Abstract
Catechols are central intermediates in the metabolism of aromatic compounds. Degradation of 4-methylcatechol via intradiol cleavage usually leads to the formation of 4-methylmuconolactone (4-ML) as a dead-end metabolite. Only a few microorganisms are known to mineralize 4-ML. The mml gene cluster of Pseudomonas reinekei MT1, which encodes enzymes involved in the metabolism of 4-ML, is shown here to encode 10 genes found in a 9.4-kb chromosomal region. Reverse transcription assays revealed that these genes form a single operon, where their expression is controlled by two promoters. Promoter fusion assays identified 4-methyl-3-oxoadipate as an inducer. Mineralization of 4-ML is initiated by the 4-methylmuconolactone methylisomerase encoded by mmlI. This reaction produces 3-ML and is followed by a rearrangement of the double bond catalyzed by the methylmuconolactone isomerase encoded by mmlJ. Deletion of mmlL, encoding a protein of the metallo-β-lactamase superfamily, resulted in a loss of the capability of the strain MT1 to open the lactone ring, suggesting its function as a 4-methyl-3-oxoadipate enol-lactone hydrolase. Further metabolism can be assumed to occur by analogy with reactions known from the 3-oxoadipate pathway. mmlF and mmlG probably encode a 4-methyl-3-oxoadipyl-coenzyme A (CoA) transferase, and the mmlC gene product functions as a thiolase, transforming 4-methyl-3-oxoadipyl-CoA into methylsuccinyl-CoA and acetyl-CoA, as indicated by the accumulation of 4-methyl-3-oxoadipate in the respective deletion mutant. Accumulation of methylsuccinate by an mmlK deletion mutant indicates that the encoded acetyl-CoA hydrolase/transferase is crucial for channeling methylsuccinate into the central metabolism.
Aromatic compounds are among the most widely distributed organic substances in nature. They are present as aromatic amino acids and as constituents of fossil fuels and lignin. Microorganisms have developed the ability to use an impressive variety of such chemical compounds as carbon and energy sources (27, 61). An extensive array of substituted aromatic structures are transformed to a few central intermediates that undergo ring cleavage (10, 29).
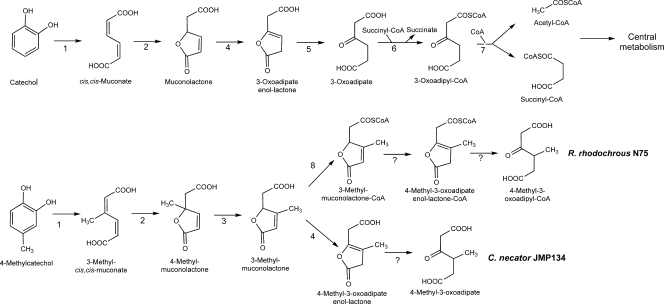
Catechol is one of the most important central intermediates in the aerobic metabolism of aromatic compounds, such as salicylate, benzoate, phenol, mandelate, and anthranilate, among others (29). This intermediate can be channeled into the Krebs cycle by ortho (intradiol) cleavage via the 3-oxoadipate pathway, which is a widely distributed route among soil bacteria (29). In this pathway, the aromatic ring is cleaved by a catechol-1,2-dioxygenase, resulting in the formation of cis,cis-muconate, which is subsequently transformed by a muconate cycloisomerase to muconolactone. This intermediate is further transformed to 3-oxoadipate-enol-lactone by a muconolactone isomerase. Subsequently, the enol-lactone is hydrolyzed by an enol-lactone hydrolase, and the resulting 3-oxoadipate is in turn channeled by 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase into the Krebs cycle (Fig. 1). However, the 3-oxoadipate pathway is not suited for the degradation of methylaromatics. If 4-methylcatechol is subjected to ortho cleavage, 4-methylmuconolactone (4-ML) accumulates (11, 35), since muconolactone isomerases require a proton at the C-4 carbon atom to catalyze the isomerization to enol-lactone (13). Most bacteria described so far mineralize methylaromatics via the alternative meta (extradiol) cleavage pathway (39, 56).
FIG. 1.
Present status of knowledge on ortho cleavage pathway for catechol (top) or 4-methylcatechol (bottom) degradation. Metabolic routes for 4-methylcatechol have been proposed for C. necator JMP134 (lower branch) and R. rhodochrous N75 (upper branch). Enzyme names are as follows: 1, catechol 1,2-dioxygenase; 2, muconate cycloisomerase; 3, 4-methylmuconolactone methylisomerase; 4, muconolactone or methylmuconolactone isomerase; 5, 3-oxoadipate enol-lactone hydrolase; 6, 3-oxoadipate:succinyl-CoA transferase; 7, 3-oxoadipyl-CoA thiolase; 8, 3-methylmuconolactone-CoA synthetase; ?, unknown enzymes.
Only two bacteria (Cupriavidus necator JMP134 [47] and Rhodococcus rhodochrous N75 [5]) have been reported to degrade 4-methylcatechol via an ortho cleavage pathway and to be capable of 4-ML mineralization. C. necator JMP134 harbors the mml gene cluster (CP000090: ReutA1502 to ReutA1508), which has been proposed to consist of seven open reading frames (ORFs) encoding enzymes and putative proteins involved in the metabolism of 4-ML (24, 46). Only 4-methylmuconolactone methylisomerase (MmlI) and methymuconolactone isomerase (MmlJ), encoded by the mmlI and mmlJ genes, respectively, have a described function (50, 53). By sequence comparison with this gene cluster, Cupriavidus necator H16 was also found to harbor a putative mml gene cluster (AY305378: PHG384 to PHG390). However, whether this cluster is functional or not remains to be elucidated.
Degradation of 4-ML in both C. necator JMP134 and R. rhodochrous N75 is initiated by MmlI (Fig. 1), which catalyzes the isomerization of 4-ML to 3-ML (6, 50). In C. necator JMP134, further degradation is accomplished by MmlJ, which by analogy with the 3-oxoadipate pathway transforms 3-ML to 4-methyl-3-oxoadipate enol-lactone (53). In addition, it has been proposed that in this strain, the enol-lactone intermediate may be transformed to 4-methyl-3-oxoadipate by a hydrolase (47). However, no typical enol-lactone hydrolase activity toward methyl-substituted muconolactones has been observed as yet (53).
In contrast, in R. rhodochrous N75, 3-ML is directly activated by a 3-methylmuconolactone-CoA synthetase, which catalyzes the synthesis of 3-ML-CoA from ATP, coenzyme A, and 3-ML (12). Unfortunately, no gene or protein sequence data related to this transformation are available. Further degradation of 3-ML-CoA has been proposed to proceed via 4-methyl-3-oxoadipyl-CoA, although details of this reaction are not available.
Recently Cámara et al. reported that Pseudomonas reinekei MT1 degrades 4-methylsalicylate via ortho cleavage of 4-methylcatechol (8). This strain harbors a gene cluster encoding a salicylate 1-hydroxylase (SalA), a catechol 1,2-dioxygenase (SalD), and a muconate cycloisomerase (SalC). Both SalD and SalC are specialized for the transformation of methyl-substituted substrates, ensuring effective funneling of methylaromatics into the ortho cleavage pathway. Additionally, P. reinekei MT1 exhibits MmlI activity (8), which indicates that methyl-substituted aromatics are degraded via 4-ML. In contrast to C. necator JMP134, which mineralizes methylaromatics, such as 4-methylphenol, mainly via a meta cleavage pathway despite the functionality of the ortho cleavage pathway (48), P. reinekei MT1 relies solely on the ortho cleavage route to mineralize methylcatechols and thus represents an ideal system with which to study this pathway in detail (8).
In this report, we describe a gene cluster encoding proteins involved in the degradation of 4-ML in P. reinekei MT1 and analyze the operonic organization and expression profile of these genes. Based on genetic data and on analysis of metabolites produced and accumulated in different deletion mutants, we were able to reconstruct the metabolic pathway encoded by this gene cluster.
MATERIALS AND METHODS
Chemicals.
4-ML, 3-ML, and 5-chloro-3-methylmuconolactone were prepared as described earlier (35, 47, 49). Methylsuccinate was obtained from Sigma-Aldrich (Steinheim, Germany).
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. P. reinekei strain MT1 was grown in minimal medium as previously described (41) with 5 mM salicylate or 4-methylsalicylate as the sole carbon source. C. necator JMP134::X, a derivative of C. necator JMP134 engineered to catabolize 4-methylbenzoate by chromosomal insertion of the xylXYZL genes, encoding a broad-substrate-range toluate 1,2-dioxygenase and a toluate dihydrodiol dehydrogenase (37), was grown in the same medium with 2.5 mM 4-methylbenzoate as the sole carbon source. Luria-Bertani (LB) medium was used as rich medium for Escherichia coli, P. reinekei, and C. necator strains. For selection of mutants, ABC medium (AB medium [18] supplemented with trace metals [22] and 20 mM citrate) was used. Antibiotics were used at the following concentrations: for E. coli, carbenicillin (Cb) (100 μg/ml), gentamicin (Gm) (10 μg/ml), tetracycline (Tc) (10 μg/ml), and spectinomycin (Sp) (100 μg/ml); for P. reinekei, Gm (200 μg/ml), Tc (15 μg/ml), and Sp (100 μg/ml); and for C. necator JMP134, Gm (20 μg/ml), Sp (100 μg/ml), and kanamycin (Km) (100 μg/ml).
Enzymatic assays.
Cell extracts of P. reinekei MT1 grown on 4-methylsalicylate were prepared as previously described (41). MmlI activity was measured by high-performance liquid chromatography (HPLC) following the transformation of 4-ML to 3-ML as reported previously (50). Activity of MmlJ was determined spectrophotometrically by measuring the transformation of 200 μM 5-chloro-3-methylmuconolactone in Tris-HCl (50 mM, pH 7.5) as previously described (54). One unit (U) was defined as μmol of product formed per minute.
Partial purification of MmlJ and N-terminal sequence determination.
MmlJ was partially purified by anion exchange chromatography using a MonoQ HR 5/5 column (GE Healthcare, Piscataway, NJ). Cells extracts were applied directly onto the column, and proteins were eluted by using a linear gradient of 0 to 0.5 M NaCl over 33 ml at a flow rate of 0.5 ml/min. MmlJ eluted at 0.37 ± 0.01 M NaCl. Aliquots of highly active fractions were subjected to SDS-PAGE and blotted onto a polyvinylidene difluoride membrane, and major protein bands with a molecular mass of ∼10 kDa were analyzed by N-terminal sequencing (32).
Fosmid library screening, sequencing, and sequence analysis.
In order to localize the mml gene cluster, part of the mmlJ gene was amplified by PCR using the degenerate primers NH3MMLIF1 and NH3MMLIR1, which were designed based on the N-terminal protein sequence of the partially purified MmlJ protein from P. reinekei MT1. Primer sequences are shown in Table S2 in the supplemental material. The 75-bp PCR product generated was cloned into the pGEM-T Easy vector (Promega, Madison, WI), transformed into E. coli Max Efficiency DH5α competent cells (Invitrogen, Carlsbad, CA), and sequenced. Based on the cloned sequence, a specific forward primer, NH3MMLIF3, was designed and used in a second PCR round with a reverse degenerate primer, NH3MMLIR4, designed from a sequence alignment of methylmuconolactone and muconolactone isomerases. The generated 125-bp fragment was cloned in the pGEM-T Easy vector, transformed into E. coli JM109 (Stratagene, La Jolla, CA), and sequenced. A previously constructed fosmid library of P. reinekei MT1 genomic DNA (8) was screened by PCR using the primers NH3MMLIF3 and NH3MMLIR7, specific for the mmlJ gene. Positive fosmid clones were purified using the FosmidMAX DNA purification kit (Epicentre, Madison, WI) and subjected to direct sequencing of upstream and downstream regions of the mmlJ gene, using the ABI Prism BigDye Terminator v1.1 ready reaction cycle sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3100 genetic analyzer (Applied Biosystems). Raw sequence data from both strands were assembled manually.
DNA and protein similarity searches were performed using the BLASTX and BLASTP programs from the NCBI website (3). Translated protein sequences were aligned with the MUSCLE software program, using default values (23). Phylogenetic trees were constructed using the MEGA4 software program (59), using the neighbor-joining algorithm (55) with p-distance correction and pairwise deletion of gaps and missing data. A total of 100 bootstrap replications were performed to test for branch robustness.
Extrachromosomal DNA extraction.
Detection of megaplasmids was attempted by pulsed-field gel electrophoresis (PFGE). P. reinekei MT1 was cultivated at 30°C in 100 ml LB medium to an A600 of 0.5. Cells were harvested by centrifugation and resuspended in SE solution (75 mM NaCl, 25 mM EDTA, pH 8). To avoid shearing of high-molecular-mass DNA, cells were mixed with an equal volume of 2% (wt/vol) low-melting-point agarose (Invitrogen). The mixture was poured into plugs, which were incubated overnight at 50°C with 0.5 mg/ml proteinase K. To inhibit the protease, the plugs were incubated in TE buffer (10 mM Tris-HCl, pH 8, 10 mM EDTA, pH 8) with 1 mM Pefabloc Sc [4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF)] (Boehringer Mannheim, Mannheim, Germany) for 2 h at 37°C. The plugs were rinsed five times with TE buffer at room temperature and stored at 4°C until used.
PFGE was performed by contour-clamped homogeneous electric field electrophoresis (CHEF) (15) using a CHEF-DRIII system (Bio-Rad, München, Germany). Multipurpose agarose (1% [wt/vol]) (Roche, Berlin, Germany) gel in TBE buffer (45 mM Tris-base, 0.5 mM boric acid, 0.1 mM EDTA) was used at 14°C for separation. Linearly increasing pulse times from 10 to 200 s were used during the total run time (24 h; 5.5 V/cm). Lambda ladder pulsed-field gel marker (New England BioLabs, Ipswich, MA) and Hansenula wingei YB-4662-VIA marker (Bio-Rad) were used as high-molecular-mass DNA standards.
Plasmid DNA extraction was performed using the QIAprep spin miniprep kit (Qiagen, Chatsworth, CA) according to the manufacturer's specifications, followed by electrophoresis on 1% agarose gels.
RT-PCR.
P. reinekei MT1 was grown overnight in minimal medium with 10 mM gluconate as a carbon source. During exponential growth (A600 = 0.7), the culture was induced by addition of 0.5 mM 4-methylsalicylate and further incubated for 1 h. After addition of 8 ml RNAprotect reagent (Qiagen), total RNA was isolated from a 12-ml aliquot using the RNeasy minikit (Qiagen), according to the manufacterer's instructions. The resulting RNA was quantified using a GeneQuant 1300 spectrophotometer (GE Healthcare) and treated with the Turbo DNase kit (Ambion, Austin, TX) to remove any DNA contamination. The reverse transcription-PCR (RT-PCR) was carried out using the ImProm-II reverse transcription system (Promega) with 1 μg of total RNA in a 20-μl reaction volume. After reverse transcription, PCR amplifications were carried out using the primer pair P01MT1/P02MT1, P1MT1/P2MT1, P3MT1/P4MT1, P5MT1/P6MT1, P7MT1/P8MT1, P9MT1/P10MT1, or P11MT1/P12MT1 (see Table S2 in the supplemental material) in a 25-μl total reaction mixture containing 1 μl of cDNA, 50 pmol of each primer, 50 μM (each) deoxynucleoside triphosphates, 1 mM MgCl2, 5 U of Taq DNA polymerase, and 1× reaction buffer supplied by the manufacturer. The temperature program was as follows: initial denaturation at 95°C for 5 min, and 30 cycles of 30 s at 95°C, 30 s at 60°C, and 60 s at 72°C, with a final extension step at 72°C for 10 min. Negative control reactions were carried out in the same way, excluding reverse transcriptase from the reaction mixtures.
For the detection of transcripts of C. necator JMP134::X, cells were grown in minimal medium with 10 mM fructose as a carbon source. During exponential growth (A600 = 0.7), the culture was supplemented with 4-methylbenzoate (0.5 mM) and incubated for 1 h. Total RNA extraction and reverse transcription were performed as described above for P. reinekei MT1 using the primer pair P1J134/P2J134 or P3J134/P4J134.
Amplification products (5 μl) were separated on 1% agarose gels after mixing with 1 μl of SYBR Safe DNA gel stain (Invitrogen).
Construction and testing of lacZ reporter fusions.
The presence of promoter regions was determined with lacZ reporter fusions in pKGWP0, a broad-range vector which was constructed as follows. The low background activity LacZ cassette and the multiple cloning site from plasmid pTZ110 (58) were amplified using the pTZ110LacZFW and pTZ110LacZRV primers and cloned into pCR8/GW/TOPO (Invitrogen) to yield pTOPO-MCS-LacZ. The LacZ cassette and the multiple cloning site sequence were transferred from pTOPO-MCS-LacZ to the gateway-compatible and broad-host-range pKGW vector (33) by recombination-based transfer using the Gateway LR Clonase II enzyme mix (Invitrogen) according to the manufacturer's instructions. The integrity of the resulting pKGWP0 vector was confirmed by sequencing.
Putative promoter regions were fused to the lacZ reporter gene of pKGWP0 as follows. A 258-bp PCR product comprising the bp 12 to 269 region upstream of the translational start site of the mmlL gene of P. reinekei MT1 was amplified with the primers PmHydMT1FW and PmHydMT1RV. Similarly, a 366-bp PCR product comprising the bp 12 to 377 region upstream of the translational start site of mmlC was amplified using the primers PmACAT_FW and PmACAT_RV. The amplified fragments were blunt end cloned into the StuI restriction site of pKGWP0, forming the plasmids pm_mmlL and pm_mmlC, respectively. The lacZ fusion of the putative promoter region of the mmlL gene of C. necator JMP134 was constructed by introducing a 332-bp PCR product comprising the sequence immediately upstream of the translational start site of mmlL. This region was amplified with the primers PmmlLFWEcoRI and PmmlLRVBamHI for insertion into the EcoRI/BamHI site of plasmid pTZ110 to generate plasmid pTZpm_mmlLJMP134. The cassette containing lacZ fused to the upstream region of mmlL from plasmid pTZpm_mmLJMP134 was amplified using the pTZ110LacZFW and pTZ110LacZRV primers and cloned into pCR8/GW/TOPO (Invitrogen) to yield pTOPO-pm_mmLJMP134-lacZ. This cassette was transferred from pTOPO-pm_mmLJMP134-lacZ to the pKGW vector by recombination-based transfer using the Gateway LR Clonase II enzyme mix to give pm_mmlLJMP134. The integrity of the constructs was verified by PCR and sequencing. Plasmids harboring the putative promoter regions were transferred to P. reinekei MT1, P. reinekei MT1ΔmmlL, P. reinekei MT1ΔmmlC, and C. necator JMP134::X by biparental mating using E. coli S17λpir as a donor strain. Transconjugants were selected in minimal medium supplemented with Sp. Reporter fusion assays were performed as previously described (38) using 0.5 mM 4-methylsalicylate, 4-methylbenzoate, 4-ML, or 3-ML as an inducer. Activities are expressed in Miller units and were determined after 4 h of induction.
Construction of deletion mutants.
mmlC, mmlD, mmlK, and mmlL gene deletion mutants were constructed with the previously described Flp-Flp recombinase target (FRT) recombination strategy (31). Briefly, PCR fragments upstream and downstream of the targeted genes (∼700 bp) were amplified with primer pairs carrying restriction sites (PstI-BamHI and BamHI-Acc65l, respectively) and cloned into the PstI-Acc65l restriction site of the pEX18Ap vector, forming the pABmml plasmid series. Subsequently, a 1.8-kb BamHI fragment from the pS858 plasmid carrying a Gmr-green fluorescent protein (GFP) cassette was cloned into the BamHI restriction site formed to give the pAGBmml plasmid series. The resulting constructs and the suicidal plasmids used for the construction of the different mutants are listed in Table S1 in the supplemental material.
These suicide plasmids were transferred independently into P. reinekei MT1 by biparental mating using E. coli S17λpir as a donor strain. The transconjugants generated by single crossover were selected on ABC medium supplemented with Gm, and merodiploids were resolved by additional plating on ABC medium supplemented with 5% sucrose. Deletion of the Gmr-GFP cassette was achieved by conjugation of the Flp-expressing pBBFLP plasmid (19) into the resulting strains by biparental mating using E. coli CC118λpir (30) as a donor and selection on ABC medium containing Tc. Plasmid pBBFLP was cured by streaking strains on ABC medium supplemented with 5% sucrose. The integrity of all mutants was verified by growth on ABC medium supplemented with different antibiotics, PCR amplification, and sequencing of regions flanking the deleted genes.
Complementation of MT1ΔmmlL mutant.
The MT1ΔmmlL deletion mutant was separately complemented with the mmlL genes from P. reinekei MT1 and C. necator JMP134. The mmlL gene from P. reinekei MT1 was amplified using the primers PmZnHydXbaIF and ZnHydSacIR, which introduce XbaI and SacI restriction sites, respectively, and cloned into the SacI-XbaI restriction site of the pBS1 vector (4) to give pBS1mmlLMT1. The mmlL gene from C. necator JMP134 was PCR amplified with the primers mmlLFW and mmlLRV and cloned using the pCR8/GW/TOPO cloning kit (Invitrogen) to form pTOPOmmlLJMP134. Subsequently, the insert was transferred to pBS1 by recombination-based transfer of the PCR product using the Gateway LR Clonase II enzyme mix (Invitrogen), according to the manufacturer's instructions, to give pBS1mmLJMP134. The integrity of both pBS1mmlL plasmids was confirmed by sequencing. Both plasmids were transferred independently to the MT1ΔmmlL deletion mutant by biparental mating using E. coli S17λpir (20) as a donor. Transconjugants were selected by plating on ABC medium supplemented with Gm.
Transformation of substrates and identification of metabolites.
For preparation of resting cells, wild-type P. reinekei MT1 and mutants were grown in minimal medium with salicylate (5 mM) as a carbon source at 30°C. During late exponential growth, cells were harvested by centrifugation and washed with 50 mM phosphate buffer (pH 7.4). Cells were suspended in the same buffer (A600 = 3.0) and supplemented with 1 mM 4-methylsalicylate, 10 mM glucose, and trace salts (22). Cell suspensions (three replicates) were incubated at 30°C and 150 rpm. After appropriate time intervals, aliquots were centrifuged and the cell-free supernatants were analyzed by HPLC and 1H nuclear magnetic resonance (NMR) spectroscopy.
In order to verify the chemical structure of the metabolite accumulated by the mutant MT1ΔmmlL, the metabolite was extracted after acidification to pH 3 from the cell-free supernatant (30 ml) with five times 20 ml ethyl acetate. Extracts were dried over MgSO4, evaporated to dryness on a rotary evaporator, and dissolved in 0.7 ml d6-acetone. Further samples for 1H NMR spectroscopy were prepared by addition of 140 μl of D2O water to 560 μl of cell-free supernatants.
Analytical methods.
HPLC was performed with a Lichrospher SC 100 RP8 reversed-phase column (125 by 4.6 mm; Bischoff, Leonberg, Germany). Methanol-H2O containing 0.1% (vol/vol) H3PO4 was used an as eluent at a 1-ml/min flow rate. The column effluent was monitored simultaneously at 210, 260, and 280 nm with a diode array detector (Shimadzu, Duisburg, Germany). Typical retention volumes were as follows: methanol-H2O, 58:42; 4-methylsalicylate, 5.0 ml; methanol-H2O, 10:90; 4-ML, 5.2 ml; 3-ML, 4.7 ml.
One-dimensional and two-dimensional correlation spectroscopy (COSY) 1H NMR spectra were recorded at 300 K on Avance DPX 300 and DMX 600 NMR spectrometers (Bruker, Bremen, Germany). The center of the suppressed water signal (δ = 4.80 ppm) was used as an internal reference. The concentrations of the accumulated metabolites in the samples were estimated by comparison of the average of the integrals of the resonance lines of the protons H2/H6 (δ = 7.49 ppm) and H3/H5 (δ = 7.84 ppm) of 4-chlorobenzoate with the integral of the resonance lines of the protons of the methyl groups of 4-methyl-3-oxoadipate (δ = 1.13 ppm) or methylsuccinate (δ = 1.10 ppm), respectively. 4-Chlorobenzoate was added to a final concentration of 1 mM.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was deposited in the DDBJ/EMBL/GenBank databases under the accession number GQ141876.
RESULTS
P. reinekei MT1 contains 4-methylmuconolactone methylisomerase and methylmuconolactone isomerase activities.
Previous analyses have shown that P. reinekei MT1 degrades 5-methyl- and 4-methylsalicylate exclusively via an ortho cleavage route and is able to transform 4-ML into 3-ML, indicating the presence of a 4-methylmuconolactone methylisomerase (MmlI) (8). Further transformation of 3-ML by cell extracts was not observed (8). However, analysis for muconolactone isomerase activity revealed the presence of such activity (255 U/g of protein) in P. reinekei MT1 cell extracts, similar to the situation in C. necator JMP134 (53). The N-terminal sequence of the partially purified enzyme (MLYCVEMTVSIPRRIPLDEVERIKAAXKERAID) differs from that of the previously characterized muconolactone isomerase of this strain (CatC of the 3-oxoadipate pathway) (9) in 22 out of the 32 determined residues. This suggests the induction of a methylmuconolactone isomerase (MmlJ) in P. reinekei MT1, responsible for the reversible rearrangement of the double bond of 3-ML to form 4-methyl-3-oxoadipate enol-lactone.
Identification and analysis of ORFs involved in 4-methylmuconolactone degradation in P. reinekei MT1.
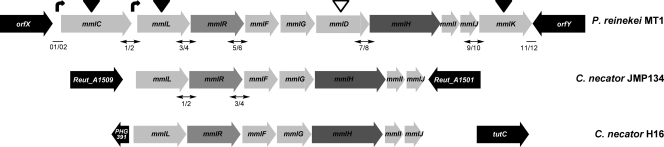
To obtain further insights into genes and proteins involved in the metabolism of methylmuconolactones in P. reinekei MT1, the region surrounding the mmlJ gene, encoding the methylmuconolactone isomerase, was analyzed as outlined in Materials and Methods. An overall 11.6 kb containing 12 ORFs was retrieved. Sequence comparison with the mml clusters present on chromosome 1 of C. necator JMP134 (24, 46) and on megaplasmid pHG1 of C. necator H16 showed the presence of seven orthologous genes probably involved in the degradation of 4-ML (Fig. 2). The ORFs were designated mml by analogy with the mml genes of C. necator JMP134. The putative activities encoded by these genes are summarized in Table 1.
FIG. 2.
Comparison of the mml gene cluster of P. reinekei MT1 with those of C. necator JMP134 and C. necator H16. Genes encoding catabolic enzymes, transcriptional regulators, and putative transporters are indicated in light gray, gray, and dark gray, respectively. Genes framing the mml gene clusters are indicated in black. Genes essential for growth on 4-methylsalicylate are indicated with black triangles, whereas those that are dispensable are indicated with unfilled triangles. Numbers below the arrows indicate the primer pairs utilized to assess transcription of intergenic regions. Double-headed arrows represent intergenic regions transcribed during growth on 4-methylsalicylate (MT1) or 4-methylbenzoate (JMP134::X), and lines indicate those regions not transcribed. Experimentally determined promoter regions are indicated by curved arrows.
TABLE 1.
ORFs and genes of the mml gene cluster of P. reinekei MT1 and surrounding regions
| Gene | Gene product |
Related gene productb |
||||
|---|---|---|---|---|---|---|
| Size (aaa) | Putative function | Name (size [aa]) | Organism | % aa identity | Accession no. (reference) | |
| orfX | 277 | Itaconyl-CoA hydratase | (275) | Pseudomonas aeruginosa PAO1 | 63 | NP_249569 |
| (278) | Pseudomonas sp. L1 | 53 | AAX86477 | |||
| mmlC | 398 | Thiolase | (398) | Burkholderia cenocepacia MC0-3 | 67 | ACA96001 |
| PaaE (401) | Pseudomonas fluorescens | 45 | ABF82237(21) | |||
| mmlL | 297 | Hydrolase | (296) | C. necator H16 | 79 | AAP86139 |
| OPHC2 (324) | Pseudomonas pseudoalcaligenes | 26 | CAE53631 (16) | |||
| mmlR | 300 | Transcriptional regulator, LysR type | (304) | C. necator H16 | 71 | AAP86138 |
| mmlF | 230 | 3-Oxoadipyl-CoA transferase α-subunit | (232) | C. necator H16 | 72 | AAP86137 |
| PcaI (231) | P. putida PRS2000 | 68 | AAA25922(44) | |||
| mmlG | 222 | 3-Oxoadipyl-CoA transferase β-subunit | (220) | C. necator H16 | 70 | AAP86136 |
| PcaJ (218) | Acinetobacter baylyi ADP1 | 59 | AAC37147(28) | |||
| mmlD | 295 | Acyl-CoA thioesterase | (303) | Methylocella silvestris BL2 | 32 | ACK50807 |
| mmlH | 429 | Transporter | (428) | C. necator JMP134 | 68 | AAZ60871 |
| MucK (426) | A. baylyi ADP1 | 30 | AAC27117(62) | |||
| mmlI | 107 | 4-Methylmuconolactone methylisomerase | MmlI (113) | C. necator JMP134 | 70 | AAZ60870(50) |
| mmlJ | 92 | Methylmuconolactone isomerase | (91) | C. necator H16 | 71 | AAP86133 |
| MmlJ (91) | C. necator JMP134 | 65 | AAZ60869(53) | |||
| mmlK | 422 | Acetyl-CoA hydrolase/transferase | (430) | Burkholderia sp. H160 | 58 | EEA04061 |
| (442) | A. caccae L1-92 | 36 | ABA39275(14) | |||
| orfY | 320 | Transcriptional regulator, LysR type | (311) | Burkholderia glumae BGR1 | 44 | YP_002907790 |
| CnmA (310) | P. putida JLR11 | 35 | AAW80266(7) | |||
aa, amino acids.
The gene product with the highest amino acid sequence identity and the most closely related gene product of validated function are given.
PFGE of total DNA and plasmid DNA extraction of P. reinekei MT1 gave no indication of the presence of plasmids in this strain, which suggests that the region harboring these mml genes is located on the chromosome, as in strain JMP134, and not on a plasmid as in strain H16.
Only proteins encoded by the mmlI and mmlJ genes have a proven function in C. necator JMP134 (50, 53). The mmlI gene encodes MmlI, a unique enzyme belonging to the MmlI protein family (PF09448). The predicted enzyme of P. reinekei MT1 shares 70% and 69% of sequence identity with MmlI of C. necator JMP134 and with the predicted MmlI protein of C. necator H16, the only homologues currently available from public databases. Phylogenetic analysis indicated that the mmlJ gene product of P. reinekei MT1 is most closely related to the mmlJ gene products of C. necator JMP134 and H16 but only distantly related to muconolactone isomerases encoded in 3-oxoadipate pathway gene clusters (see Fig. S1 in the supplemental material). The mmlL gene encodes a putative metal-dependent hydrolase, which belongs to the metallo-β-lactamase superfamily (cl00446). At the sequence level, the most closely related enzyme (only 26% identity) with proven function is the organophosphorus hydrolase (OPHC2) of Pseudomonas pseudoalcaligenes C2-1, which catalyzes the hydrolysis of phosphoester bonds (see Fig. S2 in the supplemental material) (16). The proteins encoded by the mmlF and mmlG genes are most closely related to those encoded by the mmlF and mmlG genes of C. necator JMP134 (72% and 68% identity, respectively) and H16 (72% and 70% identity, respectively) (see Fig. S3 in the supplemental material). However, they also share significant sequence identity with 3-oxoadipyl CoA transferases of proven function, such as the one from Pseudomonas putida PRS2000 (68% and 65% identity, respectively), which is part of the 3-oxoadipate pathway (44). This suggests that the mmlF and mmlG gene products have 3-oxoadipyl-CoA transferase activity and act on 4-methyl-3-oxoadipate, forming 4-methyl-3-oxoadipyl-CoA, by analogy with the 3-oxoadipate pathway. The mmlH gene encodes a putative transporter of the major facilitator superfamily (cd06174), which could be responsible for internalization of extracellular muconolactones or dicarboxylic acids, and mmlR encodes a putative LysR-type transcriptional regulator.
The organization of these seven genes both in C. necator strains and in P. reinekei MT1 is remarkably similar, except that in P. reinekei MT1 an ORF termed mmlD is located between the mmlG and mmlH genes. The mmlD gene encodes a putative acyl-CoA thioesterase which has up to 31% identity to TesB proteins, such as those from P. putida KT2440 (17) or E. coli K-12 (40), which have been described to catalyze the cleavage of C6-C18 carbon fatty acid CoA thioesters and of short acyl-CoA compounds (see Fig. S4 in the supplemental material).
The regions upstream of mmlL and downstream of mmlJ in P. reinekei MT1 differ significantly from those of both C. necator strains. Only in strain MT1, mmlL is preceded by an ORF termed mmlC, which encodes a putative protein of the thiolase family (cd00751). Members of this family catalyze the reversible thiolytic cleavage of 3-ketoacyl-CoA into acyl-CoA. Therefore, MmlC belongs to a broad protein family, which also contains 3-oxoadipate CoA thiolases, such as the enzyme from Pseudomonas knackmussii B13 (34) with which it shares 42% sequence identity (see Fig. S5 in the supplemental material). This indicates that MmlC may function as a thiolase transforming 4-methyl-3-oxoadipyl-CoA into methylsuccinyl-CoA and acetyl-CoA.
An additional ORF, termed mmlK, is located downstream of the mmlJ gene in P. reinekei MT1. This gene encodes a putative acetyl-CoA hydrolase/transferase with 36% identity to 4-hydroxybutyrate CoA transferase of Anaerostipes caccae (see Fig. S6 in the supplemental material) (14).
Genes of the mml cluster form a single operon and are induced in the presence of 4-ML and 3-ML.
The operonic structures of the mml gene clusters from P. reinekei MT1 and C. necator JMP134 were determined by RT-PCR using total RNA isolated from both strains, induced with 4-methylsalicylate and 4-methylbenzoate, respectively. The transcription of intergenic regions, considered of sufficient length to harbor a promoter, was assessed for seven regions in P. reinekei MT1 and for two in C. necator JMP134 (Fig. 2). Amplification products were obtained for five out of the seven assessed intergenic regions in P. reinekei MT1 and for both intergenic regions in C. necator JMP134 (see Fig. S7 in the supplemental material). An absence of mRNA comprising the orfX-mmlC and mmlK-orfY intergenic regions of P. reinekei MT1 indicates that the regions defined as mml clusters form single operons in both P. reinekei MT1 and C. necator JMP134 (Fig. 2).
Since RT-PCR analysis suggests the presence of promoters upstream of mmlC in P. reinekei MT1 and upstream of mmlL in C. necator JMP134, lacZ transcriptional fusions of intergenic regions upstream of mmlL and mmlC in strain MT1 and of the intergenic region upstream of mmlL in strain JMP134 were constructed and provided in trans to P. reinekei MT1 or C. necator JMP134::X. β-Galactosidase assays showed an approximately 10- to 20-fold increase in LacZ activity after incubation with 4-methylsalicylate (tested only in P. reinekei MT1), 4-methylbenzoate (tested only in C. necator JMP134), 4-ML, or 3-ML (Table 2), which indicates the functionality of all three putative promoters in their native background. To identify the nature of the inducer, transcriptional fusions of lacZ and intergenic regions upstream of mmlL and mmlC of strain MT1 were also introduced in P. reinekei MT1ΔmmlL and P. reinekei MT1ΔmmlC, which are incapable of mineralizing 4-methylsalicylate via 4-ML due to deletions in the mmlL and mmlC genes and quantitatively accumulate 3-ML or 4-methyl-3-oxoadipate, respectively (see below). Expression of the lacZ fusions was observed only in the MT1ΔmmlC background, indicating 4-methyl-3-oxoadipate is the inducer of the mml gene cluster.
TABLE 2.
β-Galactosidase activity resulting from expression of promoter fusions in P. reinekei MT1 and C. necator JMP134a
| Strain tested | β-Galactosidase activity with inducer |
||||
|---|---|---|---|---|---|
| None | 4-Methylbenzoate | 4-Methylsalicylate | 4-ML | 3-ML | |
| MT1(pm_mmlC) | 90 ± 15 | ND | 940 ± 160 | 1000 ± 50 | 980 ± 100 |
| MT1(pm_mmlLMT1) | 310 ± 70 | ND | 2140 ± 80 | 2280 ± 70 | 2270 ± 130 |
| JMP134::X(pm_mmlLJMP134) | 3.0 ± 0.2 | 62 ± 8 | ND | 52 ± 3 | 58 ± 8 |
| MT1ΔmmlL(pm_mmlC) | 86 ± 23 | ND | ND | 157 ± 38 | ND |
| MT1ΔmmlL(pm_mmlLMT1) | 36 ± 4 | ND | ND | 38 ± 3 | ND |
| MT1ΔmmlC(pm_mmlC) | 32 ± 2 | ND | ND | 620 ± 20 | ND |
| MT1ΔmmlC(pm_mmlLMT1) | 290 ± 15 | ND | ND | 5850 ± 290 | ND |
The inducers were added at the beginning of the exponential phase to a final concentration of 0.5 mM, and activity was determined after a period of 4 h. ND, not determined. Whereas P. reinekei MT1 and C. necator JMP134 are capable of mineralizing the inducers, P. reinekei MT1ΔmmlL transforms 4-ML quantitatively to 3-ML and P. reinekei MT1ΔmmlC transforms 4-ML quantitatively to 4-methyl-3-oxoadipate. Activities are expressed as Miller units.
mmlL, mmlC, and mmlK genes are essential for growth of P. reinekei MT1 on 4-methylsalicylate.
Directed deletions of mmlL, mmlC, mmlD, and mmlK from P. reinekei MT1 were performed in order to clarify the role of these genes in the degradation of 4-ML. The MT1ΔmmlL, MT1ΔmmlC, and MT1ΔmmlK mutants were unable to grow on 4-methylsalicylate as the only carbon source, whereas growth on salicylate was not affected. In contrast, deletion of mmlD had no effect on the ability of strain MT1 to grow on 4-methylsalicylate (2 mM). Both wild-type and mutant MT1ΔmmlD grew, with doubling times of 1.34 ± 0.03 h and 1.25 ± 0.08 h, respectively, on 4-methylsalicylate and with doubling times of 1.29 ± 0.08 h and 1.23 ± 0.18 h on salicylate.
Complementation of mutant MT1ΔmmlL with plasmid pBS1mmlLMT1, harboring the mmlL gene of P. reinekei MT1, was performed in order to rule out possible polar effects. Furthermore, transcomplementation with pBS1mmlLJMP134, harboring the mmlL gene of C. necator JMP134, was performed. In both cases, the ability to grow on 4-methylsalicylate was fully restored.
4-Methyl-3-oxoadipate and methylsuccinate are intermediates in degradation of 4-ML by P. reinekei MT1.
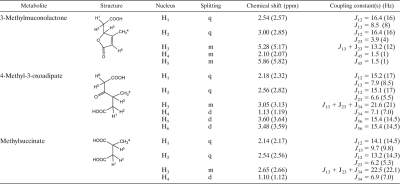
In order to determine the intermediates accumulated by the MT1ΔmmlL, MT1ΔmmlC, and MT1ΔmmlK mutants, resting cell assays were performed using 1 mM 4-methylsalicylate as a substrate. HPLC and 1H NMR analysis revealed that the mutant MT1ΔmmlL transforms 4-methylsalicylate quantitatively into 3-ML, which accumulated after 24 h up to 1.13 ± 0.08 mM (Table 3). The mutants MT1ΔmmlC and MT1ΔmmlK transform 4-methylsalicylate without accumulation of UV-absorbing metabolites. Analysis by 1H NMR spectroscopy of cell-free supernatants after complete transformation of the substrate (6 h), as well as after extended incubation (24 h), indicated that MT1ΔmmlC accumulates a single metabolite, the 1H NMR spectrum of which was essentially identical to that previously described for the dimethylester of 4-methyl-3-oxoadipate (Table 3) (47). Spiking with 4-chlorobenzoate as an internal standard showed that 4-methyl-3-oxoadipate accumulates stoichiometrically (1.18 ± 0.02 mM). 4-Methyl-3-oxoadipate was also excreted by the wild-type strain, although the amount accumulated did not exceed 0.23 ± 0.03 mM. The mutant MT1ΔmmlK accumulates two metabolites. 1H NMR analysis indicated that one of these corresponds to 4-methyl-3-oxoadipate (0.66 ± 0.03 mM). A second metabolite, observed in large amounts (0.49 ± 0.04 mM), was identified as methylsuccinate by comparison of its 1H NMR spectral characteristics with those of authentic material (Table 3).
TABLE 3.
1H NMR data of metabolites formed by P .reinekei MT1 deletion mutantsa
Chemical shift and coupling constants were calculated from representative spectra obtained from supernatants of mutant MT1ΔmmlC (4-methyl-3-oxoadipate) or MT1ΔmmlK (methylsuccinate) after incubation with 4-methylsalicylate or extract of supernatant of mutant MT1ΔmmlL (3-ML) dissolved in acetone-d6. 1H NMR data were recorded at 300 MHz. 1H NMR data previously described for 3-methylmuconolactone recorded at 80 MHz in CDCl3 (47) (top section) and those of 4-methyl-3-oxoadipate dimethylester in CDCl3 recorded at 200 MHz (47) (middle section) or of authentic methylsuccinate dissolved in the same medium and recorded at 600 MHz (bottom section) are given in parentheses. Doublets, quartets, and multiplets are abbreviated as d, q and m, respectively.
DISCUSSION
P. reinekei MT1 is the only natural isolate reported thus far to grow on methylaromatics exclusively via an ortho cleavage pathway. To achieve this, P. reinekei MT1 harbors extraordinary catabolic features. This bacterium contains, besides an ortho cleavage pathway for catechol degradation via the 3-oxoadipate pathway, a catechol 1,2-dioxygenase and a muconate cycloisomerase, which are highly specialized for the transformation of methyl-substituted substrates (8). The genes encoding these two enzymes are organized in a gene cluster, termed the sal cluster, which also comprises a gene encoding salicylate 1-hydroxylase (8). This organization ensures efficient transformation of 4-methyl- and 5-methylsalicylate to 4-ML. Further degradation of 4-ML is initiated by MmlI. This enzyme is encoded by the mml cluster, comprising 10 catabolic genes and transcribed as a single operon, with 4-methyl-3-oxoadipate acting as an inducer (Fig. 2).
Previously it was proposed that the degradation of 3-ML in C. necator proceeds via a route analogous to the 3-oxoadipate pathway with MmlJ, as the enzyme responsible for rearrangement of the double bond to form 4-methyl-3-oxoadipate enol-lactone, thus preparing the substrate for subsequent hydrolysis (47, 53) (Fig. 1). However, evidence for an enzyme performing an equivalent hydrolysis of a methylsubstituted 3-oxoadipate enol-lactone has not been reported thus far. The accumulation of 3-ML in the mutant MT1ΔmmlL indicates that the mmlL gene product probably is involved in the hydrolysis of the lactone ring and therefore that mmlL encodes a methylenol-lactone hydrolase, which is able to transform 4-methyl-3-oxoadipate enol-lactone into 4-methyl-3-oxoadipate. The accumulation of 3-ML rather than 4-methyl-3-oxoadipate-enol-lactone is explained by the reversibility of the MmlJ-catalyzed reaction, where the equilibrium favors the formation of the muconolactone (Fig. 3) (43).
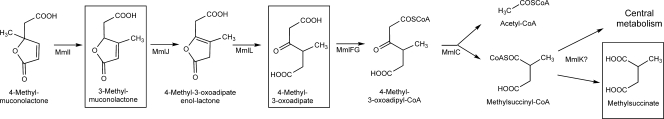
FIG. 3.
Proposed pathway for 4-ML degradation by P. reinekei MT1. Metabolites identified in the current study are depicted in boxes. MmlI, 4-methylmuconolactone methylisomerase; MmlJ, methylmuconolactone isomerase; MmlL, 4-methyl-3-oxoadipate enol-lactone hydrolase; MmlFG, 4-methyl-3-oxoadipate-CoA transferase; MmlC, 4-methyl-3-oxoadipyl-CoA thiolase; MmlK, acetyl-CoA-transferase/hydrolase.
4-Methyl-3-oxoadipate may be further metabolized by reactions identified from the classical 3-oxoadipate pathway, where 3-oxoadipate is transformed to 3-oxoadipyl-CoA by two-component 3-oxoadipate:succinyl-CoA transferases (termed PcaIJ or CatIJ). From the sequence identity with functionally characterized 3-oxoadipate:succinly-CoA transferases (34, 44), it is reasonable to assume that the mmlFG gene products are responsible for transformation of 4-methyl-3-oxoadipate into 4-methyl-3-oxoadipyl-CoA (Fig. 3). Knockout mutants of mmlF and mmlG were not generated, since pcaIJ genes, which could eventually be recruited and thus mask the mmlFG mutant phenotype, are typically observed in Pseudomonas strains.
Subsequent transformation of 3-oxoadipyl-CoA via the 3-oxoadipate pathway is catalyzed by 3-oxoadipyl-CoA thiolase, forming succinyl-CoA and acetyl-CoA (Fig. 3). 3-Oxoadipyl-CoA thiolases have been biochemically characterized for various Gram-negative bacteria, including the 3-oxoadipyl-CoA thiolases of P. knackmussii B13 (34) or P. putida PRS2000 (44). Thus far, 3-oxoadipyl-CoA thiolases of Gram-positive organisms have not been characterized, although previous analysis of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP (26) and recent genome sequencing projects show the presence of orthologous genes located in protocatechuate catabolic gene clusters of rhodococci, such as Rhodococcus jostii RHA1, whose functionality has been supported by transcriptomic and proteomic analysis (45). The close phylogenetic relationship of MmlC with PcaF of rhodococci (see Fig. S5 in the supplemental material) and the accumulation of 4-methyl-3-oxoadipate by the mutant MT1ΔmmlC support the notion that this enzyme functions as a 4-methyl-3-oxoadipyl-CoA thiolase, transforming its substrate into methylsuccinyl-CoA and acetyl-CoA (Fig. 3). Whether the accumulation of 4-methyl-3-oxoadipate, instead of the CoA thioester, is due to the action of a thioesterase such as MmlD remains to be elucidated. However, the release into the culture medium of the free acids rather than of CoA derivatives has been frequently reported (2) and has been suggested as a general strategy of bacterial cells to prevent the depletion of the intracellular CoA pool (42).
As indicated above, methylsuccinyl-CoA may be formed by MmlC during the degradation of 4-ML (Fig. 3). In fact, methylsuccinate is accumulated by the mutant MT1ΔmmlK, suggesting that methylsuccinate and/or its CoA derivative is a metabolite of 4-ML degradation. Information on the metabolic fate of methylsuccinate or methylsuccinyl-CoA is limited. Both compounds have been shown to occur as intermediates in the metabolism of 4-methylcatechol by the fungus Trichosporon cutaneum (51). In this organism, 4-methylcatechol is degraded via intradiol cleavage, but in contrast to the case with bacteria, cycloisomerization of 3-methyl-cis,cis-muconate produces 3-ML directly, thus circumventing the formation of 4-ML. The further metabolism occurs, as indicated above for P. reinekei MT1, through 4-methyl-3-oxoadipate, 4-methyl-3-oxoadipyl-CoA, and methylsuccinate. Unfortunately, no sequence information is available for either genes or proteins involved in this process (51, 52). The metabolism of methylsuccinyl-CoA proceeds via hydrolysis to the free acid, and further reactions are assumed to occur after esterification at the C-4 carbon via itaconyl-CoA and citramalyl-CoA. Methylsuccinyl-CoA has been additionally reported to be an intermediate in two pathways, the ethylmalonyl-CoA pathway for acetate assimilation in Rhodobacter sphaeroides (1, 25) and the glyoxylate regeneration cycle of Methylobacterium extorquens (36). In both cases, methylsuccinate is esterified at the C-1 carbon as an intermediate. In light of these observations, the metabolic fate of methylsuccinate in P. reinekei MT1 and whether mmlK encodes a methylsuccinyl-CoA hydrolase remain to be elucidated. A significant mechanistic difference between the 3-oxoadipate pathway and the 4-ML degradative pathway also has to be considered for future analysis. In the 3-oxoadipate activation/fission process, typically each molecule of succinyl-CoA used in activation is regenerated as soon as 3-oxoadipyl-CoA is cleaved. However, it remains unclear whether methylsuccinyl-CoA is directly used by MmlC for thiolytic cleavage of 4-methyl-3-oxoadipy-CoA or whether succinyl-CoA is independently generated and MmlK encodes a CoA transferase involved in such reactions. Biochemical characterization of enzymes encoded by the mml cluster is currently being performed in order to characterize their substrate and cofactor specificities.
In contrast to the mmlL, mmlC, and mmlK genes, the mmlD gene, which encodes a putative acyl-thioesterase, is dispensable for growth of P. reinekei MT1 on 4-methylsalicylate. It should be noted that not only the mmlD gene but also the mmlC and mmlK genes are absent from the mml clusters of C. necator JMP134 and H16. Since C. necator JMP134 has been reported to grow on 4-ML (47), the required genetic elements and their respective activities should be recruited from elsewhere on the genome. Even though 3-oxoadipyl-CoA thiolase from the 3-oxoadipate pathway is obviously not recruited to substitute for MmlC in P. reinekei MT1, it cannot be excluded that this happens in C. necator. A genome-wide analysis of both Cupriavidus strains indicated that only the genome of strain H16 encodes a thiolase with high sequence identity to MmlC (YP_840888; 64% identity). Interestingly, the gene encoding this enzyme is preceded by a gene (YP_840887) the putative gene product of which exhibits significant sequence identity (55%) with MmlK. The most closely related MmlC homologues in C. necator JMP134 are ReutA_1348 (YP_295562; 42% identity), which, based on its sequence identity and genomic context, can be assumed to be involved in polyhydroxyalkanoate formation, and ReutA_1355 (YP_295567; 43% identity). Whether these or other unrelated proteins carry out thiolytic cleavage of 4-methyl-3-oxoadipyl-CoA in C. necator JMP134 remains to be elucidated.
As mentioned above, an MmlK homologue is present in C. necator H16 but not in C. necator JMP134, which suggests that the channeling of methylsuccinly-CoA/methylsuccinate into the central metabolism proceeds by different pathways in P. reinekei MT1 and C. necator JMP134.
However, even though the mml clusters differ in the presence of the mmlC, mmlK, and mmlD genes, their organization is otherwise identical, with promoters being localized upstream of mmlL. It thus may be speculated that in order to be capable of functioning in P. reinekei MT1, an archetype mml gene cluster was complemented by additional genes. Nevertheless, it should also be noted that proteins encoded by homologous genes share only 65 to 70% sequence identity. As an example, the level of identity between methylmuconolactone isomerases (65%) resembles those between muconolactone isomerases from Pseudomonas and Cupriavidus strains (54 to 59%) rather than between muconolactone isomerases from different Pseudomonas strains (>80%). It can thus be assumed that both gene clusters diverged from a common ancestor in ancient times.
Despite the huge amount of information available from genome projects, an mml cluster with MmlI has been observed only in P. reinekei MT1, C. necator JMP134, and C. necator H16. It should be stated, however, that currently available genomes give only a highly biased overview on bacterial metabolic properties. Taking into account the widespread distribution of the 3-oxoadipate pathway at least in the Proteobacteria plus the fact that catechol 1,2-dioxygenases and muconate cycloisomerases in general exhibit significant activity with methyl-substituted substrate analogues (8, 57, 60), it can be reasoned that in the environment, a significant amount of methyl-substituted aromatics are funneled into such a route and methylmuconolactone degraders could play an important role in further funneling these intermediates into the Krebs cycle.
Supplementary Material
Acknowledgments
This work was supported by the DFG-European Graduate College 653, FONDECYT grant 1070343, Millennium Nucleus in Microbial Ecology and Environmental Microbiology and Biotechnology grant P/04-007-F, and the collaborative grant BMBF-IB/CONICYT-Chile. This study is part of the research program FONDAP 1501-0001, with funding by CONICYT to Center for Advanced Studies in Ecology & Biodiversity Program 7. Additional support from grant PBCT RED-12 is acknowledged.
We thank I. Plumeier, Christel Kakoschke, and Beate Jaschok-Kentner for meticulous technical assistance and D. Würdemann for critical reading of the manuscript.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alber, B. E., R. R. Spanheimer, C. Ebenau-Jehle, and G. Fuchs. 2006. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol. Microbiol. 61:297-309. [DOI] [PubMed] [Google Scholar]
- 2.Altenschmidt, U., and G. Fuchs. 1992. Novel aerobic 2-aminobenzoate metabolism. Purification and characterization of 2-aminobenzoate-CoA ligase, localisation of the genes on a 8-kbp plasmid, and cloning and sequencing of the genes from a denitrifying Pseudomonas sp. Eur. J. Biochem. 205:721-727. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronstein, P. A., M. Marrichi, S. Cartinhour, D. J. Schneider, and M. P. DeLisa. 2005. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J. Bacteriol. 187:8450-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce, N. C., and R. B. Cain. 1988. β-Methylmuconolactone, a key intermediate in the dissimilation of methylaromatic compounds by a modified 3-oxoadipate pathway evolved in nocardioform actinomycetes. FEMS Microbiol. Lett. 50:233-239. [Google Scholar]
- 6.Bruce, N. C., R. B. Cain, D. H. Pieper, and K.-H. Engesser. 1989. Purification and characterization of 4-methylmuconolactone methyl-isomerase, a novel enzyme of the modified 3-oxoadipate pathway in nocardioform actionomycetes. Biochem. J. 262:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caballero, A., A. Esteve-Nunez, G. J. Zylstra, and J. L. Ramos. 2005. Assimilation of nitrogen from nitrite and trinitrotoluene in Pseudomonas putida JLR11. J. Bacteriol. 187:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camara, B., P. Bielecki, F. Kaminski, V. Martins dos Santos, I. Plumeier, P. Nikodem, and D. H. Pieper. 2007. A gene cluster involved in degradation of substituted salicylates via ortho-cleavage in Pseudomonas sp. strain MT1 encodes enzymes specifically adapted for transformation of 4-methylcatechol and 3-methylmuconate. J. Bacteriol. 189:1664-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camara, B., C. Strompl, S. Verbarg, C. Sproer, D. H. Pieper, and B. J. Tindall. 2007. Pseudomonas reinekei sp. nov., Pseudomonas moorei sp. nov. and Pseudomonas mohnii sp. nov., novel species capable of degrading chlorosalicylates or isopimaric acid. Int. J. Syst. Evol. Microbiol. 57:923-931. [DOI] [PubMed] [Google Scholar]
- 10.Carmona, M., M. T. Zamarro, B. Blazquez, G. Durante-Rodriguez, J. F. Juarez, J. A. Valderrama, M. J. Barragan, J. L. Garcia, and E. Diaz. 2009. Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol. Mol. Biol. Rev. 73:71-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catelani, D., A. Fiecchi, and E. Galli. 1971. (+)-γ-Carboxymethyl-γ-methyl-Δα-butenolide. A 1,2-ring-fission product of 4-methylcatechol by Pseudomonas desmolyticum. Biochem. J. 121:89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha, C. J., R. B. Cain, and N. C. Bruce. 1998. The modified beta-ketoadipate pathway in Rhodococcus rhodochrous N75: enzymology of 3-methylmuconolactone metabolism. J. Bacteriol. 180:6668-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chari, R., C. Whitman, J. Kozarich, K. Ngai, and L. Ornston. 1987. Absolute stereochmical course of muconolactone Δ-isomerase and of 4-carboxymuconolactone decarboxylase: a 1H NMR “ricochet” analysis. J. Am. Chem. Soc. 109:5520-5521. [Google Scholar]
- 14.Charrier, C., G. J. Duncan, M. D. Reid, G. J. Rucklidge, D. Henderson, O. Young, V. J. Russell, R. I. Aminov, H. J. Flint, and P. Louis. 2006. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 152:179-185. [DOI] [PubMed] [Google Scholar]
- 15.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 16.Chu, X. Y., N. F. Wu, M. J. Deng, J. Tian, B. Yao, and Y. L. Fan. 2006. Expression of organophosphorus hydrolase OPHC2 in Pichia pastoris: purification and characterization. Protein Expr. Purif. 49:9-14. [DOI] [PubMed] [Google Scholar]
- 17.Chung, A., Q. Liu, S. P. Ouyang, Q. Wu, and G. Q. Chen. 2009. Microbial production of 3-hydroxydodecanoic acid by pha operon and fadBA knockout mutant of Pseudomonas putida KT2442 harboring tesB gene. Appl. Microbiol. Biotechnol. 83:513-519. [DOI] [PubMed] [Google Scholar]
- 18.Clark, J., and O. Maaloe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 19.de las Heras, A., C. A. Carreno, and V. de Lorenzo. 2008. Stable implantation of orthogonal sensor circuits in Gram-negative bacteria for environmental release. Environ. Microbiol. 10:3305-3316. [DOI] [PubMed] [Google Scholar]
- 20.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 21.Di Gennaro, P., S. Ferrara, I. Ronco, E. Galli, G. Sello, M. Papacchini, and G. Bestetti. 2007. Styrene lower catabolic pathway in Pseudomonas fluorescens ST: identification and characterization of genes for phenylacetic acid degradation. Arch. Microbiol. 188:117-125. [DOI] [PubMed] [Google Scholar]
- 22.Dorn, E., M. Hellwig, W. Reineke, and H.-J. Knackmuss. 1974. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch. Microbiol. 99:61-70. [DOI] [PubMed] [Google Scholar]
- 23.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erb, R. W., K. N. Timmis, and D. H. Pieper. 1998. Characterization of a gene cluster from Ralstonia eutropha JMP134 encoding metabolism of 4-methylmuconolactone. Gene 206:53-62. [DOI] [PubMed] [Google Scholar]
- 25.Erb, T. J., J. Retey, G. Fuchs, and B. E. Alber. 2008. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283:32283-32293. [DOI] [PubMed] [Google Scholar]
- 26.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs, G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 1125:82-99. [DOI] [PubMed] [Google Scholar]
- 28.Hartnett, C., E. L. Neidle, K.-L. Ngai, and L. N. Ornston. 1990. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J. Bacteriol. 172:956-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harwood, C. S., and R. E. Parales. 1996. The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 30.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 32.Junca, H., and D. H. Pieper. 2004. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ. Microbiol. 6:95-110. [DOI] [PubMed] [Google Scholar]
- 33.Karimi, M., D. Inze, and A. Depicker. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7:193-195. [DOI] [PubMed] [Google Scholar]
- 34.Kaschabek, S. R., B. Kuhn, D. Müller, E. Schmidt, and W. Reineke. 2002. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: purification and characterization of 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase. J. Bacteriol. 184:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knackmuss, H.-J., M. Hellwig, H. Lackner, and W. Otting. 1976. Cometabolism of 3-methylbenzoate and methylcatechols by a 3-chlorobenzoate utilizing Pseudomonas: accumulation of (+)-2,5-dihydro-4-methyl- and (+)-2,5-dihydro-2-methyl-5-oxo-furan-2-acetic acid. Eur. J. Appl. Microbiol. 2:267-276. [Google Scholar]
- 36.Korotkova, N., M. E. Lidstrom, and L. Chistoserdova. 2005. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J. Bacteriol. 187:1523-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ledger, T., D. H. Pieper, D. Perez-Pantoja, and B. Gonzalez. 2002. Novel insights into the interplay between peripheral reactions encoded by xyl genes and the chlorocatechol pathway encoded by tfd genes for the degradation of chlorobenzoates by Ralstonia eutropha JMP134. Microbiology 148:3431-3440. [DOI] [PubMed] [Google Scholar]
- 38.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Murray, K., C. J. Duggleby, J. M. S. Trepat, and P. A. Williams. 1972. The metabolism of benzoate and methylbenzoates via the meta-cleavage by Pseudomonas arvilla mt-2. Eur. J. Biochem. 28:301-310. [DOI] [PubMed] [Google Scholar]
- 40.Naggert, J., M. L. Narasimhan, L. DeVeaux, H. Cho, Z. I. Randhawa, J. E. J. Cronan, B. N. Green, and S. Smith. 1991. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J. Biol. Chem. 266:11044-11050. [PubMed] [Google Scholar]
- 41.Nikodem, P., V. Hecht, M. Schlomann, and D. H. Pieper. 2003. New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J. Bacteriol. 185:6790-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olivera, E. R., B. Minambres, B. Garcia, C. Muniz, M. A. Moreno, A. Ferrandez, E. Diaz, J. L. Garcia, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. U. S. A. 95:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ornston, L., and R. Stamier. 1966. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. 1. Biochemistry. J. Biol. Chem. 241:3776-3786. [PubMed] [Google Scholar]
- 44.Parales, R. E., and C. S. Harwood. 1992. Characterization of the genes encoding beta-ketoadipate: succinyl-coenzyme A transferase in Pseudomonas putida. J. Bacteriol. 174:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patrauchan, M. A., C. Florizone, M. Dosanjh, W. W. Mohn, J. Davies, and L. D. Eltis. 2005. Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Pantoja, D., R. De la Iglesia, D. H. Pieper, and B. Gonzalez. 2008. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol. Rev. 32:736-794. [DOI] [PubMed] [Google Scholar]
- 47.Pieper, D. H., K.-H. Engesser, R. H. Don, K. N. Timmis, and H.-J. Knackmuss. 1985. Modified ortho-cleavage pathway in Alcaligenes eutrophus JMP134 for the degradation of 4-methylcatechol. FEMS Microbiol. Lett. 29:63-67. [Google Scholar]
- 48.Pieper, D. H., K.-H. Engesser, and H.-J. Knackmuss. 1989. Regulation of catabolic pathways of phenoxyacetic acids and phenols in Alcaligenes eutrophus JMP 134. Arch. Microbiol. 151:365-371. [Google Scholar]
- 49.Pieper, D. H., K. Stadler-Fritsche, K. H. Engesser, and H. J. Knackmuss. 1993. Metabolism of 2-chloro-4-methylphenoxyacetate by AIcaligenes eutrophus JMP 134. Arch. Microbiol. 160:169-178. [DOI] [PubMed] [Google Scholar]
- 50.Pieper, D. H., K. Stadler-Fritzsche, H.-J. Knackmuss, K. H. Engesser, N. C. Bruce, and R. B. Cain. 1990. Purification and characterization of 4-methylmuconolactone methylisomerase, a novel enzyme of the modfied 3-oxoadipate pathway in the Gram-negative bacterium Alcaligenes eutrophus JMP 134. Biochem. J. 271:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Powlowski, J. B., and S. Dagley. 1985. β-Ketoadipate pathway in Trichosporon cutaneum modified for methyl-subsituted metabolites. J. Bacteriol. 163:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powlowski, J. B., J. Ingebrand, and S. Dagley. 1985. Ezymology of the β-ketoadipate pathway in Trichosporon cutaneum. J. Bacteriol. 163:1136-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prucha, M., A. Peterseim, and D. H. Pieper. 1997. Evidence for an isomeric muconolactone isomerase involved in the metabolism of 4-methylmuconolactone by Alcaligenes eutrophus JMP134. Arch. Microbiol. 168:33-38. [DOI] [PubMed] [Google Scholar]
- 54.Prucha, M., A. Peterseim, K. N. Timmis, and D. H. Pieper. 1996. Muconolactone isomerase of the 3-oxoadipate pathway catalyzes dechlorination of 5-chloro-substituted muconolactones. Eur. J. Biochem. 237:350-356. [DOI] [PubMed] [Google Scholar]
- 55.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 56.Sala-Trepat, J. M., K. Murray, and P. A. Williams. 1972. The metabolic divergence in the meta-cleavage of catechols by Pseudomonas putida NCIB 10015. Eur. J. Biochem. 28:347-356. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schweizer, H. P., and R. Chuanchuen. 2001. Small broad-host-range lacZ operon fusion vector with low background activity. Biotechniques 31:1258-1262. [DOI] [PubMed] [Google Scholar]
- 59.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 60.Vollmer, M. D., H. Hoier, H. J. Hecht, U. Schell, J. Groning, A. Goldman, and M. Schlomann. 1998. Substrate specificity of and product formation by muconate cycloisomerases: an analysis of wild-type enzymes and engineered variants. Appl. Environ. Microbiol. 64:3290-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe, K. 2001. Microorganisms relevant to bioremediation. Curr. Opin. Biotechnol. 12:237-241. [DOI] [PubMed] [Google Scholar]
- 62.Williams, P. A., and L. E. Shaw. 1997. mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J. Bacteriol. 179:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.