Abstract
There exist commonalities between symbiotic Sinorhizobium meliloti and pathogenic Brucella bacteria in terms of extensive gene synteny and the requirements for intracellular survival in their respective hosts. The RNA chaperone Hfq is essential for virulence for several bacterial groups, including Brucella; however, its role in S. meliloti has not been investigated. Our studies of an S. meliloti loss-of-function hfq mutant have revealed that Hfq plays a key role in the establishment of the symbiosis between S. meliloti and its host Medicago sativa. S. meliloti Hfq is involved in controlling the population density under a free-living state and affects the growth parameters and nodulation. An hfq mutant poorly colonizes the infection threads that are necessary for the bacteria to invade the developing nodule. An hfq mutant is severely impaired in its ability to invade plant cells within the nodule, which leads to the formation of small, ineffective nodules unable to fix nitrogen. In culture, the hfq mutant did not accumulate transcripts of nifA, which encodes a key regulator necessary for nitrogen fixation. Hfq may be involved in regulation of several proteins relevant to hfq mutant phenotypes. The crucial role of Hfq in symbiosis suggests that small regulatory RNAs are important for its interactions with its plant host.
The alpha subgroup of proteobacteria includes members such as Sinorhizobium meliloti, which establishes a symbiosis with certain leguminous plants, as well as members, such as Brucella abortus, that are highly pathogenic to animals and humans. Despite the very different outcomes of the chronic intracellular infections established by S. meliloti and B. abortus in their respective hosts, parallels can be drawn because in both cases the bacteria need to survive within acidic membrane compartments for a prolonged time after endocytosis (50). Comparisons of genomic sequences between brucellae and rhizobia have revealed similarities. For example, extensive gene synteny exists between chromosome I of Brucella and the genome of Mesorhizobium loti, while chromosome II shares regions of gene synteny with the pSym megaplasmids of S. meliloti. In addition, the transport and metabolic capabilities of Brucella have similarities to those of the plant symbiont (46). Genomic comparisons suggest that these two types of bacteria share a complex evolutionary history and that Brucella evolved from soil/plant-associated ancestral bacteria of the Rhizobium/Agrobacterium group (46).
The parallel between these two bacterial groups is well illustrated by the inner membrane protein BacA, which is critical for the maintenance of the chronic intracellular infections that underlie these two very diverse host-bacterial relationships (20, 35). S. meliloti bacA mutants lyse soon after being endocytosed into the plant cytoplasm (20), while B. abortus bacA mutants are defective in intracellular replication in macrophages and are unable to establish a chronic infection in BALB/c mice (35). BacA is required for the transport of certain peptides into S. meliloti (38). Furthermore, for both S. meliloti and B. abortus, loss of BacA results in a ca. 50% reduction in the very-long-chain fatty acid (27-OHC28:0 and 29-OHC30:0) content of both Sinorhizobium and Brucella lipid A (15) and in increased resistance toward the glycopeptide bleomycin (20, 35). Thus, in the context of the S. meliloti-legume symbiosis, BacA is required for transition from the free-living form of bacteria into the nonreplicating, nitrogen-fixing bacteroid form (20, 27), while in the context of B. abortus-host pathogenesis, BacA acts as a pathogenesis factor required for the long-term survival and residence of B. abortus in its intracellular niche (35, 50).
Another factor besides BacA that is required for B. abortus to establish a chronic infection is the RNA-binding Hfq protein (48, 50). Hfq also strongly contributes to stress resistance in stationary phase. A B. abortus hfq mutant shows increased sensitivity to H2O2, shows decreased survival under acidic conditions, fails to replicate in cultured murine macrophages, and is rapidly cleared from the spleens and livers of experimentally infected BALB/c mice (48). Thus, similarly to bacA mutants, B. abortus hfq mutants are not able to establish a chronic intracellular infection of BALB/c mice. The role of the homologous hfq gene in S. meliloti and its contribution to symbiosis have remained unexplored.
Hfq (HF-I), first discovered as a host factor required for Qβ phage replication in Escherichia coli, is an RNA-binding protein involved in regulation of RNA metabolism and is conserved in about half of all sequenced bacterial genomes (63, 66). Hfq is a member of the Sm and Sm-like family of proteins found in all three domains of life (45) and forms a homo-hexameric structure that binds preferentially to A/U-rich RNAs (reviewed in references 6 and 67). Hfq serves as posttranscriptional regulator that binds small RNAs (sRNA) and mRNAs and facilitates their interaction (1, 22, 37, 60). Hfq may promote duplex formation by increasing the local concentration of the sRNA and its target (7), changing the RNA structure (18, 49), and accelerating strand exchange and subsequent annealing (2). These RNA-RNA interactions mediate mRNA turnover and/or translation (1, 23, 60).
E. coli hfq null mutants have a pleiotropic phenotype that includes decreased growth rate, increased cell length, and sensitivity to UV light (64). Since Hfq is essential for efficient translation of RpoS in E. coli (44) and Salmonella (3, 8, 57), some of the phenotypic properties of an hfq mutant are due to RpoS deficiency. Hfq protein also exerts effects in an RpoS-independent manner and even can bind to the poly(A) tails of some mRNAs, stimulating poly(A) adenylation (26, 33, 43). Salmonella hfq mutants had reduced growth, had attenuated infection, and showed severe defects in invasion of epithelial cells (58). Interestingly, these phenotypes in Salmonella are largely RpoS independent (58). An RpoS homolog has not been identified in the alphaproteobacteria, the phylogenetic group to which Brucella and S. meliloti belong.
Hfq has been shown to influence the virulence of a number of other pathogenic bacteria (e.g., references 12, 14, 32, 40, 55, and 59). The parallels between Brucella pathogenesis and S. meliloti symbiosis motivated us to construct and characterize an S. meliloti loss-of-function hfq mutant and to explore its involvement in the symbiosis of S. meliloti with its host, Medicago sativa. Our results show that Hfq plays several important roles in the establishment of the S. meliloti-alfalfa symbiosis as well in aspects of the physiology of the free-living rhizobia.
MATERIALS AND METHODS
Media.
Luria-Bertani (LB) was prepared as previously described (51). S. meliloti was also grown in minimal galactose aspartate salts (GAS) medium as previously described (24, 47). Bacto agar (Gibco) was used at 1.5% for solid medium. Gentamicin (50 μg/ml for S. meliloti and 5 μg/ml for E. coli), kanamycin (25 μg/ml), neomycin (200 μg/ml), streptomycin (500 μg/ml), and tetracycline (10 μg/ml) were added when needed.
Nodulation assay.
Nodulation of alfalfa (Medicago sativa cv. Iroquois) by S. meliloti on whole plant and nodule sections was analyzed. The alfalfa seeds were surface sterilized as previously described (34) and germinated on 0.8% agar in water in the dark for 62 h. One seedling was placed on top of a petri dish plate filled with 25 ml of Jensen's agar (34). The seedlings were inoculated by 1 ml of S. meliloti cells grown to saturation in LB, washed in sterile distilled water, and resuspended to an optical density at 600 nm (OD600) of 0.05. The plates were sealed using parafilm, wrapped in aluminum foil to maintain the root in the dark, and incubated at 25°C for a maximum of 4 weeks. Microscopy was performed by using standard methods.
Molecular manipulation.
A list of strains and plasmids used in this study are given in Table S1 in the supplemental material. Cloning, subcloning, and isolation of plasmid or genomic DNAs were performed using previously described procedures (51) or using kits provided by Qiagen, Inc. (Valencia, CA). Site-directed mutagenesis was performed using a QuikChange kit (Stratagene, Cedar Creek, TX) and confirmed by sequencing.
Construction of an S. meliloti Rm1021 Hfq mutant.
Two distinct PCRs were carried out in order to amplify the sequences overlapping the hfq 5′ and 3′ ends. To make pblC60, the upstream region of hfq (including a portion of its open reading frame) was amplified from S. meliloti Rm1021 with the primers hfqL3 (5′-ATAATCATGAGAATTCCTATGTGCGCCCAG-3′) and hfqin10 (5′-TAATAAGCTTGGCATGATCCTCGAGATGGCGTG-3′). The 1,306-bp PCR product was cloned into the pGEM-T vector (Promega, La Jolla, CA). The downstream region was amplified using the primers blhfqDsfwd (5′-CGGGATGTGAGGAACATT-3′) and blhfqR2 (5′-GAATCCATCTAGACCAAGTCAGTGG-3′). The 1,567-bp PCR product was cloned into the pGEM-T vector, giving pblB58.
The hfq::lacZ transcriptional fusion was obtained by inserting a lacZ-aacC1 cassette obtained from the pAB2001 plasmid (4). This cassette was recovered by a HindIII/XhoI double digest of pAB2001 and inserted between the same sites of pC60, giving pC67. Then, the hfq::lacZ-bearing fragment was recovered through an NcoI digest and introduced in the corresponding site of pB58. Finally, the disrupted hfq gene and surrounding regions were cloned into the BamHI site of pKmob18sacB (53), a suicide vector for S. meliloti.
Transconjugants were selected as described by Schafer et al. (53). Among them, only 43% had lost the neomycin resistance carried by pKmob18sacB, which reflects a double crossing over. One of the mutants was chosen for further analysis, and the sequences surrounding the hfq gene were sequenced (data not shown) in order to ensure that none of the genes downstream or upstream were affected by a point mutation. The absence of a wild-type (WT) copy of hfq was checked by PCR and Southern blot analysis (data not shown). The mutation was then purified by transduction into the parental strain Rm1021. One of the transductants was named Smblφ500.
Disruption of the hflX open reading frame in strain Rm1021.
For disruption, a 524-bp internal fragment of the hflX open reading frame was amplified using Rm1021 genomic DNA and the oligonucleotides blhflX1 (5′-AGCTTCACCGGTCGAAGC-3′) and blhflX2 (5′-GACTGAGAAGATCGTCGACACC-3′). The PCR amplicon was cloned into the pGEM-T vector. Subsequently, the hflX-carrying HincII-NcoI fragment was transferred between the ScaI and NcoI sites of the pSUP102b vector (56), giving pblC51. pblC51 was transferred by triparental conjugation into S. meliloti Rm1021 with selection for tetracycline resistance. One of the mutants was used as a donor strain for transduction into the Rm1021 strain. The strain was named Smblφ600. The insertion was verified by PCR.
Complementing plasmids.
To make the complementing plasmid, pbl100, a fragment of 703 bp corresponding to the hfq gene and surrounding regions, was amplified from S. meliloti Rm1021 with the primers Rmhfq4 (5′-ATCATGCCGTGAACG-3′) and Rmhfq6 (5′-CAGCATCTGCTTTCC-3′). The fragment was cloned into the pGEM-T vector. The open reading frame of hfq under the control of its own promoter, defined using primer extension (data not shown), was cloned between PstI and SalI of pBBR1MCS-3 (31), creating the pbl100 plasmid. This plasmid was transferred into S. meliloti by using triparental matting. pbl100 was introduced into the strains Rm1021 and Smblφ500, creating the strains Smbl003 and Smbl503, respectively. In parallel, the strains Smbl001 and Smbl501 were obtained by conjugation of the plasmid pBBR1MCS-3 (31) into the strains Rm1021 and Smblφ500, respectively.
A derivative of pBBR1MCS-3 was constructed by deletion of 800 bp from the lacZ gene between the sites SspI and BamHI and then ligation of the vector on itself. The resulting plasmid was named pbl101 and introduced into the hfq mutant (Smblφ500) in order to follow the lacZ activity, in vivo, without the background due to the expression of lacZ from the plasmid. The strain was named Smblφ502.
RNA analysis.
Cultures were set up in 50-ml sterile Falcon tubes for microaerobic experiments. Total RNA was isolated by TRI reagent (T 9424; Sigma, St. Louis, MO). In order to identify expression of the gene induced under microaerobiosis, different sets of primers were used as follows. An internal region of nifA was amplified using the primers Sm-nifA1 (5′-CAGGAAGGTGAATTTGAGC-3′) and Sm-nifA2 (5′-CAGCGCACTGATCAACC-3′). 16S RNA was used as a control, for which the primer combination of Sm-16SL1 (5′-CAATCTAGAGTCCAGAAGAGGT-3′) and Sm-16SR1 (5′-AACATCTCACGACACGAGC-3′) was used.
Phenotypic characterization of the S. meliloti hfq mutant.
Growth characteristics were evaluated by analysis of growth curves. S. meliloti was grown on LB supplemented with antibiotics when appropriate. Cells were harvested at various densities and resuspended in the appropriate medium (LB or GAS) to an OD600 of 1.0. Then, the bacteria were inoculated into the appropriate medium at an initial OD600 of 0.1. Cell growth was determined by monitoring OD600.
In order to assay the effect of hfq mutation on swarming behavior, liquid cultures of S. meliloti, initiated from glycerol stocks, were grown at 30°C in LB broth with shaking to late logarithmic phase. After incubation, cells were pelleted and washed twice in 0.85% NaCl resuspended in saline medium to obtain a cell concentration of 1010 cells/ml. Five-microliter drops of this suspension were deposited on the surface of air-dried (10 min) plates containing the appropriate broth with 0.3% agar and allowed to dry for 10 min. The plates were then incubated for 6 days at 30°C and scored for swarming by measuring the diameter of the spreading.
β-Galactosidase assays.
hfq transcriptional expression was assayed by monitoring the β-galactosidase activity expressed from a chromosomal hfq::lacZ fusion. The enzymatic activity was measured as described by Miller (42) and expressed in Miller units.
β-Galactosidase assays in planta.
β-Galactosidase activity in vivo on nodules harvested from plants after 2, 3, or 4 weeks of culture was assayed. Nodules were treated as described by Boivin et al. (5). The wild-type strain was used as a control for the efficiency of the fixation step, which inhibits the β-galactosidase activity of the plant. For infection thread visualization, the fixation step never exceeded 2 h.
Microaerobiosis.
Cultures were set up in 50-ml sterile Falcon tubes. A pipette diameter hole was made, under sterile conditions, into the Falcon tube lid. A sterile pipette topped with a filter (Millipore) was inserted into the lid hole. The filter was connected to the gas tank by using a tube. The gas tank mixture was composed of N2 (99.5%)-O2 (0.5%), which was bubbled into 15 to 20 ml of medium containing or not containing a nitrogen source.
RESULTS
Identification of the S. meliloti hfq gene.
A search of the S. meliloti genome with the B. abortus Hfq sequence revealed that the S. meliloti gene annotated as nrfA encodes a protein 91.9% identical to the B. abortus Hfq protein (see Fig. S1 in the supplemental material). The S. meliloti Hfq/NrfA homolog has 74% similarity to NrfA (Nif regulatory factor) of Azorhizobium caulinodans (see Fig. S1 in the supplemental material) (30).
Construction and growth characteristics of the S. meliloti hfq mutant.
In order to investigate the physiological roles of hfq and to follow its expression both in the free-living stage and in planta, we disrupted the hfq gene by a partial gene deletion that also created a transcriptional fusion to a promoterless lacZ gene derived from the vector pAB2001 (4). The resulting hfq::lacZ mutation was crossed into the Rm1021 genome by a double-recombination event enabled by the use of the pKmob18sacB vector (53). One clone, designated Smbl499, was randomly chosen from the transformants for further analysis. PCR and Southern blotting were performed to confirm the partial deletion (of 60 nucleotides [nt]) of hfq and insertion of the lacZ-Gm cassette derived from the pAB2001 vector (data not shown). The hfq mutation was transduced into the parental strain Rm1021 to ensure the absence of unlinked mutations, and the resulting strain was named Smblφ500.
It was immediately evident that the loss of Hfq function affected the growth of S. meliloti, as the hfq mutant was delayed 1 to 2 days in forming colonies on LB agar in comparison to the hfq+ parental strain Rm1021. When grown in LB, the hfq mutant (Smblφ500) exhibited a doubling time of 5 h, compared to 4 h for the hfq+ parent. The maximum cell density obtained with the hfq mutant was ∼60% relative to the level for the WT when grown in LB (Fig. 1B). A similar effect was observed in minimal medium.
FIG. 1.
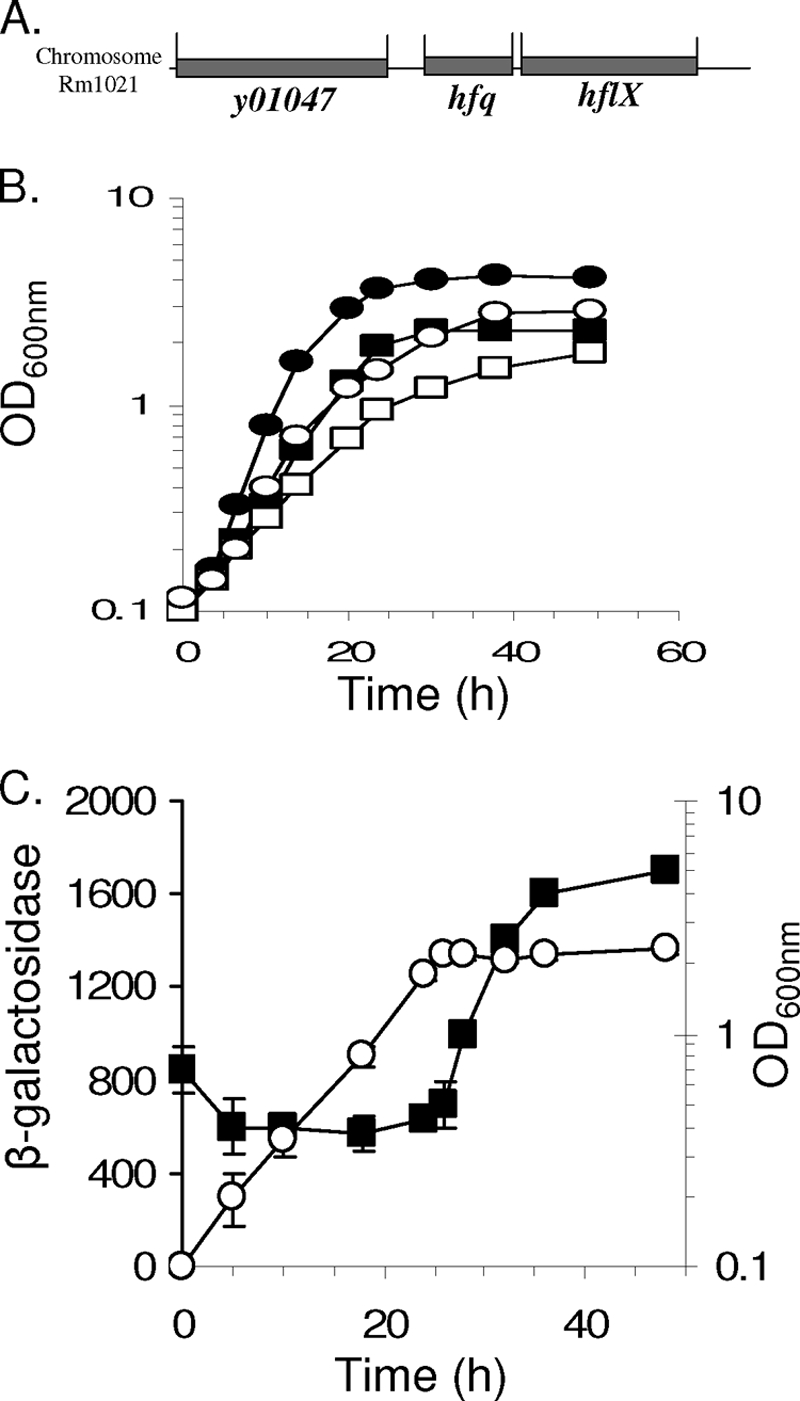
Characterization of hfq in S. meliloti. (A) Organization of the hfq locus on chromosome of S. meliloti. (B) Growth of wild-type (circles) and hfq mutant (squares) strains in LB (closed symbols) or GAS (open symbols) medium. (C) Transcriptional activity of the hfq gene. Results are shown for a time course experiment performed following the growth of the mutant by measuring the optical density of the culture at different times of the growth (open circles; expressed in OD600). Simultaneously, samples were used to assay for β-galactosidase activity resulting from the expression of the hfq::lacZ fusion. Results are expressed in Miller units, function of time, and OD600 of sample (filled squares). The error bars correspond to the variation obtained between 4 repeats of independent dilution of the sample.
hfq transcription is growth phase regulated.
Smblφ500 carrying the transcriptional fusion of hfq with the promoterless lacZ gene was grown in minimal medium, and samples were harvested at different time points and assayed for β-galactosidase activity (42). Hfq was expressed during all the stages of the growth (Fig. 1C). As a control for the background levels of β-galactosidase activity, the wild-type strain was grown simultaneously and under the same conditions. Its background level never exceeded 30 Miller units (data not shown). Relative to the levels seen in exponential phase, we observed a two- to threefold increase in β-galactosidase activity that began as the cells entered stationary phase (Fig. 1C). The same pattern of induction was observed in strain Smbl503 (complemented hfq mutant) (data not shown). Further experiments will be required to elucidate whether S. meliloti Hfq auto-controls its own expression at a translational level, as described for E. coli (65). The greater expression level of hfq during stationary phase suggests that Hfq plays a role in the stationary phase adaptation and may account, at least in part, for the lower cell density reached by the mutant than by the parental strain. It has been suggested that, during symbiosis, S. meliloti is in a physiological state closer to stationary phase than to exponential phase (28), so it seems likely that an hfq mutation might have a substantial impact on its ability to interact with its host alfalfa.
Hfq is required to establish the symbiosis between S. meliloti and alfalfa.
To investigate the role of Hfq in symbiosis, 2-day-old alfalfa seedlings were inoculated with the hfq mutant (Smblφ500) or the WT strain (Rm1021). After 4 weeks of incubation, plants inoculated with the hfq mutant reached only 2.2 ± 0.8 cm (Fig. 2A). The yellow-to-pale-green color of the leaves indicates the lack of nitrogen fixation. In contrast, the plants inoculated with the hfq+ parent strain Rm1021 reached a height of 8.1 ± 2 cm and exhibited dark green leaves (Fig. 2A). The analysis of the surrounding region of hfq in the genome of S. meliloti Rm1021 (16) revealed that, as in B. abortus (46), the gene immediately downstream of S. meliloti hfq is hflX, which encodes a putative GTP-binding protein (Fig. 1A). To test whether the impaired growth of plants was due only to a lack of Hfq and not to a polar effect on hflX, we performed a complementation experiment. A 700-bp DNA fragment carrying hfq+ under the control of its own promoter was cloned into the plasmid pBBR1-MCS3, creating pbl100. This plasmid was then introduced by triparental mating into strain Rm1021 and the hfq mutant strain Smblφ500. Seedlings were then inoculated with the strains carrying the hfq+ plasmid pbl100 and/or the control vector pbl101. At 4 weeks postinoculation, the plants inoculated with the hfq+ (complemented) strain reached a height of 7 ± 1.4 cm (Fig. 2A), showing no significant differences from the wild-type strain carrying the same plasmid. To confirm directly that the downstream hflX gene was not required for the symbiosis, an hflX mutant, Smblφ600, was constructed. This mutation had no impact on the symbiosis (data not shown), showing that the function of hflX is not essential for establishment of an effective symbiosis. Therefore, the symbiosis defect of the hfq mutant can be attributed to the mutation of hfq.
FIG. 2.
Evidence of the symbiosis defects of an hfq mutant. (A) Live alfalfa plants, 4 weeks after inoculation with the hfq mutant (panel 1), the hfq mutant carrying pbl101 (panel 2), the complemented hfq mutant (panel 3), and the WT strain (panel 4). (B) Enlarged images of nodules elicited by a plant inoculated with Rm1021, the hfq+ parent (panel 1) and the hfq mutant (panel 2).
Another indicator of normal symbiosis is the presence of leghemoglobin in the elongated nodules, which makes the nodule pink. Whereas the plants inoculated with the hfq+ parent exhibited more than 95% elongated pink nodules (Fig. 2B), more than 50 plants inoculated with the hfq mutant in distinct experiments exhibited fewer than 10% pink nodules, very few of which were elongated (Fig. 2B). Representative pink nodules were crushed and the bacteria recovered and reinoculated, but we did not observe more pink nodules on plants inoculated with these recovered hfq mutants than on plants inoculated with the parental hfq mutant.
hfq missense mutants that altered residues in the Sm1 domain that is highly conserved in bacterial Hfq proteins were constructed (see Fig. S1 in the supplemental material). Plasmids carrying mutations altering Gly35 (pbl100-G35V), Ser39 (pbl100-S39T), or Phe40 (pbl100-F40W) failed to complement the hfq mutant phenotype. No significant differences were observed between the plants inoculated with the hfq mutant and those inoculated with the derivatives carrying the mutant plasmids (data not shown). These residues form the part of β2 strand in the Hfq structure (36, 54). The β2 and β4 strands comprise the “A site,” responsible for the specificity of Hfq for adenosine nucleotides of the poly(A) RNA (36). This Gly residue, present in most of the described Hfq proteins and located in the middle of β2, is critical for maintenance of the highly distorted Sm fold (54). The Gly35 residue has been suggested to be involved in binding with ATP (61). The failure of the G35V mutant to complement and the high degree of conservation of Gly35 indicate that this residue is important for Hfq function. The Phe40 residue, which corresponds to the Phe39 residue of E. coli Hfq, has been shown to be involved in the affinity of Hfq toward the DsrA sRNA (62). If such small RNAs are involved in the symbiosis, it would not be surprising that this mutation alters the ability of hfq to complement the hfq null mutation.
hfq is expressed in planta and is required for efficient invasion through infection threads.
Expression of hfq in planta was assessed using the transcriptional fusion hfq::lacZ in the Smbl502 strain, lacking Hfq and carrying the control plasmid pbl101, as well as in the complemented strain Smbl503. β-Galactosidase expression was visualized, as previously described (5), in sections or in whole nodules harvested at 3 to 4 weeks postinoculation. β-Galactosidase activity, which correlates with hfq expression, was observed at different stages of the infection process. Three stages of the infection process include colonization of the curled root hair, invasion though infection threads, and finally release into the cytoplasm of plant cells in the infection zone of the nodule. Significant differences in the pattern of hfq expression were observed between the mutant and the complemented strains; plants inoculated with the hfq mutant (Smbl502) exhibited a high frequency of infection thread abortion.
After 4 weeks of incubation, the majority of the nodules in plants inoculated with the hfq mutant (Smbl502) failed to show any substantial staining of plant cells in the interior of the nodule (59.7% ± 10.4%), except at the surfaces of the nodules in the remnants of colonized curled root hairs and aborted root hairs (Fig. 3A). Furthermore, only 1.7% ± 1.5% of the nodules elicited by the hfq mutant were fully colonized (Fig. 3B1, left nodule). The remaining nodules (38.6%) exhibited staining of only a few invaded cells in the interior of the nodule (Fig. 3B1, right nodule). Even if some cells were invaded by the hfq mutant, these nodules did not elongate as they normally do when inoculated with a WT strain; the nodules did not differentiate, and leghemoglobin seemed to be lacking, as they appeared white. They did not enable nitrogen fixation, as the plant remained small and chlorotic. In contrast, for the plants inoculated with complemented strain (Smbl502), only 13.6% ± 5.7% of the nodules failed to exhibit substantial staining and 72% ± 7% of the nodules inoculated with the complemented strain exhibited colonization of the whole nodule (Fig. 3B2). These results further indicate that Hfq is expressed in planta and is required for an efficient colonization of the infection threads and invasion into the developing nodule. The high frequency of infection thread abortion seen with the hfq mutant evidently reduces the frequency with which the bacteria are able to invade plant cells in the infection zone of the nodule.
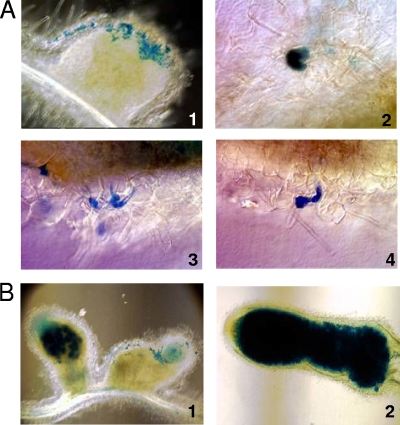
FIG. 3.
Defects of an hfq mutant during infection and nodulation. (A) Optical microscopy images showing a nodule colonized by an hfq mutant strain expressing β-galactosidase (panel 1). Pictures focusing on curled root hair colonized by the hfq mutant revealed by their β-galactosidase activity (panel 2) and abortive infection threads (panels 3 and 4). (B) Optic microscopy on hand sections of nodules inoculated with the hfq mutant (panel 1; Smbl502) and its complemented derivative (panel 2; Smbl503). The B1 panel, left nodule, represents rare nodules elicited after inoculation by the hfq mutant, which are colonized but fail to elongate; panel 1, the right nodule represents the majority of nodules observed after inoculation by the hfq mutant. β-Galactosidase activity expressed by the hfq::lacZ fusion was revealed and corresponds to the blue staining.
The severe deficiency of the hfq mutant in invading nodules was also observed in a competition experiment. When plants were inoculated with a mixture of the hfq mutant (Smblφ500) and its hfq+ parent (Rm1021) in a ratio of 1,000:1, no hfq mutant bacteria could be recovered from the nodules.
Hfq is required for efficient release and differentiation of the bacteria inside the plant cell.
To gain further insights into the nature of the symbiotic deficiency of the hfq mutant, ultrastructural analysis was performed on 4-week-old nodules by using electron microscopy. Nodules from plants inoculated with the wild-type strain showed the presence of bacteroids in most of the cells (Fig. 4A). In contrast, for plants inoculated with the hfq mutant, most of the plant cells within white nodules lacked bacteria or bacteroids and instead were filled with the starch granules that accumulate prior to the uptake of the bacteria. At a much lower frequency than in the wild type, we also saw infected plant cells that contained some bacteria or bacteroid-like cells, as shown in a representative section (Fig. 4B); we also noted occasional dividing cells (Fig. 4C). These observations are consistent with our other results indicating that the hfq mutant is much less efficient than wild type at invading plant cells and establishing the chronic intracellular infection that underlies the symbiosis.
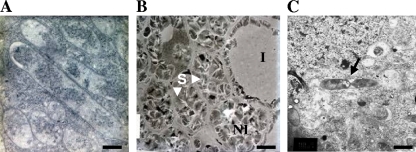
FIG. 4.
Ultrastructural evidence of hfq mutant invasion deficiency and the lack of differentiation observed using electron microscopy. (A) WT cells differentiated into bacteroids (scale bar, 1 μm). (B) hfq mutant (Smblφ500)-inoculated plants exhibit nodules with cells invaded (I) or not invaded (NI) by bacteria (scale bar, 10 μm). Noninvaded cells contain large granules of starch (S). Panel C shows a dividing cell failing to differentiate into a bacteroid (designated by an arrow) (scale bar, 1.5 μm).
Hfq is required for efficient expression of nifA and nfeB.
The hfq mutant (Smblφ500) has a Fix− phenotype since the plants inoculated were clearly deficient in fixing nitrogen. The proximal cause of the Fix− phenotype results from the combination of the deficiency of the hfq mutant in invading the root hairs and its apparent inability to differentiate into bacteroids after endocytosis. However, we were interested in whether the role of Hfq in symbiosis might extend to regulation of nitrogen fixation as well. In an attempt to address this issue, we investigated whether loss of hfq function might be affecting nifA expression. The nifA gene is an activator of nitrogen fixation in planta, so that loss of nifA expression results in the formation of small, white nodules (21). In the free-living state, the S. meliloti nifA gene can be induced under microaerobic conditions in nitrogen-free medium (9). After the parental strain Rm1021 and the hfq mutant (Smblφ500) were grown in GAS minimal medium or nitrogen-free GS minimal medium under microaerobic conditions, total RNAs were extracted and subjected to reverse transcriptase PCR (RT-PCR). nifA expression was observed only in the wild-type strain after a 4-h incubation in nitrogen-free medium and microaerobiosis. In contrast, no nifA amplification product was observed in the hfq mutant (Fig. 5), suggesting that a deficiency in nifA expression in an hfq mutant may also contribute to the Fix− phenotype.
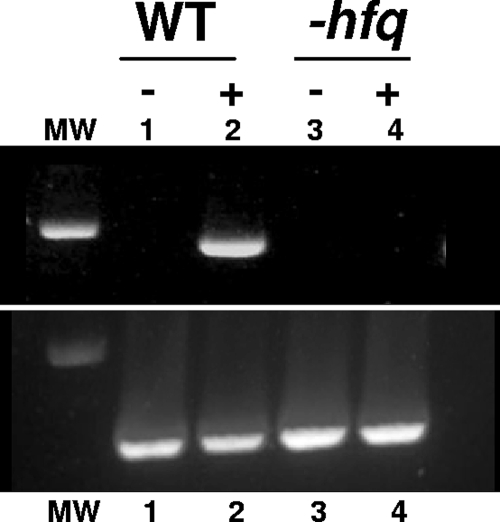
FIG. 5.
Analysis of nifA gene expression using RT-PCR. RNA were extracted from the wild-type strain Rm1021 (lanes 1 and 2) and the hfq mutant Smblφ500 (lanes 3 and 4) before (−; lanes 1 and 3) or after (+; lanes 2 and 4) 4 h of incubation in nitrogen-free medium and microaerobiosis. An equal amount of RNA was used to perform the RT-PCR, and an equal volume of the amplification product was loaded onto the gel. The nifA gene is expressed only 4 h after incubation in WT strains (upper panel). The lower panel shows RT-PCR amplification of 16S RNA, used as a control. MW, molecular weight marker.
NifA controls the expression of nfeB (nodule formation efficiency), which, in S. meliloti, is expressed in nitrogen fixation zone III (17). nfeB expression in the hfq mutant was therefore analyzed. Plasmids carrying nfeB translational fusions with uidA (17) were transferred in the Rm1021 and hfq strains (Smblφ500) by triparental mating. The resulting strains were inoculated on 2-day-old alfalfa seedlings and incubated. At 4 weeks postinoculation, the nodules were stained for β-glucuronidase activity. In the wild-type background, nfeB is specifically expressed in the nitrogen fixation zone (Fig. 6A). In contrast to what was found for Rm1021, in the hfq mutant (Smblφ500), nfeB was not expressed in most nodules, while in rare nodules, aberrant nfe expression was observed (Fig. 6B).
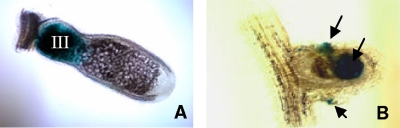
FIG. 6.
Optical microscopy images of hand sections of nodules from plants inoculated with the wild-type strain (A) or the hfq mutant (B) containing the plasmid pGus2 carrying the nfeB::uidA fusion (SmblK7 or SmblK8, respectively). Expression of this fusion is revealed by the blue staining in nodule zone III in the wild-type strain and in different zones of a nodule in the hfq mutant (SmblK8), which, although colonized, fails to elongate (arrows).
Mutating Hfq reduces swarming efficiency without affecting succinoglycan synthesis.
We evaluated the physiological consequences of loss of hfq function on two processes associated with bacterial adaptation to extra- and intracellular niches, succinoglycan production, and swarming efficiency. Efficient synthesis of succinoglycan is required for the initiation and extension of the infection threads from the colonized curled root hairs (10, 29, 34). To test whether loss of Hfq function affects succinoglycan biosynthesis, the hfq mutant and its wild-type parent were plated on LB plates containing Calcofluor, which allows this exopolysaccharide to be visualized under UV light. No significant differences were observed between the different strains (data not shown), an observation suggesting that the role of Hfq in infection thread invasion is due to its regulation of a process or processes other than succinoglycan biosynthesis. We also noted that the hfq mutants induced the curling of root hairs as efficiently as the wild-type strain. Since, both root hair curling and nodule formation require Nod factors, our observations suggest that loss of hfq function has little, if any, effect on Nod factor synthesis.
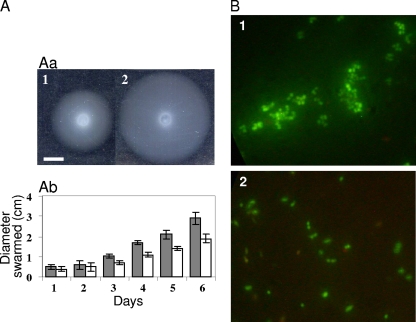
To assay its ability to swarm, the hfq mutant and its wild-type parent were spotted at the surface of a swarming plate. The spreading was measured (Fig. 7A) every day during 6 days. Results show that the hfq mutant is retarded in swarming (Fig. 7A). Interestingly, a nifA mutation reduces the ability of S. meliloti to swarm (21). Further experiments will be needed to decipher whether the swarming retardation phenotype in a nifA mutant is controlled or not by Hfq. We noted an additional phenotype where bacterial cultures were maintained in GAS medium for 1 month. The hfq mutant cells aggregated, whereas the wild-type cells did not (Fig. 7B). The aggregation was not associated with loss of viability. Taken together, our results suggest that hfq mutants may be at a disadvantage in adapting to new environments, including the various environments that the bacterium encounters in the course of establishing symbiosis.
FIG. 7.
hfq regulates physiological responses to changes in environment. (A) The S. meliloti hfq mutant swarms less efficiently than the wild-type strain. (Aa) Five-day colonies from the hfq mutant Smblφ500 (panel 1) and the WT (panel 2) strains were plated on a swarming plate (0.3% agar) and incubated at 30°C. The white bar represents 0.5 cm. (Ab) Results of swarming for the WT (gray bars) and the hfq mutant (white bars) strains. Colony progression was measured every day. The bars represent the means of results from 3 experiments. (B) When hfq mutant cultures (panel 1) in GAS medium were maintained for 1 month, aggregates were observed. Similar aggregates were not observed in WT cells (panel 2).
DISCUSSION
Our study reveals that the RNA chaperone Hfq, which plays diverse regulatory roles in many free-living bacteria (25, 66) and is needed for the virulence of numerous pathogens (e.g., references 12, 14, 32, 40, 55, and 59), is critically required for S. meliloti to establish a nitrogen-fixing symbiosis with its host plant alfalfa. This study was motivated in part by the phylogenetic relationships between the rhizobiaceae and the brucellae (46), both of which establish chronic intracellular infections within acidic membrane compartments in their respective hosts (50). Furthermore, our finding that the S. meliloti BacA protein is important for the chronic intracellular infection phase of symbiosis had previously led us to discover that B. abortus BacA is specifically required for the chronic phase of pathogenesis in the BALB/c mouse model (35). Since, like BacA, B. abortus Hfq is specifically required for the chronic phase of pathogenesis, we were curious to test whether Hfq might play an important role in the interactions of S. meliloti with its plant host.
Hfq affects at least three aspects of the symbiosis: (i) the ability of the rhizobia to invade the developing nodule through host-derived infection threads, (ii) the capability of the rhizobia to invade plant cells within the nodule and establish the chronic intracellular infection that underlies the symbiosis, and (iii) the ability of the bacteria to express various functions necessary for nitrogen fixation. Our study shows that the role of Hfq goes far beyond the regulation of nitrogen fixation that had been previously observed in A. caulinodans (30) and Rhodobacter capsulatus (13) and reveals additional parallels between pathogenesis and symbiosis. As discussed below and in more detail in our accompanying paper (3a), Hfq also plays important roles in regulating numerous responses of free-living S. meliloti as well.
In the S. meliloti strain Rm1021-alfalfa symbiosis, infection thread initiation, elongation, and extension require that the rhizobia be able to synthesize the exopolysaccharide succinoglycan. Mutants, such as the exoY strain, that are unable to synthesize succinoglycan form small white Fix− nodules (29, 34), because most of the colonized curled root hairs fail to initiate infection threads and those infection threads that are initiated then abort (10). The hfq mutant is able to synthesize succinoglycan, which allows the bacteria to initiate infections threads and stimulate their elongation. Nevertheless, a large proportion of the infection threads do abort before the release of the bacteria within the plant cells in the infection zone of the nodule. The infection thread abortion phenotype of the hfq mutant may be due to the fact that, while in the infection thread, S. meliloti is being subjected to a prolonged oxidative burst released by its host (52). Like the oxidative burst generated by mammalian cells in response to pathogens, this oxidative stress consists of at least superoxide and hydrogen peroxide, both of which are deleterious to cell survival. The prolonged burst is a chronic stress for S. meliloti, with both superoxide and hydrogen peroxide being readily detectable in nodules several weeks after the initial infection (52). Our accompanying study shows that Hfq function is important for S. meliloti to survive superoxide and hydrogen peroxide. The hfq mutant's ability to spread inside the infection thread may also be impaired by its increased doubling time and possibly by its decreased ability to swarm.
Despite its reduced ability to invade developing nodules through infection threads, the S. meliloti hfq mutant is endocytosed into the cytoplasm of plant cells in a significant fraction of the alfalfa nodules. Normally, the rhizobia stop dividing after endocytosis and then differentiate into bacteroids, a process that includes rounds of endoreduplication (19, 29, 41). However, in our ultrastructural studies of the nodules infected by the hfq rhizobia, many fewer bacteroid-like cells could be observed than with the wild type, and in some cases, we even observed what appear to be dividing bacteria. Thus, Hfq may also affect symbiosis by regulating the differentiation of the bacteria into bacteroids.
Because the hfq mutant is so inefficient in establishing a chronic intracellular infection, we were unable to examine whether Hfq regulates functions required for nitrogen fixation in the context of the nodule, a possibility suggested by the earlier observations that Hfq regulates nitrogen fixation in A. caulinodans (30) and R. capsulatus (13). However, by taking advantage of the fact that S. meliloti genes specifically involved in nitrogen fixation can be induced in free-living bacteria under microaerophilic conditions in nitrogen-free medium, we were able to show that Hfq also regulates the expression of nifA and nfeB. While nfeB is probably under the control of nifA, nifA is essential for several cellular processes and nitrogen fixation. nifA mutants fail to fix nitrogen and elicit small, ineffective white nodules (21). The interior of these nodules is reminiscent of a hypersensitive response characteristic of noncompatible host-pathogen interaction and is likely due to alteration in expression of several genes in signal transduction, cellular metabolism, growth, and development (21). As a consequence, disruption of nifA influences multiple cellular processes, such as growth, swarming, and extracellular protein production (21). hfq mutants not only failed to express nifA but showed phenotypes strikingly similar to those of nifA mutants, such as lower levels of swarming behavior, alteration in protein accumulation (as shown in the second study), and reduced ability to nodulate on the host. Thus, it seems likely that Hfq is important not only for infection thread invasion and for differentiation into bacteroids but for the induction of nitrogen-fixation functions as well.
Hfq has been shown to serve as an RNA chaperone that interacts with the sRNAs at single-stranded AU-rich regions and helps the sRNAs to pair with their target mRNAs (22, 66). Although Hfq can exert regulatory roles that are independent of sRNAs, the multiple crucial roles of Hfq in establishing S. meliloti symbiosis with its legume host strongly suggests the involvement of sRNAs in regulating multiple aspects of symbiosis. The requirement for a functional Sm1 domain, previously shown to be needed for RNA interaction of Hfq, is also consistent with the idea that regulatory S. meliloti sRNAs play an important role in its interactions with its plant host. Such a regulatory strategy would allow the bacteria to react quickly and at lower energetic cost to the stresses and sudden variations in their environments that they experience during the establishment of the symbiosis (39); as recently reviewed by Waters and Storz (69), sRNA-mediated regulation has several advantages compared to protein-regulated regulation during stress responses. Recent comparative genome analysis and prediction studies have suggested the existence of a number of sRNAs in S. meliloti, and some have been experimentally verified (11, 65, 68), but their regulatory roles in bacterial physiology and symbiosis has not yet been investigated. Expression of several of these predicted sRNAs in S. meliloti has been confirmed by Northern blots and microarray analysis. The expression patterns of these sRNAs show differential expression with respect to the stage of growth (e.g., early, exponential, or stationary). Furthermore, expression of several of the sRNAs is regulated by environmental stresses, such as heat, salinity, H2O2, alteration in pH, nutrients, and surfactants, etc. (68). The phenotypes of the S. meliloti hfq mutant suggest the involvement of multiple sRNAs during both the free-living and the symbiotic lifestyles of the bacteria.
The accompanying paper describes a proteomic analysis that identified 55 proteins whose levels are significantly altered in the free-living S. meliloti hfq mutant, most of which are involved in cell metabolism or stress resistance (3a). In enteric bacteria, Hfq has been shown to regulate the alternate sigma factors RpoS, which is involved in stationary-phase adaptation, and RpoE, which controls the envelope stress response. Although no RpoS or RpoS-like protein has been found in any of the sequenced alphaproteobacteria, this study shows that loss of Hfq function reduces the expression of the alternate sigma factors RpoE1, RpoE2, RpoE3, and RpoE4 (3a). Thus, some of the phenotypes of the hfq mutants are an indirect consequence of Hfq modulating a key regulatory protein, such as the RpoE sigma factor.
In conclusion, we have shown that Hfq plays crucial roles in mediating symbiosis by helping the bacteria adapt to extra- and intracellular niches during its interaction with the host plant cells. The involvement of sRNAs in symbiotic interactions in an Hfq-dependent manner certainly warrants further investigation. Parallel studies for identification of sRNAs in B. abortus would enable better understanding of these key regulatory elements during intracellular survival, virulence, and symbiosis.
Supplementary Material
Acknowledgments
We thank S. Georgeault, C. Monnier, M. Uguet, and M. C. Savary for technical assistance.
This work was supported by National Institutes of Health grant GM31010 (to G.C.W.), MIT Center for Environmental Health Sciences NIEHS P30 ES002109, the Centre National de la Recherche Scientifique, and the Ministère de la Recherche et de l'Education Nationale. L.B.-B. was supported by Region Bretagne. G.C.W. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 14 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10:134-139. [DOI] [PubMed] [Google Scholar]
- 2.Arluison, V., S. Hohng, R. Roy, O. Pellegrini, P. Regnier, and T. Ha. 2007. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 35:999-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang, I. S., J. G. Frye, M. McClelland, J. Velayudhan, and F. C. Fang. 2005. Alternative sigma factor interactions in Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol. Microbiol. 56:811-823. [DOI] [PubMed] [Google Scholar]
- 3a.Barra-Bily, L., C. Fontenelle, G. Jan, M. Flechard, A. Trautwetter, S. P. Pandey, G. C. Walker, and C. Blanco. 2010. Proteomic alterations explain phenotypic changes in Sinorhizobium meliloti lacking the RNA chaperone Hfq. J. Bacteriol. 192:1719-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 5.Boivin, C., S. Camut, C. A. Malpica, G. Truchet, and C. Rosenberg. 1990. Rhizobium-meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10:125-133. [DOI] [PubMed] [Google Scholar]
- 7.Brescia, C. C., P. J. Mikulecky, A. L. Feig, and D. D. Sledjeski. 2003. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA 9:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cebolla, A., and A. J. Palomares. 1994. Genetic regulation of nitrogen fixation in Rhizobium meliloti. Microbiologia 10:371-384. [PubMed] [Google Scholar]
- 10.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Val, C., E. Rivas, O. Torres-Quesada, N. Toro, and J. I. Jimenez-Zurdo. 2007. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol. Microbiol. 66:1080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53:345-354. [DOI] [PubMed] [Google Scholar]
- 13.Drepper, T., K. Raabe, D. Giaourakis, M. Gendrullis, B. Masepohl, and W. Klipp. 2002. The Hfq-like protein NrfA of the phototrophic purple bacterium Rhodobacter capsulatus controls nitrogen fixation via regulation of nifA and anfA expression. FEMS Microbiol. Lett. 215:221-227. [DOI] [PubMed] [Google Scholar]
- 14.Fantappiè, L., M. M. E. Metruccio, K. L. Seib, F. Oriente, E. Cartocci, F. Ferlicca, M. M. Giuliani, V. Scarlato, and I. Delany. 2009. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun. 77:1842-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, G. P., A. Datta, J. Baumgartner, R. M. Roop, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. U. S. A. 101:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lalaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Rodriguez, F. M., and N. Toro. 2000. Sinorhizobium meliloti nfe (nodulation formation efficiency) genes exhibit temporal and spatial expression patterns similar to those of genes involved in symbiotic nitrogen fixation. Mol. Plant Microbe Interact. 13:583-591. [DOI] [PubMed] [Google Scholar]
- 18.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, K. E., H. Kobayashi, and G. C. Walker. 2008. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42:413-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazebrook, J., A. Ichige, and G. C. Walker. 1993. A Rhizobium-meliloti homolog of the Escherichia-coli peptide antibiotic transport protein sbma is essential for bacteroid development. Genes Dev. 7:1485-1497. [DOI] [PubMed] [Google Scholar]
- 21.Gong, Z., J. Zhu, G. Yu, and H. Zou. 2007. Disruption of nifA Gene influences multiple cellular processes in Sinorhizobium meliloti. J. Genet. Genomics 34:783-789. [DOI] [PubMed] [Google Scholar]
- 22.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399-404. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 24.Gouffi, K., V. Pichereau, J. P. Rolland, D. Thomas, T. Bernard, and C. Blanco. 1998. Sucrose is a nonaccumulated osmoprotectant in Sinorhizobium meliloti. J. Bacteriol. 180:5044-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guisbert, E., V. A. Rhodius, N. Ahuja, E. Witkin, and C. A. Gross. 2007. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 189:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajnsdorf, E., and P. Regnier. 2000. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. U. S. A. 97:1501-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichige, A., and G. C. Walker. 1997. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J. Bacteriol. 179:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamet, A., S. Sigaud, G. Van de Sype, A. Puppo, and D. Herouart. 2003. Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant Microbe Interact. 16:217-225. [DOI] [PubMed] [Google Scholar]
- 29.Jones, K. M., H. Kobayashi, B. W. Davies, M. E. Taga, and G. C. Walker. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaminski, P. A., and C. Elmerich. 1998. The control of Azorhizobium caulinodans nifA expression by oxygen, ammonia and by the HF-I-like protein, NrfA. Mol. Microbiol. 28:603-613. [DOI] [PubMed] [Google Scholar]
- 31.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. 4 new derivatives of the broad-host-range cloning vector pbbr1mcs, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 32.Kulesus, R. R., K. Diaz-Perez, E. S. Slechta, D. S. Eto, and M. A. Mulvey. 2008. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 76:3019-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Derout, J., M. Folichon, F. Briani, G. Deho, P. Regnier, and E. Hajnsdorf. 2003. Hfq affects the length and the frequency of short oligo(A) tails at the 3 ′ end of Escherichia coli rpsO mRNAs. Nucleic Acids Res. 31:4017-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium-meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 36.Link, T. M., P. Valentin-Hansen, and R. G. Brennan. 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U. S. A. 106:19292-19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 40:93-113. [DOI] [PubMed] [Google Scholar]
- 38.Marlow, V. L., A. F. Haag, H. Kobayashi, V. Fletcher, M. Scocchi, G. C. Walker, and G. P. Ferguson. 2009. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masse, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 40.McNealy, T. L., V. Forsbach-Birk, C. W. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 187:1527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mergaert, P., T. Uchiumi, B. Alunni, G. Evanno, A. Cheron, O. Catrice, A. E. Mausset, F. Barloy-Hubler, F. Galibert, A. Kondorosi, and E. Kondorosi. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U. S. A. 103:5230-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Mohanty, B. K., V. F. Maples, and S. R. Kushner. 2004. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 54:905-920. [DOI] [PubMed] [Google Scholar]
- 44.Muffler, A., D. D. Traulsen, D. Fischer, R. Lange, and R. HenggeAronis. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigma(s) subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen, J. S., A. Boggild, C. B. F. Andersen, G. Nielsen, A. Boysen, D. E. Brodersen, and P. Valentin-Hansen. 2007. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA 13:2213-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. U. S. A. 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rains, D. W., L. N. Csonka, D. Le Rudulier, T. P. Croughan, S. S. Yang, S. J. Stavarek, and R. C. Valentine. 1982. Osmoregulation by organisms exposed to saline stress: physiological mechanisms and genetic manipulation, p. 238-302. In A. San Pietro (ed.), Biosaline research: a look to the future. Plenum Publishing Corp., New York, NY.
- 48.Robertson, G. T., and R. M. Roop. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690-700. [DOI] [PubMed] [Google Scholar]
- 49.Rolle, K., M. Zywicki, E. Wyszko, M. Z. Barciszewska, and J. Barciszewski. 2006. Evaluation of the dynamic structure of DsrA RNA from E-coli and its functional consequences. J. Biochem. 139:431-438. [DOI] [PubMed] [Google Scholar]
- 50.Roop, R. M., II, G. T. Robertson, G. P. Ferguson, L. E. Milford, M. E. Winkler, and G. C. Walker. 2002. Seeking a niche: putative contributions of the hfq and bacA gene products to the successful adaptation of the brucellae to their intracellular home. Vet. Microbiol. 90:349-363. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Santos, R., D. Herouart, S. Sigaud, D. Touati, and A. Puppo. 2001. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant Microbe Interact. 14:86-89. [DOI] [PubMed] [Google Scholar]
- 53.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia-coli plasmids PK18 AND PK19—selection of defined deletions in the chromosome of corynebacterium-glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 54.Schumacher, M. A., R. F. Pearson, T. Moller, P. Valentin-Hansen, and R. G. Brennan. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 21:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma, A. K., and S. M. Payne. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62:469-479. [DOI] [PubMed] [Google Scholar]
- 56.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic-analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 57.Sittka, A., S. Lucchini, K. Papenfort, C. M. Sharma, K. Rolle, T. T. Binnewies, J. C. Hinton, and J. Vogel. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63:193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35:217-228. [DOI] [PubMed] [Google Scholar]
- 60.Storz, G., J. A. Opdyke, and A. X. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 61.Sukhodolets, M. V., and S. Garges. 2003. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry 42:8022-8034. [DOI] [PubMed] [Google Scholar]
- 62.Sun, X., and R. M. Wartell. 2006. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry 45:4875-4887. [DOI] [PubMed] [Google Scholar]
- 63.Sun, X., I. Zhulin, and R. M. Wartell. 2002. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 30:3662-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsui, H. C. T., H. C. E. Leung, and M. E. Winkler. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia-coli-K-12. Mol. Microbiol. 13:35-49. [DOI] [PubMed] [Google Scholar]
- 65.Ulve, V. M., E. W. Sevin, A. Cheron, and F. Barloy-Hubler. 2007. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics 8:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525-1533. [DOI] [PubMed] [Google Scholar]
- 67.Valentin-Hansen, P., J. Johansen, and A. A. Rasmussen. 2007. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 10:152-155. [DOI] [PubMed] [Google Scholar]
- 68.Valverde, C., J. Livny, J. P. Schluter, J. Reinkensmeier, A. Becker, and G. Parisi. 2008. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics 9:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.