isolated.
In order to determine whether insertions influence phage assembly and infection efficiency, lysogenic bacterial cultures were induced by heat shock. The titers of the lysates on strain Rm41 were determined, and the lysates were examined by electron microscopy. Results are summarized in Fig. 1C and 2.
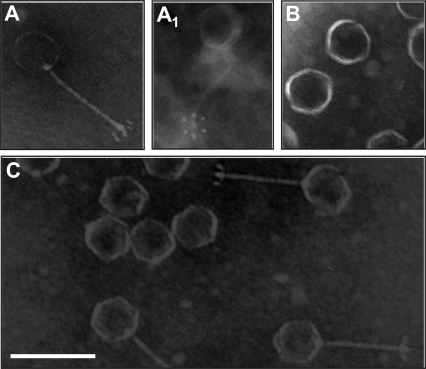
FIG. 2.
Transmission electron micrographs of phages negatively stained with PTA. (A and A1) The wild-type 16-3 phage consists of an isometric head connected to a long, flexible noncontractile tail tube. At the distal end of the tail, a baseplate structure with six small fibers is observed. (B) Representative insertion mutants showing head structures without tails. (C) A mixture of wild-type 16-3 phage and tail-less insertion mutants. The scale bar represents 100 nm.
The phage carrying a mutation at position 9286 (mutations are hereinafter referred to by position numbers) did not show any obvious differences in titer and plaque morphology from the parent strain. We could detect only very few PFU (<103 PFU/ml versus the wild-type phage yield of 1010 PFU/ml) from lysates of the mutant prophages carrying insertions 10151, 10680, 11890, 12247, 12862, 13552, 13945, 14001, 14318, 17526, and 18260. These phage particles resulted in Kms lysogenic bacteria, and no insertion could be detected by PCR, suggesting that all of the plaque-forming phages were revertants lacking the mutation. Thus, the above-listed mutations blocked the appearance of infectious phage particles.
To determine whether all ORFs represent functional genes and to exclude the possibility that an insertion influences only the expression of downstream genes by polar effects, genetic complementation experiments were carried out. Plasmid derivatives harboring different fragments of the tail region (Fig. 1D) were introduced into lysogenic bacteria containing one of the insertional mutations in the 16-3cti3 phage chromosome described above. Phage progeny from heat-induced single-step lysates were plated onto two different strain samples: (i) an Rm41 lawn was used as a control to detect wild-type recombinants, and (ii) a lawn of Rm41 containing the given subfragment of the tail region was used to observe plaque-forming mutant phage particles which assembled due to complementation and still carried the lethal mutation. All of the mutations could be complemented by the subfragment that carries the given ORF (Fig. 1D), suggesting that ORFs represent functional genes essential for phage viability.
Insertions in the tail region result in tail-less phage particles.
In order to determine in which stage phage assembly is blocked for the insertion mutants, the lysates from the heat-induced lysogenic bacteria described above were investigated by electron microscopy. Particles were collected by CsCl step gradient centrifugation, and the purified phages were stained negatively with PTA. Analysis of the wild-type phage showed that the 16-3 virion has an isometric head about 55 nm in diameter and a flexible, noncontractile tail about 97 nm long, ending in a baseplate to which six club-shaped spikes are attached (Fig. 2). Because of these characteristics, the 16-3 phage can be classified in the family Siphoviridae. The morphology of the phage carrying mutation 9286 was indistinguishable from that of the wild type, suggesting that insertion in the 5′ part of ORF017 has no any effect on phage assembly. The other insertions in ORF017 (insertions 10151 and 10680), as well as insertions in ORF018 (insertions 11890 and 12247), ORF020 (insertion 12862), ORF021 (insertion 13552), hI (insertions 13945, 14001, and 14318), and hII (insertions 17526 and 18260), resulted in serious assembly defects. On electron micrographs, only head structures lacking any tail structure could be detected for all of these mutants except that carrying insertion 12247, indicating that the affected genes are essential for phage tail formation (Fig. 2). We have also established that the heads of these mutants contained DNA (data not shown). In contrast to the mutant with insertion 11890, the lysate from the mutant with insertion 12247 contained no head structures, although both mutations were localized in ORF018. By analyzing the DNA sequence, an additional ORF (ORF018a) that was not recognized earlier was localized in this region (Fig. 1C). The putative new gene (CDS 11503. .12171) started in ORF017 and covered the 5′ end of ORF018. It is possible that insertion 11890 blocks only the production of gp018a and that insertion 12247 arrests the production of gp018 protein, which is essential for head assembly.
DISCUSSION
Recently, the DNA sequence of the entire 16-3 phage genome (accession number DQ500118) was determined. Homologues of a major portion of the 16-3 late genes (from ORF007 to ORF023) have been found in the S. medicae WSM419 genome, but no other related genes (or proteins) in the databases for these putative genes were detected. The majority of information in this area is largely speculative and is based primarily on sequence comparisons with completed phage genomes. In order to know more about the tail genes and the determinants of host range, we isolated mutant phages and analyzed them with respect to infection efficiency and morphology.
Based on morphological characteristics, phage 16-3 belongs to the Siphoviridae family. Insertional mutations in ORF017, ORF020, ORF021, hI, and hII resulted in the development of head structures only. No attached tails were observed, and the mutant particles were unable to infect the host. Therefore, we concluded that all of these ORFs represent functional genes essential for tail formation. In the case of insertion 12247 in ORF018, no virions were detected in lysates; therefore, gp018 must be essential for phage head formation or must have a chaperone-like function important for at least head assembly.
Our results suggest the existence of an additional gene, ORF018a, that is also essential for tail formation. Mutation 11890 is located in the middle of ORF018a and at the same time at the very beginning of ORF018 (Fig. 1C). Since another mutation (12247) influenced only ORF018 and resulted in a more severe defect, we hypothesize that the insertion at 11890 blocks the production of gp018a but not gp018. It is possible that either (i) the coding region for gp018 starts after the insertion or (ii) the insertion allows the production of a truncated but still functional protein.
The mutant with insertion 9286 (which is 7 codons downstream of the predicted start codon of ORF017) could infect the host with the same efficiency as the wild-type phage, and no obvious differences in plaque morphology and virion morphology were observed. This may indicate that the N-terminal part of gp017 is not essential or that translation begins at a start codon distal to the insertion.
Insertions 13945 and 14001 were localized upstream of the first ATG of the coding frame of gene hI, supporting our earlier prediction that this gene has an unusual upstream start codon (32).
In the light of the ever-expanding pool of completed phage genome sequences, the organization of the structural genes of phage 16-3 classifies it among the lambdoids. In 1998, Lucchini et al. (22) proposed that the gene map of phage λ could be used to predict gene functions in other, related phages. The λ tail genes are subgrouped according to the distinct phases of tail morphogenesis (17). Genes v to t are responsible for tail tube formation. Genes h to j code for proteins involved in the initiator complex necessary to launch tail assembly, among which tape measure protein (H) regulates tail length. The stf and tfa genes encode nonessential long tail fibers (17). Considering the pattern of λ tail assembly, we may speculate that ORF017, ORF018a, ORF020, ORF021, hI, and hII of the 16-3 phage encode proteins that form the initiator complex and that, as in λ, a mutational block in any one of these genes will restrict tail assembly (17).
To investigate phage-host recognition, we previously isolated receptor mutant bacteria and host range phage mutants and identified the phage receptor-forming protein, RkpM, and the HI protein, involved in host recognition (32). Besides rkpM, we have recently described a second host gene, rkpY, of which a special allele blocked infection with the wild-type phage but allowed the isolation of host range mutants (25). Here, we identify a third host gene, rkpZ, that also influences infection with phage 16-3. rkpZ mutants show chain length modification of KR5 antigen (36). Contrary to the roles of RkpM and RkpY, the role of RkpZ in phage adsorption/infection may be indirect, since a Tn5 insertional mutant (AT313), as well as a spontaneous mutant (strain GH4180), was suitable for the isolation of host range phage mutants. It is likely that RkpZ protein does not take part in 16-3 receptor formation but influences phage adsorption through its effect on capsular polysaccharide production.
All of the 16-3 host range mutations isolated so far resulted in amino acid residue substitutions in protein HI and in the newly identified host interaction protein HII. Host range mutant phages affected in genes hI and hII show different host specificities. Strain GH4046, an rkpM bacterial mutant, was a suitable host for the isolation of hI mutants but not for the isolation of hII mutants. In contrast, both hI and hII mutant phages could be isolated on rkpZ (strain GH4180) and rkpY (strain PP4073) receptor mutants.
We suppose that this difference may be caused by the conspicuously different capsular polysaccharide surfaces of the bacterial mutants. Both rkpZ and rkpY mutants possess an altered capsule, while rkpM mutants produce no detectable KR5 antigen (25, 32, 36). Earlier results indicated the role of this structure in 16-3 phage adsorption. On purified bacterial envelope, adsorption could be abolished by β-glucosidase and β-glucuronidase but not by protease K treatment (34). The proportion of wild-type phages adsorbed by the rkpM mutant GH4046, which produced no KR5 antigen, was below 50%, but the proportion adsorbed by strains that showed altered antigen production (PP4073 and GH4180) was above 95% (V. Deák, unpublished results). Based on these data, it seems likely that KR5 is involved in initial polysaccharide-controlled phage binding, which is followed by a secondary binding step that is protein (RkpM and RkpY) dependent. Presumably, hII host range mutants penetrate more efficiently into the altered S. meliloti 41 capsule than the wild-type phages (and reach the proteinaceous receptor), while host range mutations in hI may result in stronger interactions between phage and bacterial protein partners than those occurring in the presence of wild-type hI. In other words, h109, h843, and h182 mutants are affected in protein-polysaccharide binding while h5, h105, and h842 are affected in protein-protein binding.
The bioinformatic analyses of the corresponding protein sequences support this idea. The C-terminal part of the HII protein shows strong homology to sugar interaction proteins displaying a parallel-beta-helix structure (e.g., Bacillus phage PHI29 preneck appendage protein [Protein Data Bank identification number 3GQ8]) (42). Proteins with parallel beta-helices come under a distinctive subfamily of beta-sheet proteins, first identified among the pectate lyases used by Erwinia species to infect plant cells (43). Bradley et al. proposed that the function of the beta-helical fold is to generate a long lateral surface for reading the sequences of polysaccharides (1). Beta-helices are also characteristic of bacteriophage T4 short tail fiber and bacteriophage P22 tail spike proteins (for a review, see reference 40).
We also analyzed the previously described HI protein by using HHpred. According to these results, the C-terminal part of HI (from amino acid residue 500 to 703) shows strong homology to different types of cell adhesion and fusion proteins which have a common beta-sandwich-folded fibronectin type III domain crucial for protein-protein interactions (13). We note that host range mutations result in the substitution G588D or G588V within this domain (Table 1).
We provide direct evidence (obtained by N-terminal amino acid sequencing) that the newly identified host interaction protein HII is present in the phage particle. The host range missense mutation in phage h109 results in a temperature-sensitive allele of hII. Temperature shift experiments showed that neither adsorption nor the early life cycle was temperature sensitive. Moreover, preheating at the restrictive temperature did not block the adsorption of h109 phage particles. Thus, elevated temperature does not alter previously formed protein complexes. Based on these results, it seems likely that the part of the HII protein affected by the ts mutation not only is crucial for the assembly of phage tails but also is involved directly in the protein-polysaccharide communication in the adsorption process.
By using bacteriophage λ as a model, it may be possible to assign a function to the protein encoded by ORF017, which is the largest ORF in the 16-3 genome. It is located at the same position as the h tail tape measure protein gene of λ. By in silico analysis of gp017, an endolysin domain (between amino acids 409 and 497) and a glycine-rich region (between amino acids 716 and 797 there are 26 glycine residues) were predicted. The endolysin enzymes found in many double-stranded DNA phages function to cleave the glycosidic beta-1,4 bonds between the N-acetylmuramic acid and the N-acetylglucosamine of the peptidoglycan in bacterial cell walls. It is also a widespread pattern for the cell wall-degrading activity to be associated with structural proteins in phages. Kenny et al. characterized the Tal2009 protein at the tip of the tail of lactococcal phage Tuc2009 and found it to have cell wall-degrading activity (18). Piuri and Hatfull identified a peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein (31). The glycine-rich region encoded by ORF017 is consistent with this hypothesis, since this region is the target of the proteolytic cleavage, which activates the endolysin activities of other tail proteins (17, 18, 31). These findings suggest that given its location and sequence features, ORF017 may encode a tape measure protein. These kinds of proteins are responsible for determining tail length in lambdoids (17, 29).
Acknowledgments
We thank A. Holczinger and S. Semsey for advice in preparation of the manuscript. We are grateful to L. Seress and R. Gábriel for their help in electron microscopy and to C. Sántha, M. Miklósvári, and J. Keidl for their skillful technical assistance.
This work was supported by grant OTKA T038377.
Footnotes
Published ahead of print on 14 January 2010.
REFERENCES
- 1.Bradley, P., L. Cowen, M. Menke, J. King, and B. Berger. 2001. BETAWRAP: successful prediction of parallel beta-helices from primary sequence reveals an association with many microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 98:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Biotechniques 5:376-379. [Google Scholar]
- 3.Cannon, S. B., L. Sterck, S. Rombauts, S. Sato, F. Cheung, J. P. Gouzy, X. Wang, J. Mudge, J. Vasdewani, T. Schiex, M. Spannagl, E. Monaghan, C. Nicholson, S. J. Humphray, H. Schoof, K. F. X. Mayer, J. Rogers, F. Quetier, G. E. Oldroyd, F. Debelle, D. R. Cook, B. A. Roe, C. D. Town, S. Tabata, Y. Van de Peer, and N. D. Young. 2006. Legume evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. U. S. A. 103:14959-14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csiszovszki, Z., Z. Buzas, S. Semsey, T. Ponyi, P. Papp, and L. Orosz. 2003. immX immunity region of Rhizobium phage 16-3: two overlapping cistrons of repressor function. J. Bacteriol. 185:4382-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallmann, G., F. Marincs, P. Papp, M. Gaszner, and L. Orosz. 1991. The isolated N-terminal DNA binding domain of the c repressor of bacteriophage 16-3 is functional in DNA binding in vivo and in vitro. Mol. Gen. Genet. 227:106-112. [DOI] [PubMed] [Google Scholar]
- 6.Dallmann, G., P. Papp, and L. Orosz. 1987. Related repressor specificity of unrelated phages. Nature 330:398-401. [Google Scholar]
- 7.Dorgai, L., F. Olasz, M. Berenyi, G. Dallmann, A. Pay, and L. Orosz. 1981. Orientation of the genetic and physical map of Rhizobium meliloti temperate phage 16-3. Mol. Gen. Genet. 182:321-325. [Google Scholar]
- 8.Dorgai, L., I. Papp, P. Papp, M. Kalman, and L. Orosz. 1993. Nucleotide sequences of the sites involved in the integration of phage 16-3 of Rhizobium meliloti 41. Nucleic Acids Res. 21:1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorgai, L., G. Polner, E. Jonas, N. Garamszegi, Z. Ascher, A. Pay, G. Dallmann, and L. Orosz. 1983. The detailed physical map of the temperate phage 16-3 of Rhizobium meliloti 41. Mol. Gen. Genet. 191:430-433. [DOI] [PubMed] [Google Scholar]
- 10.Dreiseikelmann, B., R. Eichenlaub, and W. Wackernagel. 1979. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim. Biophys. Acta 562:418-428. [DOI] [PubMed] [Google Scholar]
- 11.Erdei, S., B. Dudas, L. Orosz, and E. Duda. 1982. Identification of structural proteins of Rhizobium meliloti temperate phage 16-3. J. Gen. Virol. 62:145-152. [DOI] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, J. S., Z. Yu, K. L. Maxwell, and A. R. Davidson. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359:496-507. [DOI] [PubMed] [Google Scholar]
- 14.Galibert, F., T. Finan, S. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. Davis, S. Dreano, N. Federspiel, R. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. Hyman, T. Jones, D. Kahn, M. Kahn, S. Kalman, D. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. Peck, T. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. Vorholter, S. Weidner, D. Wells, K. Wong, K. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 15.Ganyu, A., Z. Csiszovszki, T. Ponyi, A. Kern, Z. Buzas, L. Orosz, and P. P. Papp. 2005. Identification of cohesive ends and genes encoding the terminase of phage 16-3. J. Bacteriol. 187:2526-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Katsura, I. 1990. Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 26:1-18. [DOI] [PubMed] [Google Scholar]
- 18.Kenny, J. G., S. Mc Grath, G. F. Fitzgerald, and D. van Sinderen. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kereszt, A., E. Kiss, B. Reuhs, R. W. Carlson, A. Kondorosi, and P. Putnoky. 1998. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkpK gene encodes a UDP-glucose dehydrogenase. J. Bacteriol. 180:5426-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss, E., A. Kereszt, F. Barta, S. Stephens, B. L. Reuhs, A. Kondorosi, and P. Putnoky. 2001. The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol. Plant Microbe Interact. 14:1395-1403. [DOI] [PubMed] [Google Scholar]
- 21.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 22.Lucchini, S., F. Desiere, and H. Brüssow. 1998. The structural gene module in Streptococcus thermophilus bacteriophage phi Sfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology 246:63-73. [DOI] [PubMed] [Google Scholar]
- 23.Olasz, F., L. Dorgai, P. Papp, E. Kosa, and L. Orosz. 1985. On the site-specific recombination of phage 16-3 of Rhizobium meliloti: identification of genetic elements and att recombinations. Mol. Gen. Genet. 201:289-295. [Google Scholar]
- 24.Orosz, L., and T. Sik. 1970. Genetic mapping of rhizobiophage 16-3. Acta Microbiol. Acad. Sci. Hung. 17:185-194. [PubMed] [Google Scholar]
- 25.Pálvölgyi, A., V. Deák, V. Poinsot, T. Nagy, E. Nagy, I. Kerepesi, and P. Putnoky. 2009. Genetic analysis of the rkp-3 gene region in Sinorhizobium meliloti 41: rkpY directs capsular polysaccharide synthesis to KR5 antigen production. Mol. Plant Microbe Interact. 22:1422-1430. [DOI] [PubMed] [Google Scholar]
- 26.Papp, I., L. Dorgai, P. Papp, E. Jonas, F. Olasz, and L. Orosz. 1993. The bacterial attachment site of the temperate Rhizobium phage 16-3 overlaps the 3′ end of a putative proline tRNA gene. Mol. Gen. Genet. 240:258-264. [DOI] [PubMed] [Google Scholar]
- 27.Papp, P. P., and V. N. Iyer. 1995. Determination of the binding sites of RepA, a replication initiator protein of the basic replicon of the IncN group plasmid pCU1. J. Mol. Biol. 246:595-608. [DOI] [PubMed] [Google Scholar]
- 28.Papp, P. P., T. Nagy, S. Ferenczi, P. Elo, Z. Csiszovszki, Z. Buzas, A. Patthy, and L. Orosz. 2002. Binding sites of different geometries for the 16-3 phage repressor. Proc. Natl. Acad. Sci. U. S. A. 99:8790-8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen, M., S. Ostergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 30.Petrovics, G., P. Putnoky, B. Reuhs, J. Kim, T. A. Thorp, K. D. Noel, R. W. Carlson, and A. Kondorosi. 1993. The presence of a novel type of surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol. Microbiol. 8:1083-1094. [DOI] [PubMed] [Google Scholar]
- 31.Piuri, M., and G. F. Hatfull. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putnoky, P., V. Deák, K. Békási, A. Pálvölgyi, A. Maász, Z. Palágyi, G. Hoffmann, and I. Kerepesi. 2004. H protein of bacteriophage 16-3 and RkpM protein of Sinorhizobium meliloti 41 are involved in phage adsorbtion. J. Bacteriol. 186:1591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putnoky, P., E. Grosskopf, D. T. C. Ha, G. B. Kiss, and A. Kondorosi. 1988. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. J. Cell Biol. 106:597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putnoky, P., and A. Kondorosi. 1986. Two gene clusters of Rhizobium meliloti code for early essential nodulation functions and a third influences nodulation efficiency. J. Bacteriol. 167:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Putnoky, P., G. Petrovics, A. Kereszt, E. Grosskopf, D. T. C. Ha, Z. Banfalvi, and A. Kondorosi. 1990. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J. Bacteriol. 172:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuhs, B. L., M. N. Williams, J. S. Kim, R. W. Carlson, and F. Cote. 1995. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Semsey, S., I. Papp, Z. Buzas, A. Patthy, L. Orosz, and P. Papp. 1999. Identification of site-specific recombination genes int and xis of the Rhizobium temperate phage 16-3. J. Bacteriol. 181:4185-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thatte, V., D. E. Bradley, and V. N. Iyer. 1985. N conjugative transfer system of plasmid pCU1. J. Bacteriol. 163:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weigele, P. R., E. Scanlon, and J. King. 2003. Homotrimeric, beta-stranded viral adhesins and tail proteins. J. Bacteriol. 185:4022-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, M. N., R. I. Hollingsworth, S. Klein, and E. R. Signer. 1990. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J. Bacteriol. 172:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang, Y., P. G. Leiman, L. Li, S. Grimes, D. L. Anderson, and M. G. Rossmann. 2009. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoder, M. D., N. T. Keen, and F. Jurnak. 1993. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260:1503-1507. [DOI] [PubMed] [Google Scholar]