Abstract
In Bacillus subtilis cells, the GTP level decreases and the ATP level increases upon a stringent response. This reciprocal change in the concentrations of the substrates of RNA polymerase affects the rate of transcription initiation of certain stringent genes depending on the purine species at their transcription initiation sites. DNA microarray analysis suggested that not only the rrn and ilv-leu genes encoding rRNAs and the enzymes for synthesis of branched-chain amino acids, respectively, but also many genes, including genes involved in glucose and pyruvate metabolism, might be subject to this kind of stringent transcription control. Actually, the ptsGHI and pdhABCD operons encoding the glucose-specific phosphoenolpyruvate:sugar phosphotransferase system and the pyruvate dehydrogenase complex were found to be negatively regulated, like rrn, whereas the pycA gene encoding pyruvate carboxylase and the alsSD operon for synthesis of acetoin from pyruvate were positively regulated, like ilv-leu. Replacement of the guanine at position 1 and/or position 2 of ptsGHI and at position 1 of pdhABCD (transcription initiation base at position 1) by adenine changed the negative stringent control of these operons in the positive direction. The initiation bases for transcription of pdhABCD and pycA were newly determined. Then the promoter sequences of these stringent operons were aligned, and the results suggested that the presence of a guanine(s) and the presence of an adenine(s) at position 1 and/or position 2 might be indispensable for negative and positive stringent control, respectively. Such stringent transcription control that affects the transcription initiation rate through reciprocal changes in the GTP and ATP levels likely occurs for numerous genes of B. subtilis.
Stringent control is one of the most important adaptations which help bacteria survive under harsh conditions. Of the various occasions when stringent control results from the synthesis of GDP 3′-diphosphate (ppGpp) from GTP, which is catalyzed by the RelA protein associated with ribosomes, the most prominent is the repression of stable RNA synthesis (4). This control also affects the expression of certain genes, including genes involved in amino acid biosynthesis. However, Bacillus subtilis and Escherichia coli use different strategies for stringent transcription control (19). ppGpp may not inhibit B. subtilis RNA polymerase directly, whereas ppGpp decreases rRNA promoter activity by directly inhibiting E. coli RNA polymerase.
GTP and ATP are well-known gauges of the general energetic capacity and energy charge of cells, respectively. In B. subtilis, the GTP and ATP levels decrease and increase during a stringent response (e.g., amino acid starvation), respectively (20, 24, 31, 48), as shown in Fig. S1 in the supplemental material. The changes are mediated by ppGpp synthesized by RelA (52) during a stringent response, probably through its inhibition of IMP dehydrogenase, the first enzyme in the pathway leading to the biosynthesis of GTP (24). The inhibition of GTP synthesis results in a decrease in the GTP level and accumulation of IMP, which is also a precursor of ATP, resulting in the increase in the ATP level. These reciprocal changes in the ATP and GTP levels also occur when B. subtilis cells are treated with decoyinine (19, 25, 48), a GMP synthase inhibitor (46), even in a relA-deficient strain because the inhibition is not mediated by ppGpp (48).
The decrease in the GTP level during a stringent response can be sensed by the CodY protein, a GTP-binding repressor of many genes, such as dpp encoding dipeptide permease (42) and ilv-leu encoding the enzymes for synthesis of branched-chain amino acids (41), that are normally quiescent when cells are grown in a nutrient-rich medium (14, 28, 34). CodY also functions as a transcriptional activator of certain genes, such as ackA involved in acetate formation (39). Thus, lowering the GTP concentration inactivates the CodY protein, leading to derepression or deactivation of these genes. Another mechanism underlying stringent control has recently emerged. The reciprocal changes in the concentrations of GTP and ATP, which are substrates of RNA polymerase, can be sensed through modulation of the rate of initiation of transcription of several stringent genes, such as rrn encoding rRNAs (19) and ilv-leu (20, 48). This CodY-independent mechanism involves the base, guanine or adenine, at the transcription initiation site (position 1 or 2; the base at position 1 is the transcription initiation base), which is related to negative and positive stringent control of rrn and ilv-leu, respectively. The downregulation and upregulation of the rrn and ilv-leu promoter activities are always correlated with the decrease and increase in the intracellular GTP and ATP concentrations, respectively. The two mechanisms underlying B. subtilis stringent transcription control have been determined for the ilv-leu operon (see Fig. S1 in the supplemental material).
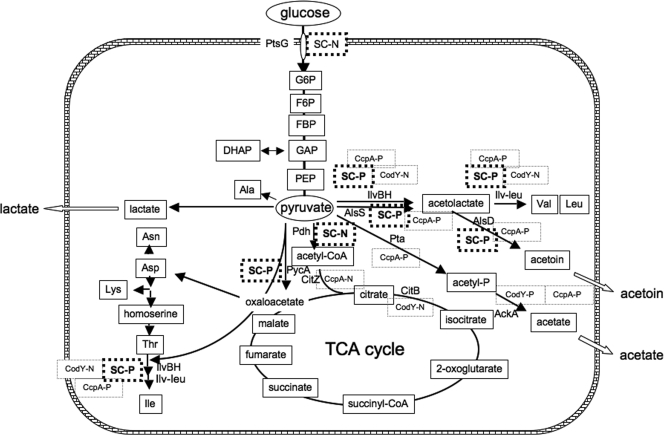
In order to answer an interesting question, whether such CodY-independent stringent control occurs in the regulation of various operons, including catabolic and anabolic operons, we performed a DNA microarray analysis using wild-type and ΔcodY strains grown with and without decoyinine. This analysis suggested that in addition to the genes for protein synthesis, such as rrn, many metabolic operons, including operons involved in critical stages of metabolic regulation, might also be under stringent control, and tens of these operons might be subject to CodY-independent stringent control. Among these operons, we focused on the genes involved in glucose and pyruvate metabolism (Fig. 1) in whose regulation a catabolite control protein (CcpA) and CodY are known to be the main proteins involved. Among the operons involved in this metabolism, only the ptsGHI (10, 45) and pdhABCD (9, 13) operons encoding the glucose-specific phosphoenolpyruvate:sugar phosphotransferase system (PTS) and the pyruvate dehydrogenase complex, respectively, were severely downregulated. In contrast, not only ilv-leu (11, 49) but also pycA encoding pyruvate carboxylase (5) and alsSD (35) involved in synthesis of acetoin from pyruvate were substantially upregulated. We demonstrated that these operons were actually under CodY-independent stringent control. The guanines located at positions 1 and 2 of the transcription initiation sites of ptsGHI and pdhABCD are essential for the CodY-independent negative stringent control, whereas adenines are located at position 1 or 2 of the transcription initiation sites of the pycA and alsSD operons, as they are in ilv-leu.
FIG. 1.
Intermediates in carbon metabolism and CcpA and CodY regulation and CodY-independent stringent control of the genes involved in this metabolism. The glucose and pyruvate metabolism described in the text includes glycolysis; the formation of acetyl-CoA, oxaloacetate, and alanine; the secretory pathways for acetoin, acetate, and lactate; and the synthesis of branched-chain amino acids. CcpA-N and CcpA-P indicate negative and positive regulation by CcpA, respectively, and CodY-N and CodY-P indicate negative and positive regulation by CodY, respectively. The ilv-leu (40) and citB (16) genes are known to be repressed by CodY, whereas the ackA gene is activated (39). Also, the citZ gene is repressed by CcpA (17), whereas the ilv-leu (41, 49), alsSD (51), pta (33), and ackA (50) genes are activated by CcpA. SC-N and SC-P indicate negative and positive CodY-independent stringent control, respectively. Abbreviations: TCA, tricarboxylic acid; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate.
MATERIALS AND METHODS
Bacterial strains and construction of strains.
The B. subtilis strains used in this work are listed in Table 1. Strain FU808 was constructed by transformation of strain 1A765 with chromosomal DNA of strain PS37 to spectinomycin resistance (60 μg/ml) on plates containing tryptose blood agar base (Difco) with 10 mM glucose (TBABG). The presence of ΔcodY in the resulting transformant was confirmed by the appearance in the ΔcodY strain of a PCR product that was 250 bp shorter than that obtained for the codY+ strain, as described previously (28). Disruption of the gid gene, present in the ΔcodY strain, did not affect expression of the target genes in this work. To construct transcriptional promoter-lacZ fusion strains with ptsGHI, pycA, alsSD, and pdhABCD, the promoter regions comprising nucleotides −55 to 26 and −47 to 167 were amplified using primer pairs PTS-F/PTS-R, PYC-F/PYC-R, ALS-F/ALS-R, and PDH-F/PDH-R (see Table S1 in the supplemental material), respectively, and DNA of strain 168 as the template. The PCR products were trimmed with XbaI and BamHI and then ligated with the XbaI-BamHI arm of plasmid pCRE-test2 (27). The ligated DNAs were used for transformation of E. coli strain DH5α to ampicillin resistance (50 μg/ml) on Luria-Bertani medium plates (36). Correct construction of the fusions in the resulting plasmids was confirmed by DNA sequencing. The plasmids carrying the promoter regions of ptsGHI, pycA, alsSD, and pdhABCD were linearized with PstI and then used for double-crossover transformation of strains 1A765, 1A766 (relA1), and FU808 (ΔcodY) to chloramphenicol resistance (5 μg/ml) on TBABG plates, which produced strains FU906, FU1025, and FU1029, strains FU977, FU1023, and FU1027, strains FU937, FU1026, and FU1041, and strains FU1019, FU1021, and FU1031, respectively.
TABLE 1.
B subtilis strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| 168 | trpC2 | 1 |
| 1A765 (= BR16)a | trpC2 lys | 47 |
| 1A766 (= BR17)a | trpC2 lys relA1 | 47 |
| PS29 | trpC2gid::spec | 38 |
| PS37 | trpC2gid::spec ΔcodY | 38 |
| FU807 | trpC2 lys relA1 gid::spec ΔcodY | This study |
| FU808 | trpC2 lys gid::spec ΔcodY | This study |
| FU906 | trpC2 lys amyE::[cat PptsG(−55/26)-lacZ] | This study |
| FU1025 | trpC2 lys relA1 amyE::[cat PptsG(−55/26)-lacZ] | This study |
| FU1029 | trpC2 lys gid::spc ΔcodY amyE::[cat PptsG(−55/26)-lacZ] | This study |
| FU1019 | trpC2 lys amyE::[cat PpdhA(−47/167)-lacZ] | This study |
| FU1021 | trpC2 lys relA1 amyE::[cat PpdhA(−47/167)-lacZ] | This study |
| FU1031 | trpC2 lys gid::spc ΔcodY amyE::[cat PpdhA(−47/167)-lacZ] | This study |
| FU977 | trpC2 lys amyE::[cat PpycA(−55/26)-lacZ] | This study |
| FU1023 | trpC2 lys relA1 amyE::[cat PpycA(−55/26)-lacZ] | This study |
| FU1027 | trpC2 lys gid::spc ΔcodY amyE::[cat PpycA(−55/26)-lacZ] | This study |
| FU937 | trpC2 lys amyE::[cat PalsS(−55/26)-lacZ] | This study |
| FU1026 | trpC2 lys relA1 amyE::[cat PalsS(−55/26)-lacZ] | This study |
| FU1041 | trpC2 lys gid::spc ΔcodY amyE::[cat PalsS(−55/26)-lacZ] | This study |
| FU934 | trpC2 lys amyE::[cat PptsG (−55/26)(G2A)-lacZ | This study |
| FU1042 | trpC2 lys amyE::[cat PptsG (−55/26)(G1A)-lacZ] | This study |
| FU1045 | trpC2 lys amyE::[cat PptsG (−55/26)(G1A, G2A)-lacZ] | This study |
| FU1044 | trpC2 lys amyE::[cat PpdhA (−47/167)(G1A)-lacZ] | This study |
| FU1060 | trpC2 lys amyE::[cat PpycA (−55/26)(A1G)-lacZ] | This study |
| FU1061 | trpC2 lys amyE::[cat PalsS (−55/26)(A1G)-lacZ] | This study |
Strain obtained from the Bacillus Genetic Stock Center (Columbus, OH).
To construct strains FU934, FU1042, and FU1045 carrying the ptsGHI promoter-lacZ fusion with an adenine substitution(s) at position 1 and/or 2 in the ptsGHI promoter (guanine at position 2 changed to adenine [G2A, GA], guanine at position 1 changed to adenine [G1A, AG], and guanine at positions 1 and 2 changed to adenine [G2A G1A, AA]), the promoter regions (nucleotides −55 to 26) were amplified using primer pairs PptsG-F2/PptsG-GA-R, PptsG-F2/PptsG-AG-R, and PptsG-F2/PptsG-AA-R (see Table S1 in the supplemental material), respectively, and chromosomal DNA of strain 168 as the template. The PCR products trimmed with XbaI and BamHI were cloned into plasmid pCRE-test2 (27) in E. coli strain DH5α, as described above. Correct construction of the fusions in the resulting plasmids was confirmed by DNA sequencing. The plasmids that had a base substitution at position 1 and/or 2 (GA, AG, and AA) were each linearized with PstI and then used for transformation of strain 1A765, resulting in strains FU934, FU1042, and FU1045, respectively.
To construct strain FU1044 carrying a pdhABCD promoter-lacZ fusion with adenine substituted for guanine at position 1 in the pdhABCD promoter (G1A), the upstream and downstream parts of the promoter region (nucleotides −47 to 167) were separately amplified with two primer pairs (PpdhA-A1-F/PpdhA-A1-R and PpdhA-A2-F/PpdhA-A2-R) (see Table S1 in the supplemental material) using chromosomal DNA of strain 168 as the template. Next, the two PCR products were mixed, and extension reactions were carried out without any primer. PCR with the resultant fragment as the template and primers PpdhA-A1-F and PpdhA-A2-R was performed to amplify the combined DNA fragment, which was then trimmed with XbaI and BamHI and cloned into plasmid pCRE-test2 (27) in E. coli strain DH5α, as described above. Correct base substitution was confirmed by DNA sequencing. The constructed plasmid was linearized with PstI and then used for transformation of strain 1A765, resulting in strain FU1044.
To construct strains FU1060 and FU1061 carrying pycA and alsSD promoter-lacZ fusions with guanine substituted for adenine at position 1 in the pycA and alsSD promoters, the promoter regions (nucleotides −55 to 26) were amplified using primer pairs PpycA-F2/PpycA-G-R and PalsS-F2/PalsS-G-R (see Table S1 in the supplemental material), respectively, and chromosomal DNA of strain 168 as the template. The PCR products were trimmed with XbaI and BamHI and were cloned into plasmid pCRE-test2 (27) in E. coli strain DH5α, as described above. Correct construction of the fusions in the resulting plasmids was confirmed by DNA sequencing. The plasmids with the base substitutions at position 1 in the pycA and alsSD promoters were linearized with PstI and then used for transformation of strain 1A765, resulting in strains FU1060 and FU1061, respectively.
Cell growth and β-Gal assay.
The lacZ-fusion strains were grown at 30°C overnight on TBABG plates containing the appropriate antibiotic(s), chloramphenicol (5 μg/ml) and/or spectinomycin (60 μg/ml). The cells were inoculated into 100 ml of minimal medium comprising 0.4% glucose, 0.2% glutamine, and 50 μg/ml tryptophan (MM medium) (57) supplemented with a mixture of 16 amino acids (glutamine, histidine, tyrosine, and asparagine were omitted) (2) at an optical density at 600 nm (OD600) of 0.08 and then incubated at 37°C. When the OD600 reached approximately 0.5, 45-ml aliquots of the culture were harvested, and the cells were centrifuged at 25°C (2,000 × g for 10 min). The cells were suspended in 45 ml of MM medium supplemented with the amino acid mixture described above with and without lysine, and then the cultures were incubated further. During incubation before and after cell resuspension, 1-ml aliquots of the culture were withdrawn at 15-min intervals at most, and the β-galactosidase (β-Gal) activity in crude cell extracts was measured spectrophotometrically as described previously (56).
When the effect of decoyinine on lacZ expression in the fusion strains was examined, a culture was divided into two 45-ml portions when the OD600 of the culture reached approximately 0.5, and then decoyinine (500 μg/ml) was added to one of the portions. The β-Gal activity was monitored as described above.
DNA microarray analysis.
DNA microarray analysis using microarrays consisting of 3,941 protein genes was performed as described previously (57). Cells of strains 1A765 and FU808 were grown in two 200-ml cultures using MM medium supplemented with the 16 amino acids described above at 37°C until the OD600 was 0.7, and then decoyinine was added to one of the cultures at a final concentration of 500 μg/ml. After cultivation for an additional 15 min, the cells were harvested by centrifugation, and then total RNA was extracted and purified for synthesis of cDNA labeled with a fluorescent dye (Cy5 for the sample to which decoyinine was added and Cy3 for the control).
Metabolome analysis.
The in vivo concentrations of metabolites, including nucleotides, were determined by capillary electrophoresis-mass spectrometry (CE-MS) after extraction with methanol, essentially as described previously (43). Cells of strain 1A765 were subjected to decoyinine treatment as described above. The cells were collected on a Millipore Isopore membrane filter (HTTP; pore size, 0.4 μm; diameter, 47 mm). Each of the cell-bearing membranes was put into a heat-sealable plastic bag containing 2 ml of ice-chilled methanol, and then the cells were detached from the membrane and the bag was sealed. The sealed bags were incubated at 70°C for 1 h. After the cell suspensions were centrifuged to remove cell debris, the supernatants were treated with chloroform to remove lipids. After the upper layer was subjected to ultrafiltration (Millipore Ultrafree-MC; 5,000 nominal molecular weight limit [NMWL]), the filtrates were freeze-dried. After the pellets were dissolved in distilled water, samples were subjected to CE-MS to identify the metabolites in them and to measure the concentrations of the metabolites, as described previously (43). (This metabolome analysis was carried out by Human Metabolome Technologies, Inc., Japan.) The in vivo molar concentrations of metabolites were calculated by assuming that the aqueous volume of 1 OD600 unit (OD600 × ml) corresponded to 0.83 μl (8).
Primer extension analysis.
Primer extension analysis was performed as described previously (55). RNA samples were prepared as described previously (57) using cells of strain 1A765 that had been grown in MM medium containing the 16-amino-acid mixture described above. Reverse transcription using an RNA sample was initiated with primers PptsG-R, PpdhA-R, and PpycA-R for the ptsGHI, pdhABCD, and pycA transcripts, respectively (see Table S1 in the supplemental material), which had been labeled at the 5′ end by using a Megalabel kit (Takara-Bio, Kyoto, Japan) and [γ-32P]ATP (MP Biomedicals). A template for the dideoxy sequencing reaction for ladder preparation starting from each of the same end-labeled primers was prepared by PCR using primer pairs PptsG-F1/PptsG-R, PpdhA-F/PpdhA-R, or PpycA-F/PpycA-R (see Table S1 in the supplemental material) and DNA from strain 168 as the template.
Northern analysis.
Total RNA was extracted from cells of strain 1A765 and purified as described previously (57). The RNA was electrophoresed in a glyoxal gel and then transferred to a Hybond-N membrane (GE Healthcare) (36). The RNA transferred to the membrane was stained with 0.003% methylene blue to check its quality before hybridization. To prepare probes for detection of the ptsGHI and pycA transcripts, the corresponding products amplified by PCR using primer pairs LptsG-F/LptsG-R, ptsG-F/ptsG-R, ptsH-F/ptsH-R, and pycA-F/pycA-R (see Table S1 in the supplemental material) and chromosomal DNA of strain 168 as the template were labeled with a BcaBEST labeling kit (Takara-Bio, Kyoto, Japan) and [α-32P]dCTP (MP Biochemedicals). Hybridization and transcript detection were carried out as described previously (36).
RESULTS
Stringent control of the genes involved in glucose and pyruvate metabolism.
Amino acid starvation and exposure of cells to decoyinine result in a decrease in the GTP level and an increase in the ATP level, which depend on and are independent of RelA, respectively (15, 20, 24, 48). Each of these treatments triggers CodY-dependent and CodY-independent stringent control of many genes, including the ilv-leu genes (48). In order to find genes that are under stringent transcription control, we performed a DNA microarray analysis with strains 1A765 (lys) and FU808 (lys ΔcodY) which had been grown on MM medium with the amino acid mixture described above to the mid-logarithmic phase and incubated for 15 min with and without decoyinine (decoyinine treatment) in the wild-type and ΔcodY genetic backgrounds. We detected hundreds of genes that are under negative and positive stringent control triggered by decoyinine addition (>3-fold change in the wild-type background level) (see the DNA microarray data deposited in the KEGG Expression Database [http://www.genome.jp/kegg/expression]). As expected, the rpl and rps genes coding for ribosomal proteins derived from the large and small subunits, respectively (22), were severely downregulated. Of the operons involved in glucose and pyruvate metabolism shown in Fig. 1, only the ptsGHI (10, 45) and pdhABCD (9, 13) operons were severely downregulated upon decoyinine treatment. In contrast, ilv-leu (11, 48), pycA (5), and alsSD (35) were upregulated substantially. Table 2 shows the expression ratios (expression with decoyinine/expression without decoyinine) for these genes that were upregulated and downregulated upon decoyinine treatment. Table 2 also shows that the expression ratios (expression in ΔcodY strain PS37/expression in wild-type strain PS29) obtained in a previous DNA microarray analysis (28). In addition to expression of the genes in the ilv-leu operon, a direct target of CodY, ptsG expression also changed more than 2-fold upon codY disruption. However, we do not know whether the ptsG gene is a direct or indirect target of CodY. For stringent control, the expression ratios of the upregulated and downregulated genes in the wild-type background in general were higher and lower than the expression ratios in the ΔcodY background, respectively. This could be explained by relief from CodY repression, as observed for ilv-leu. Also, some enhancement of the CodY-independent stringent control of these genes might occur in the wild-type background, as previously observed for ilv-leu (48).
TABLE 2.
DNA microarry analysis of CodY-dependent and -independent stringent control induced by decoyinine addition
| Genea | Ratio of expression with decoyinine to expression without decoyinine in the CodY+ backgroundb | Ratio of expression with decoyinine to expression without decoyinine in the CodY− backgroundb | CodY−/CodY+ expression ratiob,c |
|---|---|---|---|
| alsS | 21 | 7.5 | 1.0 |
| alsD | 13 | 7.8 | 0.54 |
| pdhA | 0.13 | 0.09 | 0.70 |
| pdhB | 0.10 | 0.06 | 0.54 |
| pdhC | 0.13 | 0.16 | 0.73 |
| pdhD | 0.16 | 0.31 | 0.75 |
| ilvB | 4.8 | 1.7 | 38 |
| ilvN | 5.9 | 1.9 | 44 |
| ilvC | 8.0 | 1.9 | 49 |
| leuA | 8.7 | 1.8 | 47 |
| leuB | 15 | 1.6 | 46 |
| leuC | 24 | 1.50 | 46 |
| leuD | NDd | ND | ND |
| ptsG | 0.01 | 0.03 | 0.38 |
| ptsH | 0.60 | 0.24 | 0.81 |
| ptsI | 0.45 | 0.33 | 0.70 |
| pycA | 3.1 | 2.2 | 0.96 |
Of the genes that are involved in glucose and pyruvate metabolism (Fig. 1), those whose expression was altered more than 3-fold upon addition of decoyinine in the wild-type background are included. The ptsHI genes are also included.
The standard deviations of the expression ratios were less than 15%.
The expression ratios were obtained by a DNA microarray analysis performed previously (28).
ND, the expression signal of leuD was not detectable due to unknown technical problems with our DNA microarray analysis.
Changes in the in vivo concentrations of the intermediates of glucose and pyruvate metabolism upon decoyinine addition.
In addition to the results of the DNA microarray analysis described above (Table 2), the results of a metabolome analysis of the changes in the in vivo concentrations of the intermediates of glucose and pyruvate metabolism (Fig. 1) upon decoyinine treatment in the CodY+ background of strain 1A765 (lys) that was part of a previous metabolome analysis (48) are shown in Table 3. In this analysis we were able to determine the in vivo concentrations of only some of the intermediates of carbon metabolism shown in Fig. 1, those listed in Table 3. Upon decoyinine addition, the concentrations of GTP and ATP decreased and increased, respectively. But ppGpp was undetectable when decoyinine was added, in contrast to its detection upon lysine starvation (48). The only intermediates whose concentrations decreased more than 2-fold were fructose-1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP) in the glycolytic pathway, which likely resulted from the shutdown of the PTS for glucose transport triggered by decoyinine treatment. On the other hand, the compounds whose concentrations increased more than 2-fold were amino acids derived from oxaloacetate (Val, Thr, Ilv, Leu, Asp, and Lys), and the increase in the Asp concentration was the most prominent increase. This could be explained by the induction of pycA and ilv-leu under stringent control triggered by addition of decoyinine. Moreover, the concentrations of citrate and 2-oxoglutarate in the tricarboxylic acid (TCA) cycle also increased more than 2-fold, and the concentration of isocitrate also increased. This might be explained by the finding that CcpA could not repress the citZ gene encoding citrate synthase that condenses acetyl coenzyme A (acetyl-CoA) with oxaloacetate (17). (The complex consisting of CcpA and P-Ser-HPr was not formed well with a low in vivo concentration of FBP [7], which resulted from strong negative stringent control of ptsGHI.) Actually, citZ expression increased 2.4-fold upon decoyinine addition in the wild-type background as determined by the DNA microarray analysis described above.
TABLE 3.
Metabolome analysis to determine the change in in vivo metabolite concentrations after decoyinine addition
| Compound | In vivo concn (mM) without decoyinine additiona | In vivo concn (mM) with decoyinine additiona |
|---|---|---|
| Glucose-6-phosphate | 6.3 | 7.7 |
| Fructose-6-phosphate | 2.8 | 3.6 |
| Fructose-1,6-bisphosphate | 4.5 | 0.8 |
| Dihydroxyacetone phosphate | 0.5 | 0.2 |
| Glyceraldehyde 3-phosphate | 46 | 36 |
| Phosphoenolpyruvate | 22 | 20 |
| Lactate | 270 | 410 |
| Acetyl-CoA | 7.7 | 7.3 |
| Citrate | 1.7 | 7.1 |
| Isocitrate | 0.3 | 0.4 |
| 2-Oxoglutarate | 0.5 | 1.4 |
| Succinate | 54 | 38 |
| Fumarate | 4.3 | 4.2 |
| Malate | 1.7 | 2.6 |
| Ala | 150 | 190 |
| Val | 9.9 | 40 |
| Homoserine | 3.9 | 3.7 |
| Thr | 21 | 44 |
| Ile | 17 | 35 |
| Leu | 11 | 31 |
| Asn | 0.8 | 1.4 |
| Asp | 25 | 200 |
| Lys | 12 | 21 |
| GTP | 0.4 | 0.19 |
| ATP | 3.1 | 6.6 |
| ppGpp | NDb | ND |
The standard deviations of the values were less than 20%.
ND, not determined.
Negative stringent control of the ptsGHI operon.
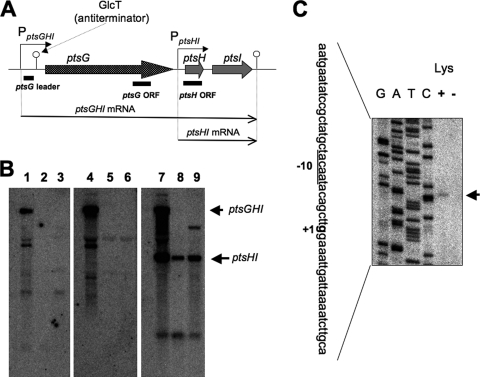
DNA microarray analysis indicated that the ptsGHI operon was severely downregulated upon decoyinine addition, suggesting that it might be under strong negative stringent control. Transcription and regulation of the ptsGHI operon were studied previously by Stülke et al. (45). As shown in Fig. 2A, there are the two transcription promoters for the ptsGHI operon; one of these promoters is PptsGHI for ptsGHI transcription, and the other is PptsHI for ptsHI transcription. GlcT, an antiterminator belonging to the BglG/SacY family, binds to a ribonucleic antiterminator upstream of ptsG in the presence of glucose. This prevents formation of an overlapping transcriptional terminator. Northern analysis of the pts transcripts with the corresponding probes for the ptsG leader between PptsGHI and the terminator, the ptsG open reading frame (ORF), and the ptsH ORF (Fig. 2B) indicated that the transcription from PptsGHI was severely downregulated upon lysine starvation (Fig. 2B, lanes 2, 5, and 8), as well as upon decoyinine addition (lanes 3, 6, and 9), whereas the transcription from PptsHI was downregulated only moderately. The former downregulation was likely due to CodY-independent stringent control, as verified below. The mechanism underlying the latter moderate downregulation cannot be explained properly at present. We also performed a primer extension analysis to identify the same transcription initiation guanine residue of ptsGHI (336 nucleotides upstream of the translation initiation site of ptsG) that was determined previously by Stülke et al. (45) (Fig. 2C). This analysis indicated that the transcription from this guanine was downregulated upon lysine starvation (Fig. 2C, lanes + and −).
FIG. 2.
Downregulation of ptsGHI transcription upon lysine starvation and decoyinine treatment. (A) The ptsGHI operon has two promoters, PptsGHI for ptsGHI transcription and PptsHI for ptsHI transcription (45). GlcT, an antiterminator belonging to the BglG/SacY family, binds to a ribonucleic antiterminator upstream of ptsG in the presence of glucose. This prevents formation of an overlapping transcriptional terminator. The ptsGHI and ptsHI transcription terminator is also located downstream of ptsI. (B) Northern analysis of the stringent control of ptsGHI transcription. RNA samples were prepared from cells of strain 1A765 (lys) that had been subjected to lysine starvation for 30 min (lanes 2, 5, and 8) or to decoyinine treatment (500 μg/ml) for 30 min (lanes 3, 6, and 9) and from cells that were not subjected to these stresses (lanes 1, 4, and 7). The transcripts in the left, middle, and right panels were detected with the probes for the pstG leader, ptsG ORF, and ptsH ORF (indicated in panel A), respectively, whose preparation is described in Materials and Methods. The positions of the ptsGHI and ptsHI transcripts are indicated by the arrows on the right. (C) Primer extension analysis for mapping of the 5′ end of the ptsGHI transcript. Total RNA from cells of strain 1A765 (lys) which had been subjected to lysine starvation for 30 min (lane −) and total RNA from cells of strain 1A765 which had not been subjected to this nutrient stress (lane +) were annealed with the PptsG-R primer (see Table S1 in the supplemental material), and then primer extension was performed as described in Materials and Methods. Lanes G, A, T, and C contained the products of the corresponding dideoxy sequencing reactions with the PCR product as the template, as described in Materials and Methods. The part of the nucleotide sequence of the coding strand corresponding to the ladder is shown on the left; the transcription initiation base (+1) is indicated by bold type, and the corresponding −10 region for the ptsGHI promoter is underlined. The position of the runoff cDNA is indicated by the arrow on the right.
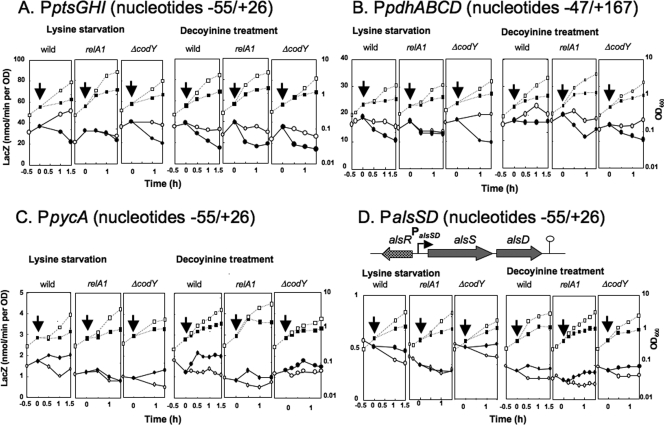
In order to characterize the stringent control of the ptsGHI promoter, the core promoter region (nucleotides −55 to 26) was placed upstream of lacZ in the wild-type, relA1, and ΔcodY backgrounds, and the β-Gal synthesis under control of the core ptsGHI promoter was monitored upon lysine starvation and decoyinine treatment (Fig. 3A). When lysine was depleted from the medium, the cells stopped growing, and β-Gal synthesis decreased in the wild-type background. (Cell growth and β-Gal synthesis were maintained for more than 1 h if lysine was not depleted.) A decrease in β-Gal synthesis was not observed in the relA1 background but was observed in the ΔcodY background. When decoyinine was added to the medium, β-Gal synthesis decreased to almost the same extent in the wild-type, relA1, and ΔcodY backgrounds. The results indicate that the negative regulation was independent of CodY and suggest that it was likely mediated by a decrease in the GTP level caused by IMP dehydrogenase inhibition by ppGpp synthesized by RelA upon lysine starvation or by GMP synthase inhibition by decoyinine.
FIG. 3.
lacZ expression under control of the promoters of the stringent operons. The core ptsGHI, pycA, and alsSD promoters (nucleotides −55 to 26), as well as the pdhABCD promoter (nucleotides −47 to 164), were placed upstream of lacZ, and each of the resultant lacZ fusions was integrated into the amyE locus of the wild-type, relA1, and ΔcodY strains. lacZ expression under control of the ptsGHI (A), pdhABCD (B), pycA (C), and alsSD (D) promoters (PptsGHI, PpdhABCD, PpycA, and PalsSD) was monitored using strains FU906, FU1025, and FU1029, strains FU1019, FU1021, and FU1031, strains FU977, and FU1023, and FU1027, and strains FU937, FU1026, and FU1041, respectively, upon lysine starvation and decoyinine treatment, as described in Materials and Methods. OD600 values and LacZ activities are indicated by squares and circles; filled and open symbols indicate treatments with and without lysine starvation and decoyinine, respectively. Arrows indicate the onset of these stresses. In the upper part of panel D, alsSD transcription, whose activation is mediated by AlsR, is shown.
Replacement of guanines at the transcription initiation sites (positions 1 and 2) of the ptsGHI operon by adenine.
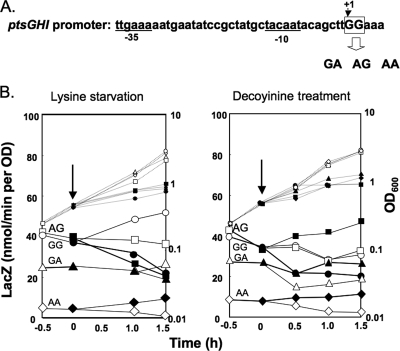
According to the results of the primer extension analyses (45) (Fig. 2C), the base at the transcription initiation site (position 1) of the ptsGHI operon was a guanine. It was reported previously that the initiating base for transcription from B. subtilis rRNA promoters is a guanine and that changes in the promoter activity always correlate with changes in the intracellular GTP concentration (19). Also, adenine at the transcription initiation site of the ilv-leu operon is critical for positive stringent control of this operon, which likely correlates with changes in the intracellular ATP concentration (20, 48). To determine if adenine substitution for guanine at positions 1 and 2 affects the negative stringent control of ptsGHI, we constructed strains FU934, FU1042, and FU1045, in which the original GG (positions 1 and 2) was replaced by GA, AG, and AA, respectively, in the core ptsGHI promoter region (nucleotides −55 to 26), which had been placed upstream of lacZ in the wild-type genetic background (Fig. 4A) .
FIG. 4.
Effect of replacement of guanines at the transcription initiation bases (positions 1 and 2) of the ptsGHI operon by adenine on negative stringent control of this operon. (A) The two guanines (GG) at the transcription initiation site (positions 1 and 2) of the ptsGHI promoter (nucleotides −55 to 26) were replaced by GA, AG, and AA, as described in Materials and Methods. The −35 and −10 regions of the ptsGHI promoter are underlined. (B) The mutant core ptsGHI promoters were placed upstream of lacZ in the relA+ and codY+ backgrounds. lacZ expression in the resulting strains (FU934 with GA, FU1042 with AG, and FU1045 with AA), as well as in strain FU906 with GG, was monitored upon lysine starvation and decoyinine treatment, as described in Materials and Methods. OD600 values (small symbols) and LacZ activities (large symbols) are indicated by circles for FU906 with GG, by squares for FU1042 with AG, by triangles for FU934 with GA, and by diamonds for FU1045 with AA; filled and open symbols indicate results obtained with and without lysine starvation and decoyinine treatment, respectively. The arrows indicate the onset of the stresses.
Although negative stringent regulation of the core ptsGHI promoter was observed upon lysine starvation in strain FU906 carrying the original GG residues (Fig. 4B, left panel), this regulation was somehow quenched in strain FU1042 (AG) and was almost completely absent in strain FU934 (GA). When the two guanines at positions 1 and 2 were replaced by adenines, resulting in strain FU1045 (AA), the lacZ expression under control of this variant of the core ptsGHI promoter was quite low, but the original negative stringent regulation was clearly converted to positive regulation. When cells of these strains were exposed to decoyinine (Fig. 4B, right panel), the original negative regulation in strain FU906 (GG) was converted to rather positive regulation in strains FU1042 (AG) and FU934 (GA). Also, the core ptsG promoter of strain FU1045 (AA) responded most positively to decoyinine treatment, although its activity was quite low. These results suggest that guanines at positions 1 and 2 in the ptsGHI transcription initiation site are indispensable for negative stringent control and that the original negative regulation was easily converted to positive stringent regulation in the order AG-GA-AA when the guanines were replaced by adenines.
Negative stringent control of the pdhABCD operon.
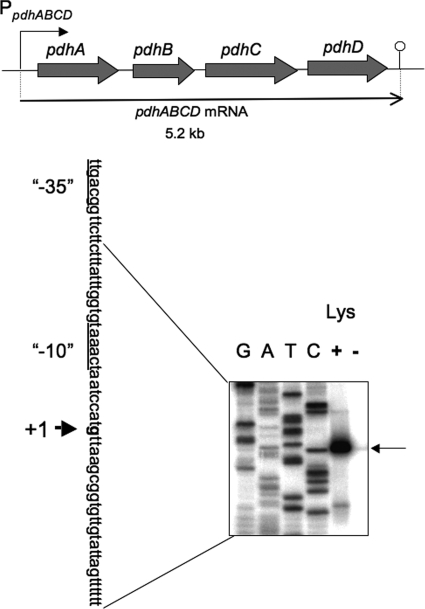
DNA microarray analysis suggested that the pdhABCD operon encoding the four subunits of the pyruvate dehydrogenase multienzyme complex (9, 13) might also be under strong negative stringent control (Table 2). This operon is transcribed as a 5.2-kb transcript from the promoter upstream of pdhA (Fig. 5) (13). The promoter for pdhABCD was designated P1, although its transcription initiation base was not determined (13). This putative promoter was found to be unable to function in lacZ fusion experiments (data not shown), so we performed a primer extension analysis to determine the transcription initiation base of pdhABCD. We found that the transcription initiation base is a guanine located 211 bases upstream of the translation initiation base of the pdhA gene (Fig. 5), which enabled us to determine its −35 and −10 regions likely recognized by σA RNA polymerase. The transcription from this guanine was severely downregulated upon lysine starvation (Fig. 5, lanes + and −). To examine this negative regulation of the pdhABCD promoter, the promoter region (nucleotides −47 to 167) was placed upstream of lacZ in the wild-type, relA1, and ΔcodY backgrounds. The β-Gal synthesis under control of the pdhABCD promoter was monitored upon lysine starvation and decoyinine treatment (Fig. 3B). A decrease in β-Gal synthesis upon lysine starvation was observed in the wild-type and ΔcodY backgrounds, but not in the relA1 background. When decoyinine was added to the medium, β-Gal synthesis decreased to almost the same extent in the wild-type, relA1, and ΔcodY backgrounds. The results indicate that this negative regulation was also independent of CodY.
FIG. 5.
Transcription of the pdhABCD operon. The genes in the pdhABCD operon encode the pyruvate decarboxylase E1α and E1β, dihydrolipoamide acetyltransferase E2, and dihydrolipoamide dehydrogenase E3 subunits of the pyruvate dehydrogenase multienzyme complex (PDH), respectively. Transcription of this operon results in a 5.2-kb pdhABCD mRNA (13). The 5′ end of the pdhABCD transcript was determined by primer extension analysis (lower panel). Total RNAs from cells of strain 1A765 (lys) that had been subjected to lysine starvation for 30 min (lane −) and from cells not subjected to this nutrient stress (lane +) were annealed with the PpdhA-R primer (see Table S1 in the supplemental material), and then primer extension was performed, as described in Materials and Methods. Lanes G, A, T, and C contained the products of the corresponding dideoxy sequencing reactions performed with the PCR product as the template, as described in Materials and Methods. The nucleotide sequence of the coding strand corresponding to the ladder is shown; the transcription initiation base (+1) is indicated by bold type, and the −35 and −10 regions of the pdhABCD promoter are underlined. The position of the runoff cDNA is indicated by the arrow on the right.
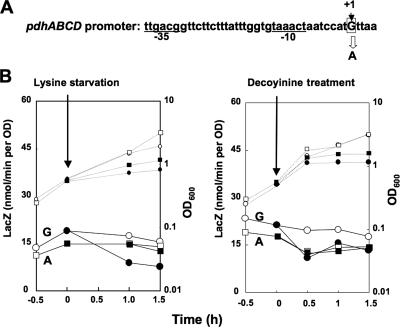
The transcription initiation base was found to be a guanine (Fig. 5). To determine if replacement of this guanine by adenine affects the negative stringent control of pdhABCD, we constructed strain FU1044 (with A) with this replacement in the pdhABCD promoter region (nucleotides −47 to 167) upstream of lacZ in the wild-type genetic background (Fig. 6A). Although negative stringent control of the pdhABCD promoter was observed upon lysine starvation in strain FU1019 with the original guanine residue (Fig. 6B, left panel), it was almost completely absent in strain FU1044 with A (Fig. 6B, left panel). When cells of these strains were exposed to decoyinine, the original negative regulation observed in strain FU1019 (with G) was completely quenched in strain FU1044 (with A) (Fig. 6B, right panel). These results suggest that the guanine at position 1 in the pdhABCD transcript is indispensable for this negative stringent control.
FIG. 6.
Effect of replacement of guanine at the transcription initiation base (+1) of the pdhABCD operon with adenine on the negative stringent control of this operon. (A) The guanine at the transcription initiation site (+1) of the pdhABCD promoter (nucleotides −47 to 167) was replaced by adenine. The −35 and −10 regions of the pdhABCD promoter are underlined. (B) The mutant pdhABCD promoter was placed upstream of lacZ in the relA+ and codY+ backgrounds. lacZ expression in the resulting strains (FU1019 [wild type] and FU1044 [mutant]) was monitored upon lysine starvation and decoyinine treatment, as described in Materials and Methods. OD600 values (small symbols) and LacZ activities (large symbols) are indicated by circles for FU1019 and by squares for FU1044; filled and open symbols indicate results obtained with and without lysine starvation and decoyinine treatment, respectively. The arrows indicate the onset of these stresses.
Positive stringent control of the pycA gene.
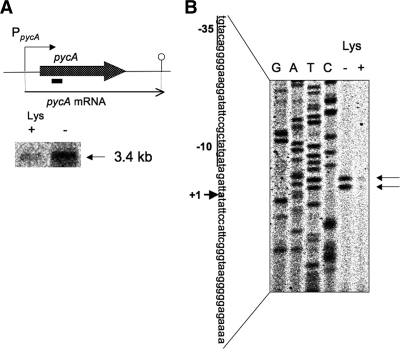
The pycA gene encodes a pyruvate carboxylase that converts pyruvate to oxaloacetic acid through CO2 fixation. This enzyme is rather constitutively synthesized and strongly activated by acetyl-CoA (5). pycA mRNA is 3.4 kb long (Fig. 7A), indicating that this gene is monocistronically transcribed. Northern analysis indicated that pycA expression was induced approximately 3-fold upon lysine starvation (Fig. 7A, lanes + and −). Primer extension analysis was performed to determine the transcription initiation base of pycA (Fig. 7B). There were two bands corresponding to adenines (positions 1 and 3); the transcription initiation adenine for the longer transcript was assigned to position 1 (30 nucleotides upstream of the translation initiation base of pycA), which enabled us to assign its −35 and −10 regions likely recognized by σA RNA polymerase. These transcripts were induced approximately 5-fold upon lysine starvation (Fig. 7B, lanes − and +).
FIG. 7.
Transcription of the pycA gene. (A) The pycA gene encodes pyruvate carboxylase. Its transcription results in a 3.4-kb monocistronic pycA mRNA. Northern analysis of the stringent control of pycA transcription was performed. RNA samples were prepared from cells of strain 1A765 (lys) that had been subjected to lysine starvation for 30 min and from cells that had not been starved. The 3.4-kb transcript was detected with the pycA probe (indicated by a bar). (B) The 5′ end of each of the pycA transcripts was mapped. Total RNAs from cells of strain 1A765 (lys) that had been subjected to lysine starvation for 30 min (lane −) and cells that had not been subjected to this nutrient stress (lane +) were annealed with the PpycA-R primer (see Table S1 in the supplemental material), and then primer extension was performed, as described in Materials and Methods. Lanes G, A, T, and C contained the products of the corresponding dideoxy sequencing reactions performed with the PCR product as the template, as described in Materials and Methods. The nucleotide sequence of the coding strand corresponding to the ladder is shown; the transcription initiation site (+1) is indicated by bold type (the upper initiation base in lane −), and another initiation base was observed in the same lane (lower base). The corresponding −35 and −10 regions of the pdhABCD promoter are underlined. The two positions of the runoff cDNA are indicated by the arrows on the right.
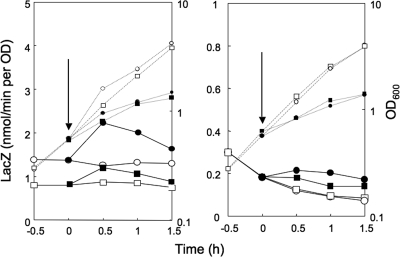
To examine the positive regulation of the pycA promoter, the core promoter region (nucleotides −55 to 26) was placed upstream of lacZ in the wild-type, relA1, and ΔcodY backgrounds. The β-Gal synthesis under control of the pycA promoter was monitored upon lysine starvation and decoyinine treatment (Fig. 3C). An increase in β-Gal synthesis upon lysine starvation was observed in the wild-type and ΔcodY backgrounds, but not in the relA1 background. When decoyinine was added to the medium, β-Gal synthesis increased to almost the same extent in the wild-type, relA1, and ΔcodY backgrounds. The results indicate that this positive regulation was also independent of CodY. Thus, the positive regulation of the presumably constitutive core pycA promoter was most likely due to the CodY-independent stringent control enhancing the transcription initiation rate, as observed for ilv-leu (20, 48).
The transcription initiation bases were found to be adenines (positions 1 and 3) (Fig. 7B). To determine if replacement of the adenine at position 1 by guanine affects the positive stringent control of pycA, we constructed strain FU1060 with this substitution in the pycA promoter region (nucleotides −55 to 26) upstream of lacZ in the wild-type genetic background. The positive stringent control of the pycA promoter that was observed upon lysine starvation in strain FU977 with the original adenine residue was slightly but significantly quenched in strain FU1060 with the guanine replacement (data not shown). When cells of these strains were exposed to decoyinine, the original positive regulation observed in strain FU977 (with A) was largely quenched in strain FU1060 (with G) (Fig. 8, left panel). These results suggest that the adenine at position 1 in the pycA transcript is involved in this positive stringent control. This partial effect of substitution of guanine for adenine at position 1 on the positive stringent response had been expected because the other transcription initiation from the adenine at position 3 appeared to occur in this mutant promoter.
FIG. 8.
Effect of replacement of adenine at each of the transcription initiation bases (position 1) of the pycA (left panel) and alsSD (right panel) operons by guanine on positive stringent control. The adenine at the transcription initiation site (+1) of the pycA and alsSD promoters (nucleotides −55 to 26) was replaced by guanine. The mutant pycA and alsSD promoters were placed upstream of lacZ in the relA+ and codY+ background. lacZ expression in wild-type strains FU977 (pycA) and FU937 (alsSD) and mutant strains FU1060 (pycA mutant) and FU1061 (alsSD mutant) was monitored upon decoyinine treatment, as described in Materials and Methods. OD600 values (small symbols) and LacZ activities (large symbols) are indicated by circles for FU977 and FU937 and by squares for FU1060 and FU1061; filled and open symbols indicate results obtained with and without decoyinine treatment, respectively. The arrows indicate the onset of decoyinine treatment.
Positive stringent control of the alsSD operon.
The alsSD genes, which comprise an operon, encode acetolactate synthase and acetolactate decarboxylase, respectively, whereas a positive regulator of AlsR, encoded by a gene divergent from alsSD, is involved in the induction of alsSD in the postexponential growth phase (35) (Fig. 3D, upper panel). It is also known that CcpA is a positive transcriptional regulator of alsSD. However, CcpA activation appears to be indirect because a catabolite-responsive element of cre is unlikely to be present in the alsSD promoter region (7, 51). Transcription of this operon starts at an adenine that is 51 bases upstream of the translation initiation base of alsS (35). DNA microarray analysis suggested that alsSD might be under CodY-independent positive stringent control. To examine this positive regulation of the alsSD promoter, the core promoter region (nucleotides −55 to 26) was placed upstream of lacZ in the wild-type, relA1, and ΔcodY backgrounds. The β-Gal synthesis under control of this promoter was monitored upon lysine starvation and decoyinine treatment (Fig. 3D). The β-Gal synthesis during the exponential growth phase without these stringent stresses, which was quite low, presumably due to a lack of positive regulation by AlsR, decreased continuously; we currently cannot explain this decrease. Thus, β-Gal synthesis upon lysine starvation was maintained in the wild-type and ΔcodY backgrounds, but not in the relA1 background, in contrast to the decrease observed without lysine starvation. When decoyinine was added to the medium, β-Gal synthesis was maintained at almost the same level in the wild-type, relA1, and ΔcodY backgrounds, in contrast to the decrease in the absence of decoyinine. The results indicate that the CodY-dependent positive regulation was also dependent on RelA upon lysine starvation and independent of RelA upon decoyinine treatment. The region upstream of nucleotide −55 is truncated in the core alsSD promoter, so it is very unlikely that AlsR or CcpA is responsible for this positive regulation. Thus, this positive regulation is most likely due to CodY-independent stringent control, as observed for ilv-leu (20, 48).
The transcription initiation base of the alsSD operon was reported to be an adenine (position 1). To determine if replacement of this adenine by guanine affects the positive stringent control of alsSD, we constructed strain FU1061 with this substitution in the alsSD promoter region (nucleotides −55 to 26) upstream of lacZ in the wild-type genetic background. The positive stringent control of the alsSD promoter, which was observed upon lysine starvation in strain FU937 with the original adenine, was slightly but significantly quenched in strain FU1061 with the guanine substitution (data not shown). When cells of these strains were exposed to decoyinine, the original positive regulation observed in strain FU937 (with A) was partially quenched in strain FU1061 (with G) (Fig. 8, right panel). These results suggest that the adenine at position 1 in the alsSD transcript is only partially involved in this positive stringent control. However, this does not necessarily mean that positive transcription regulation other than the CodY-independent positive stringent control is involved in this positive regulation, because the introduction of guanine at position 1 may cause a shift of the transcription initiation base from guanine at position 1 to other bases, such as cytosine at position 2 and adenine at position 3. Such a shift of the transcription initiation base upon introduction of the base substitution into the transcription initiation site was actually observed for ilv-leu (48).
DISCUSSION
The stringent transcription control of the ilv-leu operon was investigated in detail previously (20, 48), which revealed the molecular mechanisms underlying this control (see Fig. S1 in the supplemental material). Upon amino acid (lysine in our study [48]) starvation, the RelA protein synthesizes ppGpp, a probable inhibitor of IMP dehydrogenase. The resulting inhibition causes a decrease in the GTP level, whereas it increases the ATP level. Decoyinine treatment also induced these reciprocal changes in the GTP and ATP levels by inhibiting GMP synthase. Thus, two mechanisms underlying the positive stringent control of the ilv-leu operon in response to changes in the GTP and ATP levels have been proposed (20, 48). Mechanism 1, a CodY-dependent mechanism, involves GTP, a corepressor of CodY, so a decrease in the GTP level results in detachment of CodY from its binding sites, resulting in positive regulation of ilv-leu. In mechanism 2, a CodY-independent mechanism, the ilv-leu promoter is activated by an increase in the ATP level. In vitro transcription analysis indicated that initiation of ilv-leu transcription tolerates a decrease in the level of GTP to a level in the range of the in vivo concentrations upon lysine starvation and decoyinine addition (20, 48). Also, an increase in the ATP level actually enhances the initiation of ilv-leu transcription in vitro (20). Conservation of adenine at the transcription initiation site is critical for this positive regulation. When the adenine was replaced by guanine, the positive regulation of the core ilv-leu promoter changed to negative regulation. However, whether this adenine is located at position 1 or position 2 is unclear, because there is a difference between the two reports (20, 48).
DNA microarray analysis involving the CodY+ and CodY− strains that had been exposed and had not been exposed to decoyinine revealed that of the operons involved in glucose and pyruvate metabolism (Fig. 1 and Table 2), the ptsGHI (10, 45) and pdhABCD (9, 13) operons and the pycA (5) and alsSD (35) operons in addition to the ilv-leu operon are under negative and positive stringent control, respectively. Northern analysis showed that transcription from the ptsGHI promoter, but not transcription from the ptsHI promoter, was subject to the negative stringent control (Fig. 2). The lacZ fusion experiments revealed that the negative stringent control of the core ptsGHI promoter (nucleotides −55 to 26) was independent of CodY and mediated by ppGpp upon lysine starvation but not upon decoyinine addition (Fig. 3A). The guanine residues at the transcription initiation site (positions 1 and 2) of the ptsGHI operon were found to be indispensable for this negative regulation (Fig. 4). Similar experiments showed that the negative stringent control of the newly assigned pdhABCD promoter was independent of CodY and likely mediated by the decrease in the GTP level resulting from lysine starvation and decoyinine addition (Fig. 3B). The base substitution experiments revealed that the guanine at position 1 in the pdhABCD transcript was indispensable for the negative stringent control (Fig. 6). These results indicate that the negative stringent control of the ptsGHI and pdhABCD operons was dependent on the guanine residues at the transcription initiation site (positions 1 and 2) and was most likely triggered by the decrease in the GTP level caused by lysine starvation or decoyinine addition, as reported previously for the B. subtilis rrnB and rrnO operons (19). It is amazing that the entrance genes for glycolysis and the TCA cycle encoded by ptsGHI and pdhABCD are most likely subject to the same negative stringent control as rrn, rps, and tufA involved in protein synthesis (19, 20).
Not only the ilv-leu operon but also the pycA and alsSD operons involved in pyruvate metabolism are under CodY-independent positive stringent control caused by lysine starvation or decoyinine addition (Fig. 1 and Table 2). The lacZ fusion experiments indicated that the positive stringent control of pycA and alsSD is dependent on RelA but independent of CodY (Fig. 3C and 3D), as observed for the ilv-leu operon (20, 48). Since the core pycA and alsSD promoter regions (nucleotides −55 to 26) were placed upstream of lacZ in these fusion experiments, it is very unlikely that this positive regulation of pycA and alsSD is caused by another, unknown transcription activator because of the probable elimination of the cis-acting site upstream of the −35 promoter regions if there is any such site. Moreover, it is also unlikely that an additional global regulator, such as CodY, is involved in this positive stringent control of the core ilv-leu (48), pycA, and alsSD promoters (nucleotides −55 to 26). Thus, the promoter activity of pycA and alsSD was likely triggered by the increase in the ATP level caused by lysine starvation or decoyinine addition, as observed for ilv-leu (20). As expected, the transcription initiation bases of pycA and alsSD are adenines; the former was identified in this study (Fig. 7), and the latter was described previously (35). When these adenines at position 1 of pycA and alsSD were replaced by guanine, the base substitutions only partially affected the positive stringent control of the two promoters, which revealed the involvement of the adenines. However, this partial involvement in the positive stringent control of pycA and alsSD does not necessarily mean that other transcriptional regulation by unknown regulatory factors is involved in the positive control. This is because transcription initiation of pycA occurs at two adenines (positions 1 and 3) (Fig. 7B) and because the base substitution may have shifted the transcription initiation base, as observed for ilv-leu (48). Thus, the elaborate stringent control of pyruvate metabolism involving not only positive regulation of ilv-leu, pycA, and alsSD but also negative regulation of pdhABCD reminds us that the fate of pyruvate is very important to the cell due to its location at the center of carbon metabolism (37).
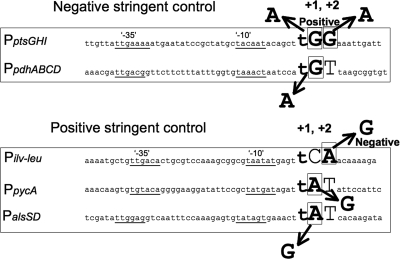
In Fig. 9, the nucleotide sequences of the promoter regions of the ptsGHI, pdhABCD, ilv-leu, pycA, and alsSD operons are aligned to determine any sequence conservation in them; the first two operons and the last three operons are under negative and positive stringent control, respectively. The GG at positions 1 and 2 of the transcription initiation site of the ptsGHI operon was changed to GA, AG, and AA. All of the base substitutions shifted the negative stringent regulation in the positive direction in the order GA-AG-AA (Fig. 4). This fact indicates that both of the guanines at positions 1 and 2 are involved in the negative stringent control of ptsGHI. Also, adenine substitution for guanine at position 1 of pdhABCD eliminated its negative regulation (Fig. 6). Guanine substitution for adenine at position 1 of pycA and alsSD partially affected the positive stringent control of these operons (Fig. 8). Moreover, we previously reported that when the adenine at position 2 of the transcription initiation site of ilv-leu was changed to guanine, the positive stringent regulation became negative stringent regulation. Interestingly, position −1 is occupied by thymine at all five transcription initiation sites (Fig. 9). These results, as well as the sequence alignment (Fig. 9), suggest that a guanine and not an adenine is located at either position 1 or 2 or at both positions for negative stringent control and that an adenine and not a guanine is located at either position 1 or 2 or at both positions for positive stringent control. If guanine and adenine are present at positions 1 and 2 of the same site, which is the case in many operons (12; T. Tojo, unpublished data) and as actually site-directed mutagenized in the case of ptsGHI, the positive stringent control and negative stringent control of such operons are assumed to be neutralized or to cancel each other out.
FIG. 9.
Alignment of the nucleotide sequences of the promoters under CodY-independent stringent control. The nucleotide sequences of the promoters of the ptsGHI and pdhABCD operons (PptsGHI and PpdhABCD) under CodY-independent negative stringent control and the nucleotide sequences of the ilv-leu, pycA, and alsSD operons (Pilv-leu, PpycA, and PalsSD) under CodY-independent positive stringent control are aligned. The transcription initiation base of the ptsGHI operon determined previously (45) was confirmed in this work (Fig. 2C). The initiation bases of the ilv-leu operon were reported previously to be cytosine at position 1 (48) and adenine at position 2 (20), a difference which has not been resolved. The initiation bases of the pdhABCD (Fig. 5) and pycA (Fig. 7B) operons were determined in this work; there are two initiation adenines for pycA, and the upstream position is designated position 1. The initiation base of the alsSD operon was determined previously (35). The sequences of the −35 and −10 regions of the promoters for these operons are underlined. The bases at positions 1 and 2 are indicated by large uppercase letters, and adenine and guanine are also indicated by bold type. The thymine at position −1 is indicated by a large lowercase letter. Conversion of guanine at position 1 and/or 2 of ptsGHI and pdhABCD to adenine alters the original negative stringent regulation to less negative or positive stringent regulation, while conversion of A at position 2 of ilv-leu to G changes the positive regulation to negative regulation (48). Moreover, conversion of adenine at position 1 of pycA and alsSD to G partially quenched the positive stringent control of these operons.
DNA microarray analysis revealed that 371 and 317 protein genes are negatively and positively regulated more than 3-fold upon decoyinine addition even in the ΔcodY genetic background. However, these genes are not always considered to be under the stringent control regulating the transcription initiation rate, as inferred from the concentrations of metabolites such as FBP, a signal compound in carbon catabolite regulation in B. subtilis (7), which are substantially changed upon decoyinine treatment, as observed in the metabolome analysis (Table 3). Thus, various transcriptional repressor and activators are thought to affect the expression of many genes when there are changes in the levels of the signal metabolites. Nevertheless, we aligned the promoter sequences of the operons, the expression of whose constituent genes was found to be altered more than 10-fold upon decoyinine addition in a DNA microarray analysis and whose transcription initiation bases are known (see Table S2 in the supplemental material). The downregulated operons were the S10 (23), str (21), pyrG (26), and pur (6) operons, whereas the upregulated operons were the metIC (3), ureABC (54), and rapA (29) operons. Also, we aligned the promoter sequences of the rrnB (P1) (44), rrnO (P1) (32), and tufA (21) operons that are downregulated and the appDFABCA (18) and ywaA (20) operons that are upregulated and are known to be subject to the stringent control mediated by modulation of the translation initiation rates (19, 20). As Table S2 in the supplemental material shows, there are adenines or guanines at positions 1 and 2 depending on whether the regulation is positive or negative. Moreover, all of the transcription initiation bases at position 1 for the P1 and P2 promoters of the 7 rrn operons are guanines, and the bases at position 2 are all thymines, except for cytosine in the case of the rrnB P2 promoter (30). However, the thymine at position −1 shown in the alignment in Fig. 9 is not well conserved in the promoter sequences shown in Table S2 in the supplemental material. This implies at least that the CodY-independent stringent control involving modulation of the transcription initiation rate might occur in numerous stringent operons other than those investigated so far by us (48; this study) and by Krásný et al. (19, 20).
The presence of either guanine or adenine in the transcription initiation site (positions 1 and 2) is likely indispensable for the negative and positive stringent control affecting the transcription initiation rate. Currently, we have no data showing that any other sequence conservation or requirement is necessary for this stringent control. Interestingly, the fact that B. subtilis and E. coli use different strategies to control the operons under stringent control might be related to the difference in the stabilities of open complexes of the two microorganisms previously noticed by Whipple and Sonenshein (53). The open complex formed with E. coli RNA polymerase before the formation of the elongation complex for transcription is stable before the formation of any phosphodiester bond, whereas the open complex formed with B. subtilis RNA polymerase in the absence of nucleotide triphosphate is unstable. This implies that a rate-limiting transcription initiation step might involve the formation of the first phosphodiester bond between the nucleotides at positions 1 and 2, which is assumed to be most affected by the concentrations of GTP and ATP, which change greatly when there is a stringent response. The details of the molecular mechanism underlying the stringent control affecting the transcription initiation rate, including the hypothesis described above, remain to be determined.
Supplementary Material
Acknowledgments
We thank Y. Sato, M. Hayasugi, H. Urabe, and Y. Kobayashi for their help with the experiments.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas and the High-Tech Research Center Project for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 14 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auger, S., W. H. Yuen, A. Danchin, and I. Martin-Verstraete. 2002. The metIC operon involved in methionine biosynthesis in Bacillus subtilis is controlled by transcription antitermination. Microbiology 148:507-518. [DOI] [PubMed] [Google Scholar]
- 4.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology Press, Washington, DC.
- 5.Diesterhaft, M. D., and E. Freese. 1973. Role of pyruvate carboxylase, phosphoenolpyruvate carboxykinase, and malic enzyme during growth and sporulation of Bacillus subtilis. J. Biol. Chem. 248:6062-6070. [PubMed] [Google Scholar]
- 6.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262:8274-8287. [PubMed] [Google Scholar]
- 7.Fujita, Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245-259. [DOI] [PubMed] [Google Scholar]
- 8.Fujita, Y., and E. Freese. 1979. Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis. J. Biol. Chem. 254:5340-5349. [PubMed] [Google Scholar]
- 9.Gao, H., X. Jiang, K. Pogliano, and A. I. Aronson. 2002. The E1β and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J. Bacteriol. 184:2780-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzy-Tréboul, G., M. Zagorec, M. C. Rain-Guion, and M. Steinmetz. 1989. Phosphoenolpyruvate:sugar phosphotransferase system of Bacillus subtilis: nucleotide sequence of ptsX, ptsH and the 5′-end of ptsI and evidence for a ptsHI operon. Mol. Microbiol. 3:103-112. [DOI] [PubMed] [Google Scholar]
- 11.Grandoni, J. A., S. A. Zahler, and J. M. Calvo. 1992. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J. Bacteriol. 174:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemilä, H., A. Palva, L. Paulin, S. Arvidsion, and I. Palva. 1990. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J. Bacteriol. 172:5052-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate, 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H. J., A. Roux, and A. L. Sonenshein. 2002. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol. Microbiol. 45:179-190. [DOI] [PubMed] [Google Scholar]
- 18.Koide, A., and J. A. Hoch. 1994. Identification of a second oligopeptide transport system in Bacillus subtilis and determination of its role in sporulation. Mol. Microbiol. 13:417-426. [DOI] [PubMed] [Google Scholar]
- 19.Krásný, L., and R. L. Gourse. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23:4473-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krásný, L., H. Tišerová, J. Jonák, D. Rejman, and H. Sanderová. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 69:42-54. [DOI] [PubMed] [Google Scholar]
- 21.Krásný, L., T. Vacík, V. Fucík, and J. Jonák. 2000. Cloning and characterization of the str operon and elongation factor Tu expression in Bacillus stearothermophilus. J. Bacteriol. 182:6114-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 23.Li, X., L. Lindahl, Y. Sha, and J. M. Zengel. 1997. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-α cluster. J. Bacteriol. 179:7046-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez, J. M., C. L. Marks, and E. Freese. 1979. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim. Biophys. Acta 587:238-252. [DOI] [PubMed] [Google Scholar]
- 26.Meng, Q., C. L. Turnbough, Jr., and R. L. Switzer. 2004. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc. Natl. Acad. Sci. U. S. A. 101:10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miwa, Y., and Y. Fujita. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 183:5877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller, J. P., G. Bukussoglu, and A. L. Sonenshein. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174:4361-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natori, Y., K. Tagami, K. Murakami, S. Yoshida, O. Tanigawa, Y. Moh, K. Masuda, T. Wada, S. Suzuki, H. Nanamiya, Y. Tozawa, and F. Kawamura. 2009. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J. Bacteriol. 191:4555-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochi, K., J. C. Kandala, and E. Freese. 1981. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 256:6866-6875. [PubMed] [Google Scholar]
- 32.Ogasawara, N., S. Moriya, and H. Yoshikawa. 1983. Structure and organization of rRNA operons in the region of the replication origin of the Bacillus subtilis chromosome. Nucleic Acids Res. 11:6301-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Presecan-Siedel, E., A. Galinier, R. Longin, J. Deutscher, A. Danchin, P. Glaser, and I. Martin-Verstraete. 1999. Catabolite regulation of the pta gene as part of carbon flow pathways in Bacillus subtilis. J. Bacteriol. 181:6889-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renna, M. C., N. Najimudin, L. R. Winik, and S. A. Zahler. 1993. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J. Bacteriol. 175:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Sauer, U., and B. J. Eikmanns. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29:765-794. [DOI] [PubMed] [Google Scholar]
- 38.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62:811-822. [DOI] [PubMed] [Google Scholar]
- 40.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 41.Shivers, R. P., and A. L. Sonenshein. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 56:1549-1559. [DOI] [PubMed] [Google Scholar]
- 42.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 43.Soga, T., Y. Ohashi, Y. Ueno, H. Naraoka, M. Tomita, and T. Nishioka. 2003. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2:488-494. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, G. C., and K. F. Bott. 1983. DNA sequence of the tandem ribosomal RNA promoter for B. subtilis operon rrnB. Nucleic Acids Res. 11:6289-6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 46.Suhadolnik, R. J. 1970. Nucleoside antibiotics, p. 96-122. John Wiley & Sons, Inc., New York, NY.
- 47.Swanton, M., and G. Edlin. 1972. Isolation and characterization of an RNA relaxed mutant of B. subtilis. Biochem. Biophys. Res. Commun. 46:583-588. [DOI] [PubMed] [Google Scholar]
- 48.Tojo, S., T. Satomura, K. Kumamoto, K. Hirooka, and Y. Fujita. 2008. Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J. Bacteriol. 190:6134-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56:1560-1573. [DOI] [PubMed] [Google Scholar]
- 50.Turinsky, A. J., F. J. Grundy, J. H. Kim, G. H. Chambliss, and T. M. Henkin. 1998. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J. Bacteriol. 180:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turinsky, A. J., T. R. Moir-Blais, F. J. Grundy, and T. M. Henkin. 2000. Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis. J. Bacteriol. 182:5611-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 53.Whipple, F. W., and A. L. Sonenshein. 1992. Mechanism of initiation of transcription by Bacillus subtilis RNA polymerase at several promoters. J. Mol. Biol. 223:399-414. [DOI] [PubMed] [Google Scholar]
- 54.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida, K., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 179:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida, K., K. Kobayashi, Y. Miwa, C. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.