Abstract
Mycobacterium tuberculosis survives in latently infected individuals, likely in a nonreplicating or dormancy-like state. The M. tuberculosis DosR regulon is a genetic program induced by conditions that inhibit aerobic respiration and prevent bacillus replication. In this study, we used a mutant incapable of DosR regulon induction to investigate the contribution of this regulon to bacterial metabolism during anaerobic dormancy. Our results confirm that the DosR regulon is essential for M. tuberculosis survival during anaerobic dormancy and demonstrate that it is required for metabolic processes that occur upon entry into and throughout the dormant state. Specifically, we showed that regulon mechanisms shift metabolism away from aerobic respiration in the face of dwindling oxygen availability and are required for maintaining energy levels and redox balance as the culture becomes anaerobic. We also demonstrated that the DosR regulon is crucial for rapid resumption of growth once M. tuberculosis exits an anaerobic or nitric oxide-induced nonrespiring state. In summary, the DosR regulon encodes novel metabolic mechanisms essential for M. tuberculosis to survive in the absence of respiration and to successfully transition rapidly between respiring and nonrespiring conditions without loss of viability.
Mycobacterium tuberculosis, a major human pathogen, infects nearly one-third of the people in the world and causes two million deaths per year (8). Most infections are latent, and a substantial number of new infections are transmitted by individuals in whom latent infections are being reactivated. Latency is a clinical term describing people that are infected with M. tuberculosis but lack symptoms of active disease. Traditionally, it has been thought that bacilli in latently infected individuals reside almost exclusively inside granulomas and mature tubercle lesions. Recent studies indicate that in latently infected individuals M. tuberculosis may also be found outside granulomas in places such as endothelial cells, fibroblasts, and adipose tissue (17, 28). The evidence for M. tuberculosis metabolic activity in vivo is more limited, but two studies by Lillebaek et al. are informative (24, 25). In these studies the researchers used detailed records of tuberculosis epidemiology and strain types in the fairly static population of Denmark. They found that strains isolated from patients thought to have reactivated disease (rather than a primary infection) were nearly identical to strains present 30 years earlier in the same geographic population. The near-identity of the strains and the fact that infections were attributed to reactivation suggest that bacteria in latently infected individuals experience little genetic change during years of latent infection. The researchers concluded that during latency, M. tuberculosis divides infrequently and is likely in a minimal metabolic state.
One approach to study the M. tuberculosis metabolic state during latent infection is to use in vitro models that mimic conditions thought to exist in vivo. Such conditions include hypoxia produced in avascular calcified granulomas (40) and nitric oxide (NO) (27) or carbon monoxide (CO) (33) produced by activated immune cells. A widely used model is the “Wayne model” pioneered by Lawrence Wayne. In this model, a low-inoculum culture is sealed in a tube with stirring and allowed to slowly consume oxygen until the culture is anaerobic, resulting in a nonreplicating and apparently dormant state (45, 46). Another model used to look at dormant M. tuberculosis is a constant-hypoxia model that maintains a 0.2% oxygen tension in culture flasks (31).
The common theme in these in vitro models used to obtain M. tuberculosis dormancy is inhibition of respiration. The DosR regulon is a set of at least 48 coregulated genes that are induced by three conditions that inhibit aerobic respiration: hypoxia, NO, and CO (42). Induction of the DosR regulon closely mirrors inhibition of respiration, indicating that control of the regulon is linked to the aerobic respiratory state of the bacilli (43). Several studies have shown that the DosR regulon is controlled by a three-component regulatory system composed of two sensor histidine kinases, DosS and DosT, and a response regulator, DosR (42). DosS and DosT both bind the respiration-impairing gases NO and CO (19, 20, 38), further supporting the hypothesis that the DosR regulon responds to, and is important during, conditions that do not allow aerobic respiration. Although the majority of the DosR-regulated genes have not been characterized, the timing of their induction combined with the conditions under which they respond suggests that they may play a role in adaptation of M. tuberculosis to its host environment. Consistent with this notion, DosR regulon genes are induced in the lungs of M. tuberculosis-infected mice (43), as well as in interferon-gamma-activated murine macrophages (34) and guinea pigs (37).
Several studies have suggested that the DosR regulon plays a role in latent infection and in persistence in animal models that resemble human infection in some respects. Leyten et al. found that latently infected humans are more likely than humans with active infections to bear T cells specific for DosR regulon antigens (23), suggesting that the regulon is expressed during latency. Two recent studies confirmed that there is an immune response to DosR regulon antigens during latent infection (4, 36). Further evidence for clinical relevance in humans comes from a study showing that M. tuberculosis in sputum expresses the DosR regulon (15). The importance of this regulon for persistence in rabbit and guinea pig models was demonstrated by data showing a 2-log decrease in recovery of a DosR mutant 2 weeks (guinea pig) and 8 weeks (rabbit) after aerosol infection (11). A DosR mutant was also found to be significantly attenuated in guinea pig infection (26), further supporting the notion that the DosR regulon is required for persistence in vivo. It should be noted that in both studies showing the DosR phenotype (11, 26), full complementation and reversion to full virulence were not observed. However, it is now known that regulation of dosR expression is quite complex. Multiple regulatory sequences exist in and upstream of Rv3134c, the gene directly upstream of dosR (8). Failure to include such a regulatory sequence in a complemented strain would likely result in misregulation of dosR and poor complementation. Studies of DosR regulon mutants for murine infection have produced inconsistent findings that vary from hypervirulent (30) to attenuated (11) and not attenuated (3, 31). When animal models are compared, it is important to remember that M. tuberculosis-induced granulomas in primates, rabbits, and guinea pigs develop caseous necrosis and are hypoxic and/or anaerobic, while M. tuberculosis induced-granulomas in mice are neither hypoxic nor anaerobic (2, 21, 41). Furthermore, M. tuberculosis divides regularly in chronic murine infections (16), in contrast to the replication during latent infections, as demonstrated in the studies of Lillebaek et al. (24, 25). Such studies underscore the significant differences between models.
A previous study with a DosR mutant in a closely related Mycobacterium bovis BCG strain showed that DosR expression is required for survival in an in vitro Wayne-like model of dormancy (5). Unexpectedly, two similar studies in M. tuberculosis did not show a strong survival defect for a DosR mutant (31, 43). The most recent study showed that there was only a modest survival defect in an H37Rv DosR mutant and concluded that the DosR regulon is a short-term phenomenon and is not responsible for the adaptation necessary to survive under primarily hypoxic conditions in vitro (31, 32).
In this study we showed that the DosR regulon is required for M. tuberculosis survival during anaerobic dormancy. We also used a combination of genetic and biochemical approaches to demonstrate that this regulon is necessary to shift away from oxygen consumption, maintain ATP levels, and balance the redox state (NAD/NADH ratio) of the cell as oxygen becomes scarce. Furthermore, we showed that the DosR regulon is necessary for optimal transition of M. tuberculosis back to aerobic growth from an anaerobic or nitric oxide-induced nonrespiring state.
MATERIALS AND METHODS
Strains and dormancy models.
Strains used in this study are described in Table 1. Cultures used in dormancy models were first grown with rapid stirring at 37°C in 250-ml sterile Erlenmeyer flasks with DTA medium composed of Dubos broth base (Difco/Becton Dickinson) supplemented with Tween 80 (0.05%), bovine serum albumin (BSA) (0.5%), and glucose (0.75%). The rapid anaerobic dormancy (RAD) model experiments were performed using sterile glass tubes (20 by 125 mm) containing stir bars (12 mm by 4.5 mm) and 16 ml DTA medium at a culture-to-headspace ratio of 0.65 (adjusted for the altitude in Denver, compared to a ratio of 0.5 for sea level) and inoculated with 360 μl culture at an optical density at 600 nm (OD600) of 0.2. Stop cock grease was applied to the threads of the glass tubes, and the tubes were sealed with phenolic caps. Cultures were stirred rapidly (200 rpm) using a rotary magnetic tumble stirrer obtained from V & P Scientific (San Diego, CA). The data below are the averages of 3 independent experiments.
TABLE 1.
Strains used in this study
Recovery in liquid media.
At various time points during the RAD model experiments, 100 μl of culture was added to 900 μl fresh DTA medium in 24-well plates. Tenfold serial dilutions were made up to a 10−10 dilution. The 24-well plates were sealed with Parafilm and stored in plastic containers to prevent evaporation. The plates were checked every week for growth. The most probable number (MPN) per milliliter was determined by multiplying the lowest dilution with visible growth by the dilution factor.
ATP measurement.
A method adapted from the method of Wayne and Hayes (45) was used for ATP measurement. At various time points during the RAD model experiments, glass tubes containing cultures were sacrificially opened inside an anaerobic chamber, and 1 ml of each culture was pelleted, frozen on dry ice, and stored at −80°C until it was processed. Frozen cell pellets were suspended in 100 μl 0.025 M HEPES buffer with 0.02% Tween 80 (pH 7.75) and added to glass tubes (12 by 75 mm) containing 40 μl chloroform. Each mixture was then heated at 80°C for 20 min. Samples were then diluted with 4.9 ml 0.025 M HEPES buffer (pH 7.75). One milliliter of a diluted sample was frozen at −80°C before it was used in a luciferase-based ATP assay. The ATP assay was performed as described by the manufacturer (Promega Enliten ATP assay system). Ten-microliter samples were added to a 96-well plate (Costar solid white), which loaded into an L-Max microplate luminometer (Molecular Devices). Enliten luciferase-luciferin reagent dissolved in sample buffer was injected immediately before the results were read. Sample luminescence was determined every 0.1 s for a total of 100 s. The data below are the averages of 3 independent experiments assayed in duplicate.
NAD and NADH measurement.
The NAD-NADH assay used was adapted from the assay of Leonardo et al. (22). The sample collection procedure was the sample collection procedure used for intracellular ATP measurement. Frozen cell pellets were suspended in 100 μl of acid (0.2 M HCl) or 100 μl of base (0.2 M NaOH) for examination of NAD or NADH, respectively, and heated at 80°C for 20 min. Samples were then neutralized with 100 μl of base (0.1 M NaOH) or 100 μl of acid (0.1 M HCl) for NAD and NADH, respectively, and snap-frozen on dry ice. The nucleotide cycling assay was adapted for a 96-well plate format. Wells were filled with 90 μl of sample and 100 μl of cycling cocktail. The master mixture for the cycling cocktail contained 2 ml 1 M bicine (pH 8.0), 2 ml 100% ethanol, 2 ml 40 mM EDTA (pH 8.0), 2 ml 4.2 mM thiazolyl blue tetrazolium bromide (MTT), and 4 ml 16.6 mM phenazine ethosulfate (PES). The mixture was incubated for 3 min at 30°C before addition to each well of 10 μl alcohol dehydrogenase (500 U/ml in 0.1 M bicine [pH 8.0]). Using a fluorimeter with plate reader capability (BioTEK Synergy HT) the absorbance at 550 nm was determined every minute for 10 min. Standards were included to determine concentrations. The data below are the averages of 3 independent experiments assayed in duplicate.
Relative oxygen tension measurement.
Cultures were set up as described above for the RAD model experiments, with addition of methylene blue (final concentration, 9 μg/ml). The optical density at 600 nm was measured every 2 h during the critical period between day 3 and day 6 after sealing. The data below are the average optical densities of 4 tubes from one representative experiment. For more precise measurement, a 2-mm O2 sensor and a TBR4100 free radical analyzer (World Precision Instruments) were used to monitor oxygen consumption. Briefly, RAD model tubes were opened sacrificially, and the O2 probe was inserted into the cultures for measurement of the dissolved oxygen. Measurements (in nA) were obtained at 0, 2, 3, 4, 5, and 6 days after tubes were sealed. To convert these measurements to percentages of oxygen, a standard curve was created using 37°C DTA medium (18% oxygen) and 37°C DTA medium containing the reagent oxyrase (0% oxygen). The data below are the averages of 3 experiments.
Recovery experiments.
After 10 and 40 days of dormancy, bacteria were exposed to oxygen and fresh medium (diluted 1:1) and allowed to resume growth. Bacterial growth was measured every day for 5 days by plating bacteria on DTA agar plates. Growth rates were expressed as the fold increases in the number of CFU compared with the number of bacteria present at day 10 or 40. The data below are the averages of 3 independent experiments.
Nitric oxide exposure.
Wild-type strain H37Rv, DorKO, and DorCO were grown in logarithmic phase for three consecutive days, after which they were diluted to obtain an OD600 of 0.1. Ten milliliters of each culture was added to a glass tube with a stir bar, and 100 μM diethylenetriamine-nitric oxide (DETA-NO) was added six times, once every 6 h. Recovery from exposure to DETA-NO was monitored by recording the OD600 after each addition of DETA-NO, as well as every 24 h after addition. Four biological replicates were used for each sample.
Microscopy.
Live-dead staining of bacilli was performed at various stages of dormancy. Cultures (1 ml) were resuspended in an equal volume of 0.025 M HEPES with 0.02% Tween 80 (pH 7.75) (HEPES was used instead of phosphate-buffered saline [PBS] to avoid 4′,6′-diamidino-2-phenylindole [DAPI] precipitation) and stained with fluorescein diacetate (final concentration, 50 μg/ml) and DAPI (final concentration, 300 nM) for 20 min at 37°C. Samples were then washed once in 0.025 M HEPES with 0.02% Tween 80 (pH 7.75) and resuspended in 1/10th the original volume to concentrate cells and obtain a density suitable for microscopy. Wet mounts were made by spotting 6.5 μl of a sample on poly-l-lysine coated slides and covering the bacteria with a coverslip. Microscopy was performed with a Nikon Eclipse TE200-U inverted microscope using a 100× phase objective with 1.5× zoom. Pictures were taken with a Nikon DS-Fil diagnostic instrument 1.0X camera and were analyzed using MetaVue software.
RESULTS
Rapid anaerobic dormancy model.
M. tuberculosis dormancy has been studied by using a variety of models, often based on a model described by Wayne and Hayes (45) and commonly referred to as the Wayne model. In these models, M. tuberculosis initially replicates until oxygen is consumed via respiration, producing an anaerobic environment in sealed test tubes. To achieve consistent and more precise results, we made minor modifications to the Wayne model. In the rapid anaerobic dormancy (RAD) model larger stir bars were used and stirring was more rapid. Also, grease was used to seal the threads of each test tube to ensure that oxygen did not slowly leak into the cultures. These changes ensured that there was even distribution of oxygen throughout the culture.
No loss of viability of wild-type M. tuberculosis was observed in the RAD model until after 40 days (Fig. 1A). The results show that there was increased survival compared with that in the original Wayne model, in which the M. tuberculosis half-life was approximately 11 days after onset of anaerobiosis (45), and contrast with the results of another approach recently reported for modeling bacilli during latent infection (31). In the defined hypoxia model, gas containing 0.2% oxygen was flushed into a culture to produce a hypoxic environment as an alternative to oxygen consumption by bacillus respiration (31). The defined hypoxia method allows only 1% of bacteria to survive after 1 week (31). The method described here allows bacteria to survive much longer.
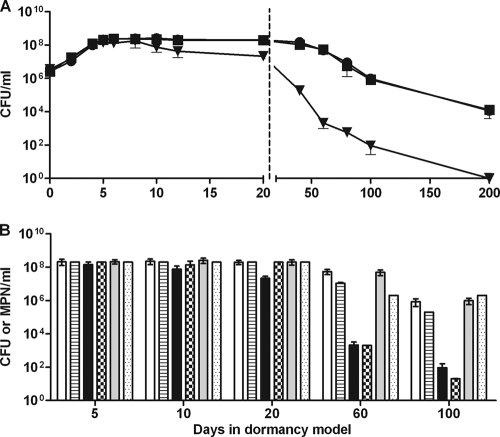
FIG. 1.
Growth and survival during anaerobic dormancy determined by recovery on solid and liquid media. Wild-type, DorKO mutant, and complemented bacteria were grown in glass tubes with stir bars and a culture-to-headspace ratio of 0.65. Cultures were stirred with magnetic stirrers. (A) CFU were counted at various times by plating samples from tubes harvested sacrificially. Squares, H37Rv; triangles, DorKO; circles, DorCO. (B) Viability assessed by serial dilution of dormant bacteria in DTA broth using the MPN technique and comparison to CFU values (39). Open bars, H37Rv on plates; bars with horizontal stripes, H37Rv in liquid medium; solid bars, DorKO on plates; checkered bars, DorKO in liquid medium; gray bars, DorCO on plates; stippled bars, DorCO in liquid medium. The values are the averages of three experiments.
DosR regulon is essential for anaerobic survival.
A mutant defective in DosR regulon induction was used to determine the role of this regulon under hypoxic and anaerobic conditions. The three-gene operon including dosR was deleted to create a DosR regulon mutant (DorKO) (3). The DorKO mutant was complemented by introduction of the entire three-gene operon, including 300 bases upstream of the structural gene, into the M. tuberculosis chromosome. Deletion of the operon as a unit allowed construction of a complemented strain (DorCO) that included all promoter elements (9).
The wild-type M. tuberculosis strain, DorKO, and DorCO initially grew at similar rates in the RAD model experiments, with doubling times of approximately 18 h (Fig. 1A). By days 7 we observed no additional growth of the wild type. The DorKO mutant bacilli, however, lost viability when they were plated on solid agar within days of becoming nonreplicating. By days 10 and 20 in the RAD model experiments, the DorKO mutant had lost 58% and 88% of its viability, respectively. The loss of mutant bacilli during early points in the model was dependent on plating on solid agar as the mutant bacilli were recovered in liquid media (see below). The recovery of wild-type bacilli remained fairly constant between days 20 and 40, and there was no loss of viability until 60 to 80 days, at which point we recovered 10% of the bacilli that were present at day 20 (Fig. 1A). After day 80, the viability of the wild type decreased gradually over time; 0.01% of the bacilli were still alive after 200 days. The DorKO mutant lost viability much faster. By day 40, we were able to recover only 0.1% of the mutant bacilli. At day 60, only 0.01% of the bacilli were recovered, at day 100 0.001% of the bacilli were recovered, and no bacilli were recovered at day 200 despite the fact that an entire culture was plated. These data demonstrate that there was a marked survival defect in a DosR regulon mutant during anaerobic dormancy.
DosR regulon mutant becomes nonculturable.
Shleeva et al. demonstrated that during dormancy M. tuberculosis becomes nonculturable on plates but not in liquid medium (39). We attempted to recover dormant M. tuberculosis in liquid media to determine if the decrease in viability observed for DorKO was a result of the inability of damaged cells to recover on solid media. Most probable numbers (MPN) were determined by recovery in liquid media on days 5, 10, 20, 60, and 100 in the RAD model and compared to the data for recovery on solid media (Fig. 1B). While DorKO lost viability when it was recovered on solid medium at days 5, 10, and 20, no loss of viability was observed when the recovery occurred in liquid. However, by days 60 and 100 the loss of viability observed for DorKO using CFU was reflected in the MPN data.
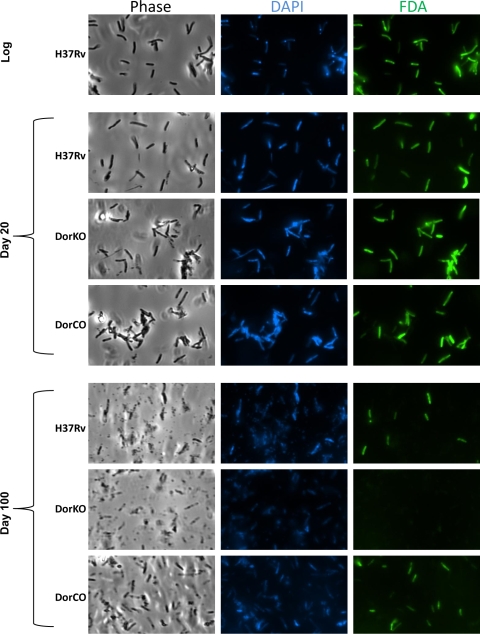
Fluorescein diacetate staining, which differentiates live mycobacteria from dead mycobacteria (18), was also used to assess bacillus viability in the RAD model. Fluorescein diacetate is not fluorescent in its native form but is modified in metabolically active cells by esterases to a fluorescent form that is visible at 490 nm (35). Figure 2 demonstrates that at day 20 in the RAD model nearly all of the cells of both the wild type and the DorKO mutant were viable even though CFU data indicated that nearly 90% of the mutant bacilli were dead. The fluorescein staining data corroborate the finding obtained by MPN analysis that mutant bacilli are not dead during early dormancy but are defective for recovery on solid medium. However, by day 100, virtually no viable DorKO bacilli were observed by microscopy, while large portions of the wild-type and complemented strain cells were intact and metabolically active. These data demonstrate that up to day 20 in the RAD model DosR regulon mutant bacilli are metabolically active but the majority of the mutant bacilli cannot recover on solid medium. However, the mutant bacilli ultimately die, as demonstrated by CFU/ml and MPN/ml data and by microscopy.
FIG. 2.
Live-dead staining during dormancy. Viability was assessed using live-dead staining in combination with the DNA stain DAPI. Cultures from log phase, day 20, and day 100 were washed in HEPES with Tween, stained with fluorescein diacetate (FDA) and DAPI for 20 min, and then washed and visualized as wet mounts. The images are representative of multiple captured images.
DosR regulon mechanisms spare oxygen consumption.
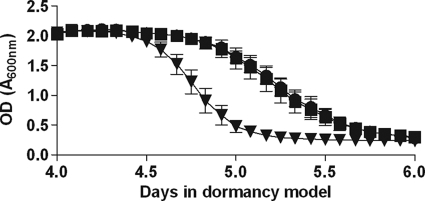
To determine the timing of oxygen depletion in the RAD model, we added methylene blue, an oxygen-sensitive dye, to cultures before they were sealed (Fig. 3; see Appendix SA in the supplemental material). In addition, we used a 2-mm oxygen probe to measure dissolved oxygen in the cultures (see Appendix SA in the supplemental material). In wild-type and DorCO cultures fading of methylene blue started after 4.5 days, and it started just before 4.5 days in DorKO cultures. The total transition time for the wild-type culture (the time between the onset of fading and the time when the culture became clear) was approximately 30 h, whereas the transition time for the DorKO culture was 16 h, about one-half that of the wild-type M. tuberculosis culture. We determined that fading of methylene blue began when the dissolved oxygen concentration fell below 3%. We observed that between days 4 and 6 the oxygen levels in the DorKO cultures were roughly 2- to 3-fold lower than those in wild-type cultures. The oxygen probe measurements were not analyzed to determine statistical significance due to the high degree of variability observed for biological replicates. Small variations in the low-density starting inoculum could result in a shift of several hours in the model. It should be noted, however, that a difference in the dissolved oxygen contents of the wild-type and DorKO cultures was apparent in all replicates. These findings demonstrate that the DosR regulon mutant bacilli consume oxygen more rapidly than the wild-type bacilli. Therefore, the DosR regulon appears to encode mechanisms that slow oxygen consumption or function in place of aerobic respiration.
FIG. 3.
Oxygen consumption (methylene blue decolorization) in cultures during adaptation to dormancy. Methylene blue (9 μg/ml) was added to dormancy model cultures before they were sealed. The optical density at 600 nm was measured every 2 h during the critical period between day 3 and day 6 after the start of the experiment. The values are the averages of four experiments. Squares, H37Rv; triangles, DorKO; circles, DorCO.
DosR regulon is required to sustain ATP levels.
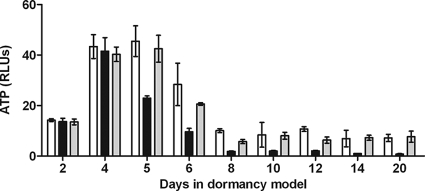
There were no detectable differences in ATP levels between the wild-type, DorKO, and DorCO strains during the first 4 days of the aerobic phase of the RAD model, and the total ATP levels increased as the cell numbers increased (Fig. 4). However, by day 5 the ATP levels in DorKO had fallen sharply (∼50%) compared to the day 4 levels, and they continued to drop steadily, reaching 1/10th of the wild-type level of ATP by day 14. The ATP levels in wild-type bacilli exhibited a more gradual decline and remained approximately 10-fold higher than the mutant levels after 20 days. A fraction of the difference between the absolute levels of ATP for the wild-type and mutant bacilli may have been because the wild-type cultures contained ∼25% (maximum) more bacilli per ml of culture and thus more ATP per ml of culture. However, 25% more bacilli in the wild-type and complemented cultures could not account for the 10-fold-higher ATP levels in the wild-type culture. Therefore, mechanisms encoded by the DosR regulon maintain ATP levels that are 1/4th the amount found in aerobic bacilli. The regulon mutant contains less than 1/40th the amount of ATP found in aerobic bacilli (Fig. 4).
FIG. 4.
ATP measurement during early dormancy. Samples from RAD model cultures were harvested at various time points in an anaerobic chamber. ATP was extracted using a chloroform heat-based method and frozen until measurements could be obtained using the Promega Enliten ATP assay system. The values are the averages of three experiments. Open bars, H37Rv; black bars, DorKO; gray bars, DorCO. RLUs, relative light units.
DosR regulon is required for redox balance.
We measured NAD and NADH levels in the RAD model cultures (Fig. 5A and B). As expected, during aerobic growth similar levels of NAD and NADH were detected in wild-type and DosR regulon mutant bacilli. However, by day 5 the NAD levels began to drop in mutant bacilli, and the NAD levels were nearly one-half those in wild-type bacilli. After this, the NAD levels in mutant bacilli continued to drop, and they were 10% of the levels in wild-type bacilli by day 20. The NADH levels in mutant bacilli were much closer to those in wild-type and complemented strains. When we examined the NAD/NADH ratio (Fig. 5C), we observed that there was a steady decline in this ratio for the DosR regulon mutant, while for the wild type the ratio initially declined but then rose again after a few days to approximately six times that for the mutant by day 20. These data indicate that in wild-type bacilli there is an early adjustment period before stable NAD levels are achieved. These data also demonstrated that the DosR regulon mutant was unable to maintain NAD levels, and thus redox balance, under anaerobic conditions.
FIG. 5.
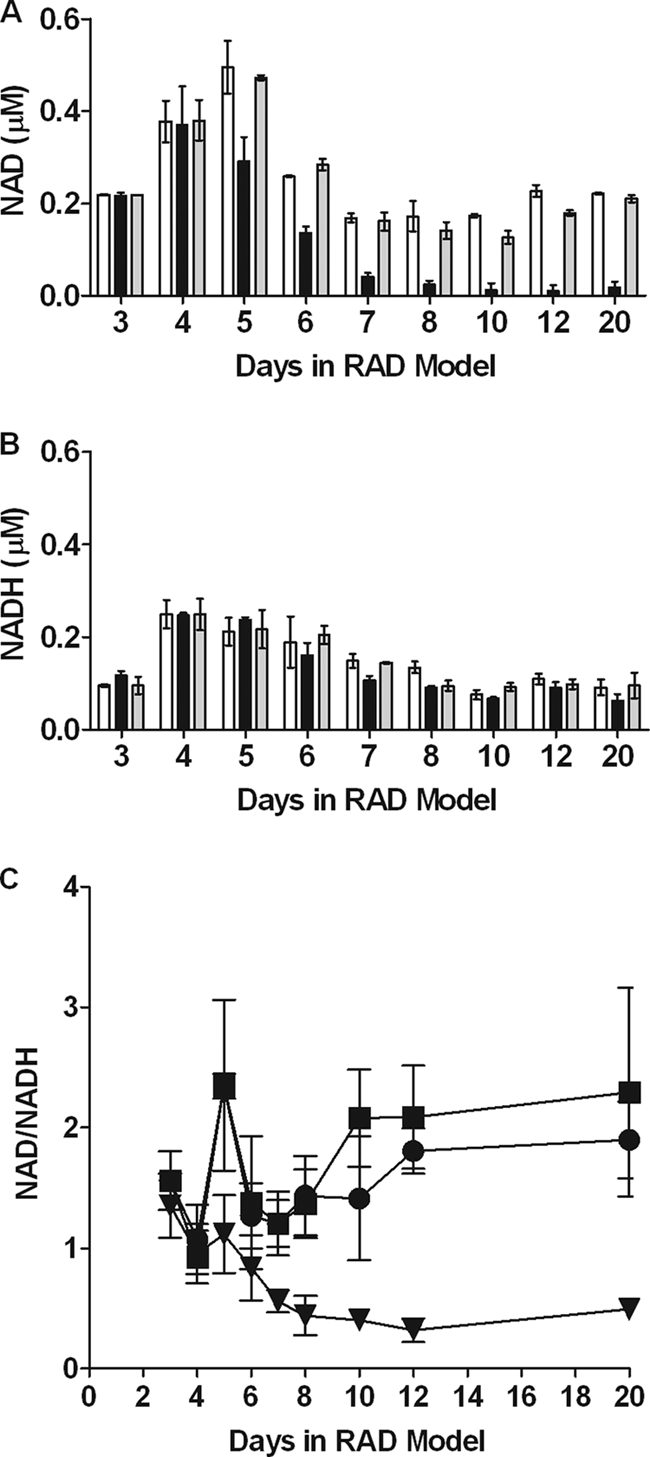
NAD and NADH levels and NAD/NADH ratio during early dormancy. Samples from dormancy model cultures were harvested at various time points in an anaerobic chamber. Nucleotides were extracted using either acid or base combined with heat and frozen prior to measurement using an alcohol dehydrogenase-based NAD-NADH cycling assay. The values are the averages of three experiments. (A and B) Levels of NAD (A) and NADH (B). Open bars, H37Rv; black bars, DorKO; gray bars, DorCO. (C) Ratio of NAD to NADH. Squares, H37Rv; triangles, DorKO; circles, DorCO.
Because the majority of DorKO mutant bacteria at day 20 were viable as determined by microscopy (Fig. 2) but could not be counted on plates (Fig. 1B), ATP, NAD, and NADH levels were expressed per ml of culture and not per CFU. Further, the amounts of NADH per ml of culture were comparable for the three strains (Fig. 5B), consistent with the similar numbers of viable bacilli in the wild-type, DorKO, and DorCO cultures demonstrated by MPN/ml data and microscopy. The total levels of nicotinamide cofactors decreased during anaerobiosis in the wild type and even more in the DosR mutant.
DosR regulon is required for rapid recovery.
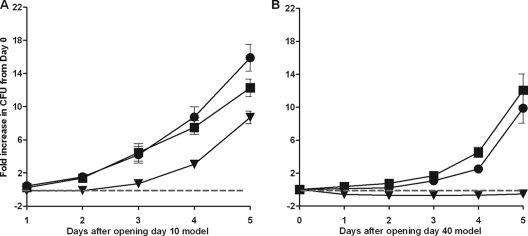
When the survival of wild-type and DosR regulon mutant bacilli was measured, the mutant took longer to recover on plates and required an extra week to produce colonies that could be counted (data not shown). Small or slowly growing colonies are not typical of this mutant. Slow colony growth occurred only after exposure to hypoxic or anaerobic conditions. These observations suggested that the regulon mutant was damaged and thus unable to recover properly. The ability to resume growth upon reintroduction of oxygen was determined at 10 and 40 days in the RAD model (Fig. 6). As early as day 10 the mutant exhibited a nearly 2-day lag, while the wild-type and complemented strains resumed growth almost immediately upon reintroduction of oxygen (Fig. 6A). After 40 days the strains that expressed the DosR regulon began to replicate within a day, although the maximal growth rate was not observed until day 3 (Fig. 6B). Not only did the mutant not resume growth for 5 days after the introduction of oxygen, but the introduction of oxygen resulted in a 50% decline in the viable count for the mutant within 3 days after the tubes were opened at day 40 (Fig. 6B). These data indicate that in a microenvironment that oscillates between respiring and nonrespiring conditions, a strain lacking the DosR regulon is at a severe disadvantage, as demonstrated in Appendix SB in the supplemental material.
FIG. 6.
Growth rate during recovery from dormancy. After 10 or 40 days of dormancy, the wild-type, DorKO, and DorCO strains were exposed to oxygen and fresh medium and allowed to resume growth. Bacterial growth was measured every day for 5 days by plating on DTA agar plates. The growth rate was calculated by determining the fold increase in the number of CFU compared with the number of bacteria present immediately after the cultures were opened at day 10 or 40. The values are the averages of three experiments. (A) Cultures opened at day 10. (B) Cultures opened at day 40. Squares, H37Rv; triangles, DorKO; circles, DorCO.
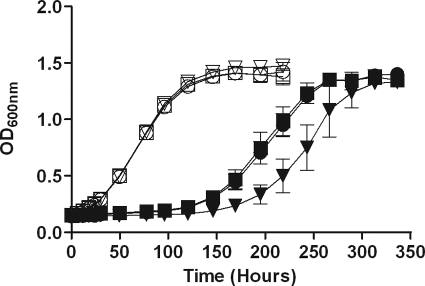
We also investigated whether the regulon mutant was defective in recovery from extended exposure to low levels of NO. We previously demonstrated that a low dose of NO blocks M. tuberculosis respiration and induces DosR regulon expression (43). Figure 7 shows the recovery of M. tuberculosis after exposure to NO delivered by administering 0.1 mM DETA-NO every 6 h. Recovery from the NO-induced nonrespiring state was slowed by nearly a day in the absence of DosR regulon expression. The lack of rapid recovery was not due to a significant loss of viable cells of the mutant after exposure to NO (see Appendix SC in the supplemental material). However, a slight loss of viability as measured by CFU was observed for the mutant after 2 days of exposure to NO, similar to the decrease in the number of CFU observed after a few days of anaerobiosis (Fig. 1A). Our previous study demonstrated that the DosR regulon was not necessary for survival after a high single dose of NO from the same donor (43). However, the current data indicate that the DosR regulon is important for recovery from a level of NO that blocks respiration over many hours, even though the regulon is not required for the acute stress response after exposure to a high level of NO. These results establish that the DosR regulon is important for adaptation to conditions that do not allow aerobic respiration, such as a lack of oxygen or NO inhibition of the respiratory chain.
FIG. 7.
DosR regulon and recovery from extended exposure to a low dose of NO. DETA-NO (100 μM) was added to aerobic growing H37Rv, DorKO, or DorCO cultures every 6 h over a 36-h period. Growth was followed by determining the OD600. Squares, strain H37Rv; triangles, DorKO; circles, DorCO; open symbols, control; filled symbols, NO added.
DISCUSSION
DosR regulon transcriptional control mechanisms have been intensively investigated in recent years as DosR-controlled genes are among the most highly expressed genes in M. tuberculosis when their expression is activated (43). However, little is known about the biological role of the encoded proteins. Here we demonstrated that deletion of DosR resulted in a 10,000-fold defect in anaerobic dormancy survival. We also report that there was eventual sterilization of the DosR regulon mutant culture. These results contrast with findings obtained with a defined hypoxia model in which the survival of a DosR regulon mutant was similar to that of the wild-type strain (31). It should be noted, however, that in the hypoxia model the viability of both the wild type and the mutant decreased 100-fold within 1 week (31), while in the model described here a loss of wild-type viability was not observed until after several weeks under anaerobic conditions. It is possible that in the hypoxia model a role for the DosR regulon is obscured by death of bacilli unrelated to the DosR regulon.
Expression of the DosR regulon first occurred under hypoxic conditions, before a measurable survival defect was observed. We suggest that induction during hypoxia is necessary to prepare the bacilli for eventual nonrespiring conditions caused by anaerobiosis, NO, or CO. Without respiration the bacilli have fewer metabolic options and, therefore, less energy to produce DosR regulon proteins. Induction of the regulon prior to inhibition of respiration allows the bacteria to transcribe and translate the DosR regulon before cellular energy diminishes and such processes become limited. Thus, M. tuberculosis preadapts for the ensuing nonrespiring state. Consistent with this, the DosR regulon is not induced indefinitely in the Wayne model of anaerobic dormancy (44). Regulon transcription gradually ceases along with most transcription in the bacilli within a few weeks after the onset of anaerobiosis. However, even though the genes are not expressed for months, their previous expression remains essential. The defect in the regulon mutant is most dramatic after 40 days compared to early stages of the model, when the genes are expressed. These data suggest that while the DosR regulon may be important for latent infection, continual expression of this regulon may not be necessary once the proteins have been made. Instead, expression may occur early to adapt to a nonrespiring state while energy is still available. The presence of DosR regulon protein during latent infection is confirmed by several in vivo studies showing that latently infected individuals exhibit strong immune responses to DosR regulon antigens (4, 23, 36).
To determine if the DosR regulon is directly involved in novel anaerobic metabolic pathways, we examined the ability of a regulon mutant to sustain markers of metabolic homeostasis. Not surprisingly, the ATP levels dropped in both the wild-type and mutant strains as soon as oxygen was depleted to a point where growth stopped. The ATP levels in wild-type bacilli stabilized at 25% of the aerobic level, while the ATP levels in the DosR regulon mutant continued to fall to 10% of the aerobic level. The lower ATP levels appear to be adequate to sustain the dormant bacilli in the nonreplicating state. As replication stops, less energy is likely required for cellular processes. During dormancy wholesale repression of global gene transcription is observed, indicating that there is downregulation of cellular activity (44).
The primary obstacle faced by obligate aerobes, such as M. tuberculosis, during anaerobiosis is the regeneration of NAD as the aerobic respiratory chain is blocked in the absence of oxygen as the terminal electron acceptor. To investigate whether DosR regulon mechanisms are involved in redox homeostasis, we measured the levels of NAD and NADH. Only small differences were observed in NADH levels between the wild-type and regulon mutant bacilli, which corresponded to the slightly lower density of bacteria in the mutant cultures. Like the ATP levels, the NADH levels fell, reaching levels that were about one-half the aerobic levels once oxygen was consumed and growth stopped. The decrease in the level of this nucleotide is likely due to lower metabolic activity; in the case of the DosR mutant the absence of DosR-controlled PncB2 also contributes to a defect in maintaining nicotinamide levels during dormancy (6). PncB2 is a nicotinate phosphoribosyltransferase that is important for cofactor salvage under dormancy conditions (6). NAD levels also dropped to about one-half the aerobic levels in the wild type and remained constant through day 20. In the DosR regulon mutant the NAD levels dropped rapidly to only 10% the wild-type level. Thus, the ratio of NAD to NADH was 6-fold less in the DosR regulon mutant, while the wild type maintained a ratio similar to that observed under aerobic conditions. Deletion of the primary redox balance regulator under anaerobic conditions in Escherichia coli (ArcAB) results in a <3-fold difference in the NAD-to-NADH ratio (1). As enzymes are highly sensitive to redox balance, a 6-fold change is a large effect that has major implications for cellular functions. Thus, the DosR regulon is central to maintaining metabolic homeostasis under anaerobic conditions. It appears that the regulon encodes mechanisms employed by M. tuberculosis to sustain anaerobic metabolism even though it does not encode many proteins used by bacteria under anaerobic conditions, such as the lactate dehydrogenases and alcohol dehydrogenases. It is unlikely that the DosR regulon encodes the primary metabolic pathways that are employed anaerobically as few of the regulon-encoded proteins are involved in central metabolic processes. Instead, it appears that regulon enzymes, such as triacylglyceride synthase (Tgs1), ferredoxin (FdxA), and phosphofructokinase B (PfkB), assist in adapting metabolic processes to anaerobic conditions.
Our results demonstrate that the regulon not only is necessary for long-term anaerobic survival but also is important for recovery from conditions that inhibit aerobic respiration for even short periods of time. Competition between the wild type and the DosR regulon mutant demonstrated that expression of the regulon conferred an advantage to bacilli cycling between aerobic and anaerobic conditions. We also found that the longer the period of anaerobiosis, the greater the defect in recovery of the regulon mutant. Thus, not only does the mutant not survive anaerobiosis well, but the bacilli that do survive are notably more fragile. M. tuberculosis in vivo is thought to experience hypoxia in the context of a granuloma (21) or fat cells (28). However, the environment of a granuloma has recently been shown to be quite dynamic (10, 13, 14). These findings suggested that M. tuberculosis may experience frequent fluctuations in oxygen availability. In such an environment, M. tuberculosis must have the capacity to quickly alter its metabolism between respiring and nonrespiring pathways which are at least partially encoded by the DosR regulon.
An anaerobic environment is just one environment that M. tuberculosis likely encounters during infection that can effectively prevent aerobic respiration. The two other factors known to induce expression of the regulon, NO (29, 43) and CO (19, 38), are also potent inhibitors of aerobic respiration via inhibition of cytochrome c oxidase (7, 12). We previously demonstrated that the very low levels of NO required to induce the DosR regulon were also sufficient to inhibit M. tuberculosis respiration and also that NO and low oxygen levels work synergistically to block respiration and induce the regulon (43). Here we demonstrate that the DosR regulon is necessary for rapid recovery from nonrespiring states resulting from exposure to either low levels of NO or anaerobiosis. Thus, expression of the DosR regulon responds to stimuli that block aerobic respiration, and, in turn, this regulon encodes mechanisms that allow adaptation to the stimuli.
Over the past several years many studies have investigated the patterns of expression and the regulatory aspects of the DosRST system. Currently, little is known about the mechanisms encoded by the highly expressed DosR regulon. Here we confirm that the DosR regulon is essential for anaerobic survival of M. tuberculosis. Without induction of this regulon, M. tuberculosis cannot maintain energy levels or sustain the necessary redox balance that is essential for metabolic processes. Thus, if during latent infection bacilli are periodically in a nonrespiring environment, the DosR regulon is essential for survival. We also demonstrate that this regulon provides a significant competitive advantage to bacilli that are transitioning between respiring and nonrespiring microenvironments. The ability to transition back to a respiring state may be important during reactivation after a latent infection, but it may also be important during active infection, especially in microenvironments where oxygen may be limited and NO and CO are produced intermittently.
Supplementary Material
Acknowledgments
This research project was funded by NIH grant RO1 AI061505 awarded to M.I.V. and by NIH grants T32 AI052066-05 and T32 AI052066-07 awarded to R.L.L. and I.L.B., respectively. Biomedical Research Internship Program for Undergraduates grant T35 HL007961-10 supported K.W.
We thank Ryan W. Honaker for assistance with the manuscript.
Footnotes
Published ahead of print on 18 December 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aly, S., K. Wagner, C. Keller, S. Malm, A. Malzan, S. Brandau, F. C. Bange, and S. Ehlers. 2006. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210:298-305. [DOI] [PubMed] [Google Scholar]
- 3.Bartek, I. L., R. Rutherford, V. Gruppo, R. A. Morton, R. P. Morris, M. R. Klein, K. C. Visconti, G. J. Ryan, G. K. Schoolnik, A. Lenaerts, and M. I. Voskuil. 2009. The DosR regulon of Mycobacterium tuberculosis and antibacterial tolerance. Tuberculosis (Edinb.) 89:310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, G. F., B. A. Thiel, M. Ota, S. K. Parida, R. Adegbola, W. H. Boom, H. M. Dockrell, K. L. Franken, A. H. Friggen, P. C. Hill, M. R. Klein, M. K. Lalor, H. Mayanja, G. Schoolnik, K. Stanley, K. Weldingh, S. H. Kaufmann, G. Walzl, and T. H. Ottenhoff. 2009. Immunogenicity of novel DosR regulon-encoded candidate antigens of Mycobacterium tuberculosis in three high-burden populations in Africa. Clin. Vaccine Immunol. 16:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boon, C., and T. Dick. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., X. Xu, K. Tahlan, C. S. Dowd, K. Pethe, L. R. Camacho, T. H. Park, C. S. Yun, D. Schnappinger, S. Ehrt, K. J. Williams, and C. E. Barry III. 2008. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J. Biol. Chem. 283:19329-19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, G. C. 2001. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta 1504:46-57. [DOI] [PubMed] [Google Scholar]
- 8.CDC. 1 June 2009, posting date. A global perspective on tuberculosis. http://www.cdc.gov/tb/events/WorldTBDay/resources_global.htm.
- 9.Chauhan, S., and J. S. Tyagi. 2008. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190:4301-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly, L. E., P. H. Edelstein, and L. Ramakrishnan. 2007. Why is long-term therapy required to cure tuberculosis? PLoS Med. 4:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Converse, P. J., P. C. Karakousis, L. G. Klinkenberg, A. K. Kesavan, L. H. Ly, S. S. Allen, J. H. Grosset, S. K. Jain, G. Lamichhane, Y. C. Manabe, D. N. McMurray, E. L. Nuermberger, and W. R. Bishai. 2009. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 77:1230-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, C. E., and G. C. Brown. 2008. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J. Bioenerg. Biomembr. 40:533-539. [DOI] [PubMed] [Google Scholar]
- 13.Cosma, C. L., O. Humbert, D. R. Sherman, and L. Ramakrishnan. 2008. Trafficking of superinfecting mycobacterium organisms into established granulomas occurs in mammals and is independent of the Erp and ESX-1 mycobacterial virulence loci. J. Infect. Dis. [DOI] [PMC free article] [PubMed]
- 14.Davis, J. M., and L. Ramakrishnan. 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136:37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garton, N. J., S. J. Waddell, A. L. Sherratt, S. M. Lee, R. J. Smith, C. Senner, J. Hinds, K. Rajakumar, R. A. Adegbola, G. S. Besra, P. D. Butcher, and M. R. Barer. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill, W. P., N. S. Harik, M. R. Whiddon, R. P. Liao, J. E. Mittler, and D. R. Sherman. 2009. A replication clock for Mycobacterium tuberculosis. Nat. Med. 15:211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Pando, R., M. Jeyanathan, G. Mengistu, D. Aguilar, H. Orozco, M. Harboe, G. A. Rook, and G. Bjune. 2000. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet 356:2133-2138. [DOI] [PubMed] [Google Scholar]
- 18.Jarnagin, J. L., and D. W. Luchsinger. 1980. The use of fluorescein diacetate and ethidium bromide as a stain for evaluating viability of mycobacteria. Stain Technol. 55:253-258. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, A., J. S. Deshane, D. K. Crossman, S. Bolisetty, B. S. Yan, I. Kramnik, A. Agarwal, and A. J. Steyn. 2008. Heme oxygenase-1 derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. [DOI] [PMC free article] [PubMed]
- 20.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U. S. A. 104:11568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenaerts, A. J., D. Hoff, S. Aly, S. Ehlers, K. Andries, L. Cantarero, I. M. Orme, and R. J. Basaraba. 2007. Location of persisting mycobacteria in a guinea pig model of tuberculosis revealed by r207910. Antimicrob. Agents Chemother. 51:3338-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leonardo, M. R., Y. Dailly, and D. P. Clark. 1996. Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 178:6013-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyten, E. M., M. Y. Lin, K. L. Franken, A. H. Friggen, C. Prins, K. E. van Meijgaarden, M. I. Voskuil, K. Weldingh, P. Andersen, G. K. Schoolnik, S. M. Arend, T. H. Ottenhoff, and M. R. Klein. 2006. Human T-cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis. Microbes Infect. 8:2052-2060. [DOI] [PubMed] [Google Scholar]
- 24.Lillebaek, T., A. Dirksen, I. Baess, B. Strunge, V. O. Thomsen, and A. B. Andersen. 2002. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J. Infect. Dis. 185:401-404. [DOI] [PubMed] [Google Scholar]
- 25.Lillebaek, T., A. Dirksen, E. Vynnycky, I. Baess, V. O. Thomsen, and A. B. Andersen. 2003. Stability of DNA patterns and evidence of Mycobacterium tuberculosis reactivation occurring decades after the initial infection. J. Infect. Dis. 188:1032-1039. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra, V., D. Sharma, V. D. Ramanathan, H. Shakila, D. K. Saini, S. Chakravorty, T. K. Das, Q. Li, R. F. Silver, P. R. Narayanan, and J. S. Tyagi. 2004. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 231:237-245. [DOI] [PubMed] [Google Scholar]
- 27.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neyrolles, O., R. Hernandez-Pando, F. Pietri-Rouxel, P. Fornes, L. Tailleux, J. A. Barrios Payan, E. Pivert, Y. Bordat, D. Aguilar, M. C. Prevost, C. Petit, and B. Gicquel. 2006. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS One 1:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohno, H., G. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5:637-648. [DOI] [PubMed] [Google Scholar]
- 30.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rustad, T. R., M. I. Harrell, R. Liao, and D. R. Sherman. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustad, T. R., A. M. Sherrid, K. J. Minch, and D. R. Sherman. 2009. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell. Microbiol. 11:1151-1159. [DOI] [PubMed] [Google Scholar]
- 33.Ryter, S. W., J. Alam, and A. M. Choi. 2006. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 86:583-650. [DOI] [PubMed] [Google Scholar]
- 34.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnurer, J., and T. Rosswall. 1982. Fluorescein diacetate hydrolysis as a measure of total microbial activity in soil and litter. Appl. Environ. Microbiol. 43:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck, S. D., H. Mueller, F. Kunitz, A. Neher, H. Hoffmann, K. L. Franken, D. Repsilber, T. H. Ottenhoff, S. H. Kaufmann, and M. Jacobsen. 2009. Identification of T-cell antigens specific for latent Mycobacterium tuberculosis infection. PLoS One 4:e5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma, D., A. Bose, H. Shakila, T. K. Das, J. S. Tyagi, and V. D. Ramanathan. 2006. Expression of mycobacterial cell division protein, FtsZ, and dormancy proteins, DevR and Acr, within lung granulomas throughout guinea pig infection. FEMS Immunol. Med. Microbiol. 48:329-336. [DOI] [PubMed] [Google Scholar]
- 38.Shiloh, M. U., P. Manzanillo, and J. S. Cox. 2008. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shleeva, M. O., G. V. Mukamolova, M. V. Telkov, T. L. Berezinskaia, A. V. Syroeshkin, S. F. Biketov, and A. S. Kaprel'iants. 2003. Formation of nonculturable Mycobacterium tuberculosis and their regeneration. Mikrobiologiia 72:76-83. (In Russian.) [PubMed] [Google Scholar]
- 40.Tsai, M. C., S. Chakravarty, G. Zhu, J. Xu, K. Tanaka, C. Koch, J. Tufariello, J. Flynn, and J. Chan. 2006. Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell. Microbiol. 8:218-232. [DOI] [PubMed] [Google Scholar]
- 41.Via, L. E., P. L. Lin, S. M. Ray, J. Carrillo, S. S. Allen, S. Y. Eum, K. Taylor, E. Klein, U. Manjunatha, J. Gonzales, E. G. Lee, S. K. Park, J. A. Raleigh, S. N. Cho, D. N. McMurray, J. L. Flynn, and C. E. Barry III. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voskuil, M. I., R. W. Honaker, and A. J. C. Steyn. 2009. Oxygen, nitric oxide, and carbon monoxide signaling, p. 119-147. In T. Parish and A. Brown (ed.), Mycobacterium: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 43.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 84:218-227. [DOI] [PubMed] [Google Scholar]
- 45.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.