Abstract
To characterize the role of the three PAS domains in KinA, the major sporulation kinase in Bacillus subtilis, we constructed a series of systematic PAS domain deletion mutants and analyzed their activities using an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible artificial sporulation induction system, which we have developed recently. The results showed that any one of the three PAS domains is sufficient to maintain the kinase activity and trigger sporulation, if not fully then at least partially, when the protein levels increase beyond a certain level.
When Bacillus subtilis cells encounter nutrient deprivation, they decide to switch from a symmetric cell division to an asymmetric cell division as a means of survival. The complex signal transduction system includes at least three sensor histidine kinases (KinA, KinB, and KinC), two phosphotransferases (Spo0F and Spo0B), and one response regulator (Spo0A). Although KinD and KinE were originally suggested to be sensor kinases involved in sporulation (11), an in vivo study demonstrated that these kinases do not feed phosphates into the phosphorelay under physiological conditions (6). Among the three sensor kinases, the major sporulation kinase is the cytoplasmic protein KinA (17). Upon nutrient limitation, KinA autophosphorylates and indirectly transfers the phosphate group to the master transcription factor Spo0A via two intermediate proteins, Spo0F and Spo0B (1). Once the activated Spo0A accumulates to a certain threshold level in a gradual manner, each of the 121 genes under the direct control of the master regulator is turned on or off at different times depending on the concentration of the regulator (5, 15). Therefore, the four-component phosphate transfer system found in B. subtilis sporulation is called a multicomponent “phosphorelay” (1, 9).
Based on the structural and functional characterization of other two-component systems, mostly in Escherichia coli, it has been defined that amino-terminal (N-terminal) and carboxy-terminal (C-terminal) domains are sensor and autokinase domains, respectively (18, 20, 25, 26). In KinA, the N-terminal sensor domain contains three PAS domains (PAS-A, -B, and -C) (14, 20, 25), and the C-terminal autokinase domain includes a dimerization and histidine phosphotransfer (DHp) domain and a catalytic ATP-binding (CA) domain (18, 25). The PAS domain was originally found in the Drosophila period clock protein (PER), the vertebrate aryl hydrocarbon receptor nuclear translocator (ARNT), and the Drosophila single-minded protein (SIM) as imperfect repeat sequences involved in protein-protein interaction (2, 16). PAS domains have been found in numerous histidine kinases of two-component systems in bacteria and are suggested to be responsible for sensing a variety of signals and regulate the activity of each of the associated histidine kinases (8, 22, 23). Thus, it has been widely believed that PAS domains of KinA could sense an as-yet-unknown sporulation signal and activate the sporulation kinase through autophosphorylation (14, 20, 25). However, the signaling molecule that is sensed by the PAS domains in KinA has never been identified. Therefore, the molecular mechanism by which KinA gets activated is still not clear.
In our recent report, using our artificial sporulation induction (ASI) system (see Fig. S1 in the supplemental material), which triggers sporulation in response to the artificial induction of KinA (6), it has been shown that the PAS-A domain of KinA, which was originally believed to be essential for signal sensing and function (14, 20, 25), is dispensable (3). It has also been suggested that the role of PAS domains in KinA is to form a stable homotetramer complex and possibly not for sensing the starvation-induced signal (3).
Here in this study, we furthered our understanding of the role of PAS domains by constructing a series of systematic PAS domain deletion mutants consisting of a different combination of domain deletions and studying them individually.
Systematic construction of KinA domain-deletion mutants and the primary assay system.
Although PAS domains appear to be used as sensor modules that sense a wide range of stimuli and regulate an associated histidine kinase (8, 22, 23), our previous results suggest that the primary role of the amino-terminal domain composed of three PAS domains in KinA is to form a stable complex as a functional kinase (3). However, it remains unclear how two of the three PAS domains (PAS-B and PAS-C) contribute to the kinase activity and whether any two of the three PAS domains are sufficient for the function. In order to address these questions, we began by constructing a mutant collection with systematic domain deletions. Thus, we generated a series of deletion mutants in which each of the PAS domains along with the CA domain was deleted systematically (Fig. 1). Each deletion mutant of KinA was fused to the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter (Phyper-spank), and then the fusion was integrated as a single copy at the amyE locus (coding for a nonessential α-amylase used for ectopic integration) by double crossover homologous recombination in the endogenous kinA null mutant as the ASI system described previously (3). To avoid the phosphate input to the phosphorelay by another sporulation kinase, KinB (10, 12, 24), the kinB null mutation was introduced into all the strains used in this study. We note that the contribution of KinC to sporulation is less than that of KinA and KinB under our standard sporulation conditions (13), and thus, the kinC null mutant was not used in this study. B. subtilis strain PY79 was used as the parental strain for all experiments (27). Details of the strains and plasmids used in this study are listed in Tables S1 and S2 in the supplemental material.
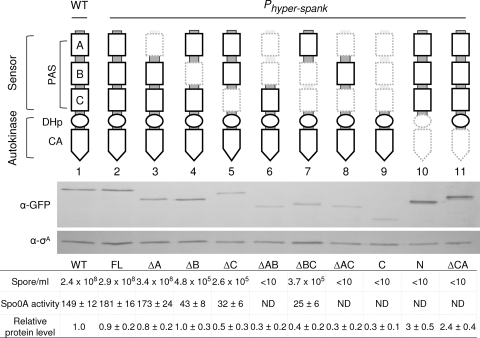
FIG. 1.
Quantitative assay for detection of the enzymatic activities of the KinA domain-deletion mutants. The schematic diagram at the top shows a domain organization of KinA. The sensor domain at the N terminus is further subdivided into three PAS domains (PAS-A, PAS-B, and PAS-C). The autokinase domain at the C terminus is further subdivided into a DHp and a CA domain (18, 25). The catalytic histidine (His405) is located within the DHp domain. Deleted portions are depicted in hatched lines in the diagrams. Genes for full-length KinA and each truncated mutant protein were fused to Phyper-spank and introduced at the amyE locus of cells containing both kinA and kinB null mutations. β-Galactosidase activities (Miller units) from PspoIIG-lacZ were measured at hour 3 after suspension as a measure of Spo0A activity. The same culture was used for the determination of sporulation efficiency. The wild-type strain (WT; lacking the IPTG-inducible construct) was used as a control. Strains used are as follows: MF3366 (WT), MF3316 (full-length KinA; FL), MF3317 (ΔPAS-A; ΔA), MF3318 (ΔPAS-B; ΔB), MF3319 (ΔPAS-C; ΔC), MF3320 (ΔPAS-AB; ΔAB), MF3321 (ΔPAS-BC; ΔBC), MF3322 (ΔPAS-AC; ΔAC), MF3323 (KinAC; C), MF3324 (KinAN; N), and MF3325 (ΔCA). Expression of full-length KinA or KinA mutant protein was examined by immunoblot analysis. Cell extracts of strains expressing GFP fusion proteins were prepared at hour 2 after suspension and were processed for immunoblotting with anti-GFP (α-GFP) antibodies. σA, a constitutively expressed protein, was used as a loading control. The protein levels were quantified and normalized to both the levels of σA and the levels of the wild-type KinA-GFP. Strains used for immunoblotting are as follows: MF3593 (WT; lane 1), MF3352 (FL; lane 2), MF3353 (ΔA; lane 3), MF3354 (ΔB; lane 4), MF3355 (ΔC; lane 5), MF3356 (ΔPAS-AB; lane 6), MF3357 (ΔPAS-BC; lane 7), MF3358 (ΔPAS-AC; lane 8), MF3359 (KinAC; lane 9), MF3360 (KinAN; lane 10), and MF3361 (ΔCA; lane 11). All experiments were performed at least three times independently. The representative data are shown. ND, not detected.
B. subtilis strains were grown in CH (casein hydrolysate) medium and were induced to sporulate by resuspension in Sterlini-Mandelstam (SM) medium (21) as described previously (3). IPTG was added at the time of resuspension at the indicated concentration to induce the synthesis of KinA or its mutants under the control of the IPTG promoter (Phyper-spank). Sporulation efficiency was measured as the number of CFU (CFU/ml) after incubation at 80°C for 10 min. To measure the autokinase activity of each mutant, we used an indirect reporter system in which the β-galactosidase gene (lacZ) is fused to the Spo0A-directed promoter, PspoIIG, as described previously (3). The strains expressing full-length and stepwise PAS domain deletions (ΔPAS-A, ΔPAS-AB, or KinAC mutants) reported previously (3) were used as controls. Assay of β-galactosidase activity was performed as described previously (3, 7).
PAS-A domain by itself and any two of the three PAS domains are sufficient to maintain partial kinase activity.
To test the functionality of each mutant, using the ASI system as described above, we performed a quantitative sporulation assay where we monitored the sporulation efficiency of each mutant under normal sporulation conditions in SM medium (21). To induce the synthesis of the proteins, we added the inducer (IPTG) at a 10 μM final concentration, at which the level of full-length KinA is comparable to the endogenous level of KinA in the sporulating wild type as defined in the previous report (3, 6).
First, sporulation efficiency of the overnight culture was measured by the production of heat-resistant CFU. The full-length KinA (depicted as FL in Fig. 1, and hereafter, similar abbreviations in parentheses denote each deletion mutant) was used as a positive control, and both the N and C terminus deletion mutants of KinA, KinAC (C), and KinAN (N), respectively, served as negative controls (3). When the inducer was added to the culture, each single PAS domain deletion mutant (ΔPAS-B [ΔB] and ΔPAS-C [ΔC]) and the PAS-BC domain deletion mutant (ΔPAS-BC [ΔBC]) exhibited reduced sporulation efficiency (∼105 range) compared to that of the full-length KinA (FL) and the functional ΔPAS-A (ΔA) mutant (3) but significantly higher levels than those in the negative controls, KinAC (C) and KinAN (N) (Fig. 1). However, the ΔPAS-AC (ΔAC) two-PAS-domain deletion mutant showed decreased sporulation efficiency similar to that of the negative controls, including the PAS-AB deletion mutant (ΔAB) (Fig. 1) (3). The deletion mutant of the catalytic ATP-binding domain (ΔCA) also displayed significantly lower sporulation efficiency, comparable to that of the negative controls (Fig. 1), indicating that the CA domain is essential for its function.
To further verify the results obtained above, we performed a β-galactosidase assay with the reporter strains containing the Spo0A-directed PspoIIG-lacZ, as described in the previous section, to measure the Spo0A∼P activity. The results obtained were consistent with the sporulation efficiency data that the Spo0A activities in the full-length KinA strain and the PAS-A deletion mutant were found to be similar to that in the wild type, while those in the other three partially functional sporulation mutants (ΔPAS-B, ΔPAS-C, and ΔPAS-BC) were relatively low (∼20% of the wild-type). Furthermore, little or no Spo0A activity was detected in the remaining strains with significantly lower sporulation efficiencies. Therefore, these results suggest that the sporulation efficiency exhibited by each strain correlates well with the Spo0A activity (Fig. 1).
In order to confirm that all the mutant proteins tested were expressed stably, we constructed strains expressing a KinA mutant fused to green fluorescent protein (GFP) under the ASI system and examined the protein levels in the cells prepared at 2 h after IPTG induction under the sporulation conditions (SM medium). Then, immunoblot analyses were performed as described previously (3, 7), using polyclonal anti-GFP (7, 19) and anti-σA (4) antibodies to detect appropriate proteins.
The protein levels of full-length ΔPAS-A and ΔPAS-B mutants were similar to the wild-type KinA level under sporulation conditions (Fig. 1). However, the protein levels of all the other PAS domain deletion mutants (ΔPAS-C, ΔPAS-AB, ΔPAS-BC, ΔPAS-AC, and KinAC) were decreased approximately 2- to 3-fold. In contrast, the protein levels of KinAN and the CA domain deletion mutant (ΔCA) were 2- to 3-fold higher than that of the wild-type control. These results suggest that the PAS domains of KinA stabilize the protein complex and possibly protect the protein from proteolytic degradations, while the C-terminal domain, possibly the CA domain, might be a target of unknown protease cleavage because the ΔCA mutant was stabilized. Based on this set of results we divided our mutants into three different groups: group I, the ΔPAS-A mutant as fully functional; group II, ΔPAS-B, ΔPAS-C, and ΔPAS-BC mutants as partially functional; and group III, ΔPAS-AB and ΔPAS-AC mutants as nonfunctional.
The mutant kinase activity is restored with a protein dosage effect.
We have provided evidence for the existence of a critical threshold, beyond which, when KinA protein level is increased, sporulation is triggered irrespective of the nutrient availability (P. Eswaramoorthy, D. Duan, J. Dinh, and M. Fujita, submitted for publication) (6). Furthermore, we have shown that the N terminus of KinA (KinAN) forms a more stable complex than does the C terminus (KinAC) and that at least the presence of two PAS domains, PAS-BC, is sufficient to form a stable homocomplex (3). Thus, based on these previous results and the results obtained in the above section, we hypothesized that by increasing the protein level, the probability of formation of the functional homocomplex with other PAS domain deletion mutants, which are not fully functional under the standard assay conditions (10 μM IPTG) as defined previously (3), would be increased. Thus, as a result of a dosage effect, the kinase activity of the partially or nonfunctional PAS domain deletion mutants would be rescued. To test these ideas, we monitored the sporulation efficiencies in all the mutants at various concentrations of the inducer under normal sporulation conditions. By confirming the previous findings, we used the strain harboring the full-length KinA construct and demonstrated that the efficiency of sporulation was increased sharply by inducing the synthesis of the protein to the threshold at around 4 μM to 8 μM IPTG concentrations (Eswaramoorthy et al., submitted for publication). Therefore, the result of full-length KinA serves as a control. The ΔPAS-A mutant, which is functional as reported (3), displayed an essentially similar trend of effectiveness on sporulation efficiencies and Spo0A activities as that observed with the full-length KinA (Fig. 2). However, group II mutants (ΔPAS-B and ΔPAS-C and ΔPAS-BC), which displayed decreased sporulation efficiencies and Spo0A activities compared to wild type at the standard IPTG concentration (10 μM), restored both sporulation efficiencies and Spo0A activities to the wild-type levels by increasing the inducer concentration to up to 40 μM (Fig. 2).
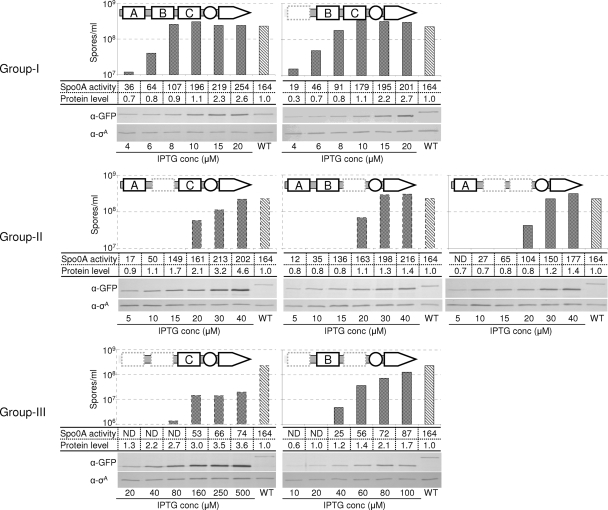
FIG. 2.
Protein dosage effect. IPTG concentration was increased gradually in order to increase the protein level of KinA or its mutants. Groups were classified based on the IPTG concentration range required to achieve wild-type efficiency of sporulation. A schematic diagram of each of the proteins is depicted at the top of each panel. The IPTG concentration used is indicated at the bottom of each panel. Sporulation efficiencies, β-galactosidase activities, and protein levels were determined as described in the legend for Fig. 1. Strains used for β-galactosidase assay are as follows: MF3316 (full-length KinA), MF3317 (ΔPAS-A), MF3318 (ΔPAS-B), MF3319 (ΔPAS-C), MF3320 (ΔPAS-AB), MF3321 (ΔPAS-BC), MF3322 (ΔPAS-AC), and MF3366 (WT). Strains used for immunoblot analysis are as follows: MF3352 (full-length KinA), MF3353 (ΔPAS-A), MF3354 (ΔPAS-B), MF3355 (ΔPAS-C), MF3356 (ΔPAS-AB), MF3357 (ΔPAS-BC), MF3358 (ΔPAS-AC), and MF3593 (WT). All experiments were performed at least three times independently, and the representative data are shown. ND, not detected.
Next, group III mutants, the ΔPAS-AB and ΔPAS-AC mutants, which displayed significantly lower sporulation efficiency and Spo0A activity at standard IPTG concentration (10 μM), were examined similarly, but with further increased concentrations of the inducer. Thus, the range of IPTG concentration was changed from 10 μM to 100 μM for the ΔPAS-AC mutant and 20 μM to 500 μM for the ΔPAS-AB mutant. The sporulation efficiency and Spo0A activity were restored to significantly higher levels at around 60 μM IPTG in the ΔPAS-AC mutant and 160 μM IPTG in the ΔPAS-AB mutant, respectively.
Finally, by immunoblot analysis, we confirmed that the protein levels of each mutant were proportional to the added concentration of IPTG, but saturated at different concentrations (Fig. 2). At the saturation point of each mutant protein, the sporulation efficiency and the Spo0A activity were restored to significantly higher levels (Fig. 2). Therefore, in support of our hypothesis, these results suggest that a structural instability of the complex with the PAS domain deletion mutant can be stabilized with a protein dosage effect. We note that the negative-control mutants, the ΔCA and KinAC (ΔPAS-ABC) mutants, produced no detectable sporulation efficiencies (<10) and Spo0A activities even at a 1 mM final IPTG concentration (data not shown). To provide further direct evidence to support our hypothesis, we plan to measure the stability of complex formation between each of the single PAS domains by using genetic and biochemical approaches.
Now, based on the set of results reported here, we predict that the order of affinity residing in each combination of the PAS domain for complex formation is as follows: ABC (wild type) ≈ BC (ΔPAS-A mutant) > AB (ΔPAS-C mutant) ≈ AC (ΔPAS-B mutant) ≈ A (ΔPAS-BC mutant) > B (ΔPAS-AC mutant) > C (ΔPAS-AB mutant). Interestingly, any one of the three PAS domains is sufficient to maintain the kinase activity and trigger sporulation, if not fully at least partially, when the protein levels increase beyond a certain level.
Taking all our previous and current results into consideration, we postulate that (i) each of the PAS subdomains has a redundant role in the complex formation and (ii) a combination of two and three PAS subdomains shows a synergistic effect to stabilize the complex formation. Based on these points, we propose that the maintenance of KinA activity might be fully dependent on the complex formation, possibly tetramerization, but not for signal sensing. While the mechanism of the increase in the level of KinA to the threshold remains elusive, our results in this and previous studies (3) might suggest a possible alternative mechanism for KinA activation in light of the unsuccessful attempts in identification of the sporulation signal.
Supplementary Material
Acknowledgments
We thank Ashlee Dravis and Karon P. Cassidy for helpful comments and discussions.
This work was supported in part by the Texas Advanced Research Program (003652-0072-2007), the Welch Foundation (E-1627), and the NSF (MCB-0920463).
Footnotes
Published ahead of print on 14 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Crews, S. T. 1998. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 12:607-620. [DOI] [PubMed] [Google Scholar]
- 3.Eswaramoorthy, P., T. Guo, and M. Fujita. 2009. In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. J. Bacteriol. 191:5358-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita, M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5:79-88. [DOI] [PubMed] [Google Scholar]
- 5.Fujita, M., J. E. Gonzalez-Pastor, and R. Losick. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita, M., and R. Losick. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 8.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 9.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441-465. [DOI] [PubMed] [Google Scholar]
- 10.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 12.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeDeaux, J. R., N. Yu, and A. D. Grossman. 1995. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:861-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, J., D. R. Tomchick, C. A. Brautigam, M. Machius, R. Kort, K. J. Hellingwerf, and K. H. Gardner. 2008. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry 47:4051-4064. [DOI] [PubMed] [Google Scholar]
- 15.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 16.Nambu, J. R., J. O. Lewis, K. A. Wharton, Jr., and S. T. Crews. 1991. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell 67:1157-1167. [DOI] [PubMed] [Google Scholar]
- 17.Perego, M., S. P. Cole, D. Burbulys, K. Trach, and J. A. Hoch. 1989. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J. Bacteriol. 171:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowland, S. L., W. F. Burkholder, K. A. Cunningham, M. W. Maciejewski, A. D. Grossman, and G. F. King. 2004. Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis. Mol. Cell 13:689-701. [DOI] [PubMed] [Google Scholar]
- 19.Rudner, D. Z., and R. Losick. 2002. A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephenson, K., and J. A. Hoch. 2001. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 98:15251-15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trach, K. A., and J. A. Hoch. 1993. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol. Microbiol. 8:69-79. [DOI] [PubMed] [Google Scholar]
- 25.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 183:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitten, A. E., D. A. Jacques, B. Hammouda, T. Hanley, G. F. King, J. M. Guss, J. Trewhella, and D. B. Langley. 2007. The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition. J. Mol. Biol. 368:407-420. [DOI] [PubMed] [Google Scholar]
- 27.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.