Abstract
Secondary transporters are workhorses of cellular membranes, catalyzing the movement of small molecules and ions across the bilayer, coupling substrate passage to ion gradients. However, the conformational changes that accompany substrate transport, the mechanism by which substrate moves through the transporter, and principles of competitive inhibition remain unclear. Here we use crystallographic and functional studies on LeuT, a model for neurotransmitter sodium symporters, to show that various amino acid substrates induce the same occluded conformational state, and that a competitive inhibitor, tryptophan, traps LeuT in an open-to-out conformation. In the Trp complex the extracellular gate residues, Arg30 and Asp404, define a second weak binding site for substrates as they permeate from extracellular solution to the primary substrate site, demonstrating how residues that participate in gating also mediate substrate permeation.
Secondary active transporters are ubiquitous integral membrane proteins that couple the potential energy stored in pre-existing ion gradients to the concentrative uptake of polar and charged molecules across the lipid bilayer (1-3). Members of the solute carrier 6 (SLC6) family of sodium-coupled transporters, also known as neurotransmitter sodium symporters (NSS), comprise one of the most widely investigated and pharmacologically important classes (4, 5). SLC6 proteins play a central role in diverse physiological processes, ranging from the maintenance of cellular osmotic pressure (6) to the reuptake of small molecule neurotransmitters in the brain (7). SLC6 dysfunction is implicated in numerous debilitating illnesses such as depression (8), obsessive-compulsive disorder (9), epilepsy (10), autism (11), orthostatic intolerance (12), X-linked creatine deficiency syndrome (13), and retinal degeneration (14). Importantly, the transport activity of these molecular machines can be inhibited by many different compounds, including tricyclic antidepressants (TCAs) (15), selective-serotonergic reuptake inhibitors (SSRIs) (15), anticonvulsants (16) and cocaine (17).
Unraveling the molecular principles that define a substrate, a molecule that can be transported, versus a competitive inhibitor, a molecule that can displace the substrate but is not itself transported, is intimately linked to the larger goal of elucidating transport mechanism and ultimately to the development of new therapeutic agents. LeuT, a prokaryotic SLC6 member (18), affords an opportunity to couple functional and structural data to uncover the molecular mechanisms of transport and inhibition. Recently, a model for noncompetitive inhibition was proposed using a combination of steady-state kinetics (19), binding, and crystallographic studies with LeuT and three TCAs (19, 20). The structures of LeuT bound to the TCA clomipramine (19), imipramine (19), or desipramine (19, 20) revealed that each of these drugs binds to LeuT in the extracellular vestibule, about 11 Å above the substrate and directly above the extracellular gating residues, R30 and D404 (19, 20), stabilizing the occluded state in a closed conformation. Zhou et al. have proposed that the TCA binding site observed in LeuT is equivalent to the TCA site in SERT and the norepinephrine transporter (NET), the therapeutic targets in humans. However, in SERT and NET TCAs are competitive inhibitors (21-23) and their binding site likely overlaps with the substrate binding site (24). Therefore, we suggest that the LeuT-TCA complexes do not provide a model for competitive inhibition of eukaryotic SLC6 transporters.
Here we show that LeuT is capable of transporting many hydrophobic amino acids and that a fundamental requirement for a molecule to be a substrate is that it must fit within the occluded substrate-binding cavity. Molecules such as tryptophan which can bind but are too large to be accommodated within the occluded state cavity are not substrates but instead are competitive, non-transportable inhibitors. Structural analysis of the LeuT-Trp complex reveals that tryptophan traps LeuT in an open-to-out conformation and unveils the movements that accompany transition from the occluded-to an open-to-out state. Molecular insights gleaned from our studies are especially relevant to transporter mechanism because many other transporter families, including SLC5 (25), have the same fold as LeuT and likely share mechanistic principles.
Substrate Screen of LeuT
To identify a competitive inhibitor of LeuT, we examined the ability of a spectrum of amino acids to displace [3H]leucine binding from purified, detergent-solubilized LeuT and inhibit [3H]leucine transport by LeuT reconstituted into lipid vesicles (Fig. 1A). We found multiple aliphatic and aromatic amino acids of varying size inhibited [3H]leucine binding and transport. We chose glycine, alanine, leucine, methionine, tyrosine, and tryptophan (Table 1) for further functional analysis. Competition binding of [3H]leucine with unlabeled amino acids (Fig. 1B, Table 1) revealed that after leucine, methionine binds the most tightly followed by alanine, tyrosine, tryptophan, and glycine. A similar trend of affinities for LeuT was observed in direct radioligand binding experiments with leucine (fig S1A), alanine (fig S1B), and methionine (fig S1C; Table 1).
Fig. 1.
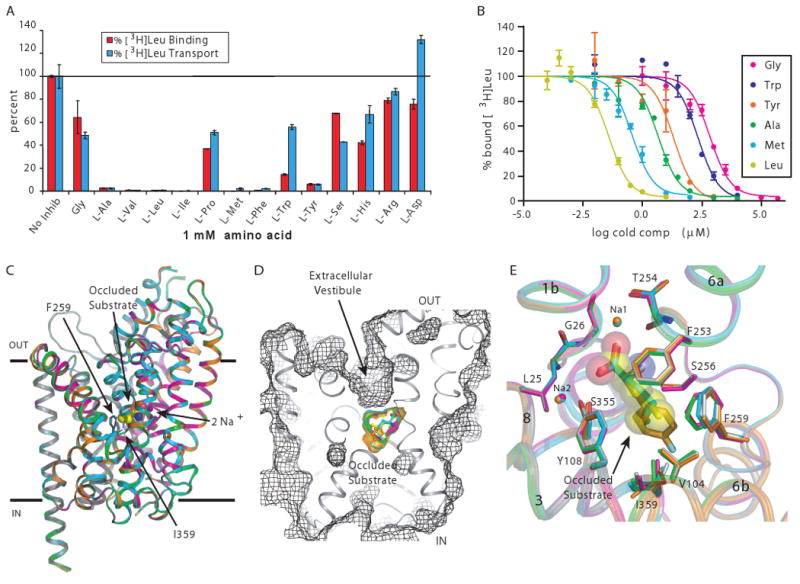
LeuT substrate screen and occluded state structures. (A) Inhibition of [3H]leucine binding (red bars) and transport (cyan bars) by L-amino acids. (B) Displacement of [3H]leucine binding by leucine (yellow), methionine (cyan), alanine (green), tyrosine (orange), tryptophan (blue), and glycine (magenta). Errors bars represent SEM of triplicate (A) or duplicate (B) measurements. (C) Superposition of the LeuT-Leu (gray), -Ala (green), -Gly (magenta), -Met (cyan), and L-4-F-Phe (orange) complexes using α-carbon positions. Shown in CPK are leucine and the two Na+ ions from the LeuT-Leu complex. Membrane boundaries are demarcated by the two solid black lines. (D) Solvent-accessible surface (depicted in mesh) illustrating the occluded state of the LeuT-substrate complexes. (E) Close-up of the substrate binding pocket, with substrates depicted as sticks. Leucine is shown in semi-transparent CPK representation. Coloring is the same as in (C).
Table 1.
Binding and Kinetic Constantsa
| Binding & Displacement Constants | ||
|---|---|---|
| L-Amino Acid (Vol in Å3)b | Kd (nM) | Ki (nM)c |
| Glycine (65) | n.d.d | 322,000 ± 36,900 |
| Alanine (90) | 512 ± 131 | 3320 ± 810 |
| Leucine (164) | 20 ± 2 | 16 ± 1 |
| Methionine (167) | 69 ± 8 | 232 ± 21 |
| Tyrosine (197) | n.d.d | 9040 ± 550 |
| L-4-F-Phenylalanine | n.d.d | 950 ± 100 |
| Tryptophan (228) | n.d.d | 64800 ± 4670 |
| Steady-State Kinetics | ||||
|---|---|---|---|---|
| L-Amino Acid (Vol in Å3)b | Km (nM) | Vmax (pmol/min/mg) | kcat (hr−1) | kcat/Km (nM−1hr−1) |
| Glycine (65) | 1910 ± 30 | 444 ± 57 | 1.58 ± 0.20 | 0.0008 |
| Alanine (90) | 583 ± 28 | 1730 ± 94 | 6.06 ± 0.30 | 0.0104 |
| Leucine (164) | 146 ± 25 | 343 ± 46 | 1.20 ± 0.20 | 0.0082 |
| Methionine (167) | 289 ± 27 | 523 ± 12 | 1.86 ± 0.04 | 0.0064 |
| Tyrosine (197) | 2830 ± 150 | 209 ± 15 | 0.74 ± 0.05 | 0.0003 |
| Tryptophan (228) | not transported | |||
| Competitive Inhibition of L-alanine Transport by L-tryptophan | |||
|---|---|---|---|
| [Trp] μM | Km (nM) | Vmax (pmol/min/mg) | Ki(μM)e |
| 0 | 665 ± 78 | 1530 ± 50 | |
| 20 | 1020 ± 230 | 1550 ± 110 | 24 ± 3 |
| 50 | 1880 ± 330 | 1610 ± 110 | |
Unless otherwise noted, the errors represent the standard error of the mean (SEM) from two or three independent experiments, each performed in duplicate or triplicate.
Volume in Å3 of amino acid as defined in (36).
K i refers to inhibition of [3H]leucine binding.
n.d., not determined.
K i refers to inhibition of [3H]alanine transport.
For a compound to be a competitive inhibitor, it must not only displace the substrate but cannot itself be transported. We previously demonstrated (18, 19) and replicated here that leucine and alanine are substrates (fig. S2, A and B). Compared to leucine, alanine is transported with a 5-fold higher turnover rate (kcat) and a 27% higher catalytic efficiency (kcat/Km; fig. S3, A and B, Table 1). We show that LeuT also catalyzes the uptake of methionine, tyrosine and glycine (fig. S2, C to E) with catalytic efficiencies roughly correlated to the inverse of substrate volume (fig. S3, C to E, Table 1).
Tryptophan is not a substrate of LeuT (fig. S4A) but rather is an inhibitor. To determine the kinetic mechanism of tryptophan inhibition, we performed steady-state kinetic experiments. With [3H]alanine as the substrate, increasing concentrations of tryptophan increased the Michaelis constant (Km) of LeuT for [3H]alanine without changing the maximum velocity (Vmax) (fig. S4b and Table 1). The corresponding Eadie-Hofstee plot (fig. S4C) exhibited nonparallel lines intersecting on the y-axis, hallmarks of competitive inhibition.
LeuT-Substrate Crystal Structures Reveal an Occluded State
To probe the atomic basis of ligand specificity, we cocrystallized LeuT with each of the six amino acids, measured x-ray diffraction data to high resolution, and solved the structures by molecular replacement. The readily-soluble, isosteric tyrosine analog, L-4-fluorophenylalanine (4-F-Phe), was used in place of tyrosine because tyrosine's low solubility limit precluded successful cocrystallization. All cocrystals diffracted to 1.8–2.3 Å resolution (table S1) and the resulting structures refined well (table S2).
The structures of LeuT in complex with each of the substrates (glycine, alanine, leucine, methionine, 4-F-Phe) are similar, with overall Cα RMSDs ranging from 0.2 to 0.3 Å (Fig. 1C) despite the 132 Å3 variation in substrate volume. All five structures adopt the same outward facing occluded state as originally seen in the LeuT-Leu complex in which access to the substrate binding pocket from both the extracellular and cytoplasmic sides of the membrane is obstructed, with access from the extracellular side being blocked by just a few residues and access from the intracellular side being blocked by ∼25 Å of tightly packed protein (18, 19) (Fig. 1, D and E). Simulated-annealing Fo-Fc omit maps (fig. S5, A-D) confirm the position of the substrates in the occluded binding pocket located in the center of the transporter, halfway across the lipid bilayer. Residues F253 and Y108 reside on “top” of the substrate, with electrostatic interactions formed by D404 and R30 layered directly above F253 and Y108. The hydroxyl of Y108 retains its two critical hydrogen bonds: one with the substrate carboxylate that helps anchor the substrate in place and the second with the amide nitrogen of L25 that stabilizes the unwound region of TM1 and bridges TM1 with TM3 (Figs. 1E and 2, A to E).
Fig. 2.
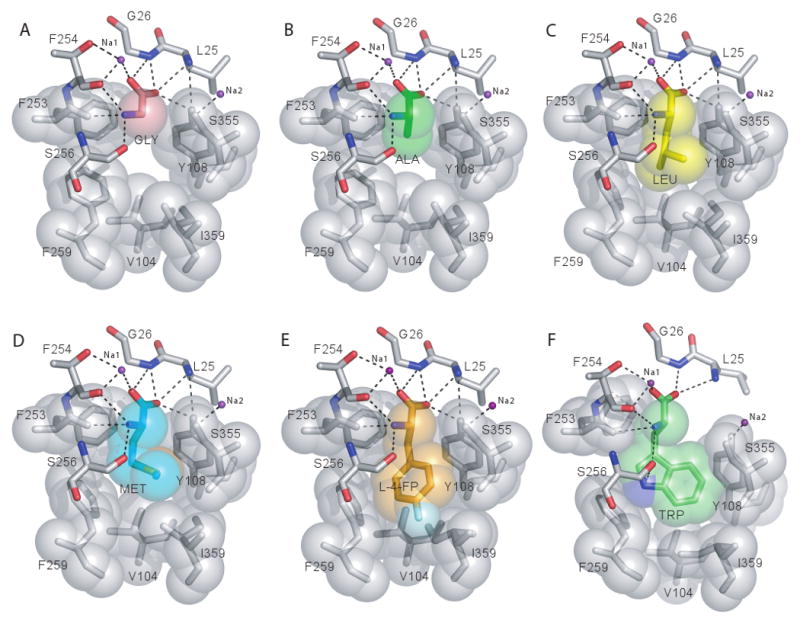
Substrate binding pocket - substrate and inhibitor interactions. Substrate binding pockets of the (A) LeuT-Gly, (B) -Ala, (C) -Leu,(D) -Met, (E) -L-4-F-Phe, and (F) -Trp complexes. Hydrogen bonds and polar interactions are illustrated by black dotted lines.
Despite overall congruence among the structures, there are differences in comparison to the Leu complex, localized primarily to F259 and I359. For the glycine and alanine complexes, the “R” groups of these ligands induce a ∼30° (χ1) torsion of F259's phenyl ring and ∼15° torsion of I359's sec-butyl moiety into the substrate binding cavity, compensating for the poor fit of the substrate to the binding pocket (Figs. 1E, 2A-B) and consistent with the weak affinity of LeuT for these two amino acids. The LeuT-Met and –Leu complexes superimpose within experimental error (Figs. 1E, 2, C and D), a finding in accord with their similar binding and transport parameters. Substituents larger than those of leucine or methionine begin to sterically hinder binding and formation of the occluded state. The structure of the 4-F-Phe complex exhibits the most pronounced differences: a ∼180° rotation of I359's sec-butyl group as well as a ∼0.5 Å outward shift in the backbone of the unwound region in TM6 (G258-A261) (Figs. 1E and 2E). The displacement of the backbone is suggestive of a strained occluded state, perhaps one that is less likely to isomerize to the open-to-in state. While speculative, this hypothesis is consistent with tyrosine's reduced turnover rate.
LeuT-Trp Complex Adopts an Open-to-Out Conformation
What are the structural principles by which tryptophan acts as a competitive inhibitor of LeuT? The LeuT-Trp complex exhibits an open-to-out conformation characterized by a widening of the extracellular vestibule and solvent accessibility to the substrate binding pocket (Fig. 3, A and B). Relative to the occluded state, in the LeuT-Trp complex there is a 9° outward rotation of a structural element comprised of TMs 1b (residues 23-38), 2a (40-54), and 6a (241-257) about an axis oriented nearly parallel to the membrane and located near the unwound regions of TMs 1 and 6 (Fig. 3C, movie M1). To accommodate movement of these helices, the highly conserved glycine-rich loop between TMs 1b and 2a slides under EL3 (233-240), the latter of which also undergoes a concerted translation along its helical axis, approximately parallel to the membrane. The EL4a helix (307-318) also undergoes an outward rotation of nearly 13° about an axis running approximately perpendicular to the plane formed by the EL4 loop (Fig. 3C, movie M1). Together, these movements widen the extracellular vestibule at the base by 3 Å, as defined by measurements between residues Y108 and F253 (Fig. 3, A, B, and E).
Fig. 3.
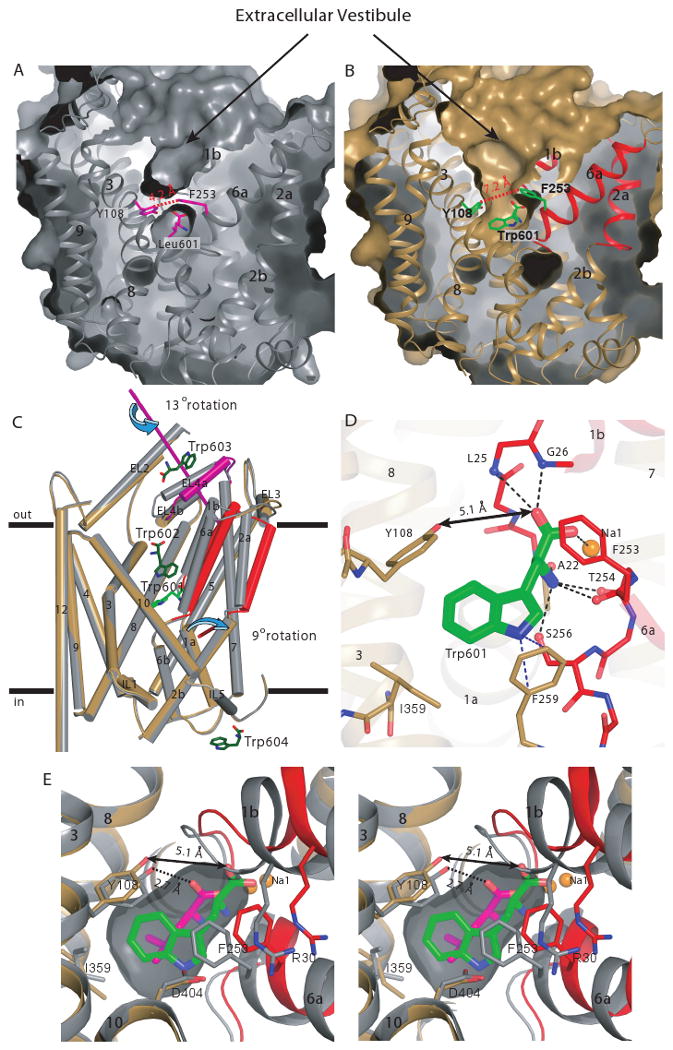
Tryptophan is a competitive inhibitor that stabilizes an open-to-outside conformation. Solvent-accessible surface of the (A) LeuT-Leu (gray) and (B) LeuT-Trp complexes (sand/red/magenta). Leucine, tryptophan, Y108, and F253 are depicted in both panels. Distances between Y108 (Cδ1) and F253 (Cξ) in each panel are shown. Helices involved in the domain shift (TM1b, 2a, and 6b) are colored red. (C) Cα superposition (depicted as cylinders) of the LeuT- Leu and LeuT–Trp complexes. Colors are the same as in (A) and (B). EL4a, an additional element involved in the domain shift, is magenta. The rotation axes of the two domains are depicted in their respective colors. The bound tryptophans are shown as stick models, with Trp601 colored bright green and the other three colored dark green. TM11 is omitted from the figure for clarity. (D) Close-up of the Cα superposition depicting the hydrogen bonding network in the substrate binding pocket of the LeuT-Trp complex. Note disruption of the critical hydrogen bond between Y108 and the carboxylate of tryptophan, indicated by a double-headed arrow. (E) Overlay (in stereoview) of the leucine and tryptophan binding sites to illustrate displacement of the ligand α-amino carboxylate group and concomitant shift in protein and sodium positions. Leucine and tryptophan are colored in magenta and green, respectively.
TM11 also undergoes a substantial displacement in the LeuT-Trp complex. Concomitant with the outward rotation of neighboring TM6a, TM11 shifts inward by ∼2 Å in the region around W467, while the indole side-chain of W467 on the interior face of the helix rotates 90° about χ2 relative to that in the occluded state structures. The space vacated by rotation of this indole ring and the outward movement of TM6a is partly occupied by the alkyl chain of a β-octylglucoside molecule (fig. S6).
Like substrates, the amino group of Trp601 forms hydrogen bonds with oxygen atoms in TMs 1b and 6a (Figs. 2F and 3D). Similar coordination exists between the carboxylate of Trp601 and the backbone amide nitrogens of TM1b (L25 and G26) as well as Na1 (Figs. 2F and 3D); Na2 is also present in the Trp complex and is coordinated in a similar manner as the occluded state complexes. The indole ring is accommodated in the binding pocket and, with only minor adjustments to the sidechain geometry of I359 and a slight rotation of Y108, engages in nonpolar interactions with these residues (Fig. 3E). Furthermore, the indole ring nitrogen of Trp601 is within ∼3.2 Å of the phenyl ring face of F259, forming an edge-to-face aromatic interaction (Figs. 2F and 3D).
Unlike the substrate-bound complexes, the LeuT-Trp structure reveals significant differences that define the molecular basis of competitive inhibition. The α-amino and α-carboxylate substituents of Trp601 are shifted by ∼2 Å relative to the corresponding positions of these atoms in the substrate-bound, occluded-state complexes (Fig. 3E). Trp601's rigid indole ring acts like a strut that braces the binding pocket open, with the α-amino and carboxylate groups maintaining their conserved interactions with TMs 1 and 6 (Fig. 3D) and the distal edge of the indole ring lodged against TMs 3 and 8. This mode of binding prevents the extracellular vestibule from closing and adopting the occluded state (Fig. 3, A and B). Specifically, the hydrogen bond observed in the occluded state between the substrate and Y108 hydroxyl does not form with Trp601 (Figs. 2F, 3, D-E), and the extracellular gate residue F253 on TM6a is ∼3.0 Å farther away from Y108, resulting in a solvent-accessible channel to the binding pocket (Fig. 3B).
Second Ligand Binding Site Is Unique to the Open-to-Out Conformation
Electron density maps revealed unanticipated density for a second Trp molecule (Trp602) at the base of the extracellular vestibule. Trp602 contacts TM10 and its α-substituents form an ionic bridge between residues D404 and R30 of the extracellular gate (Fig. 4A). The amino group also makes a hydrogen bond with the side chain hydroxyl of T409, while the indole nitrogen hydrogen bonds with the carbonyl oxygen of G408, located in a stretch of π-helix between M403 and V412 (Fig. 4A).
Fig. 4.
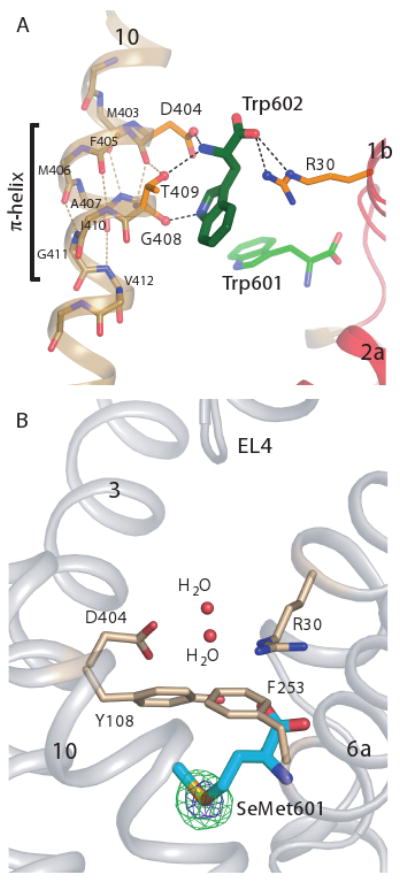
A second Trp molecule is bound between R30 and D404 of the extracellular gate only in the open-to-outside conformation. (A) Trp602 bound in the extracellular vestibule of LeuT, residing between D404 and R30, flanked by the π-helix in TM10. (B) Extracellular vestibule of the LeuT-SeMet complex. Anomalous difference Fourier map (contoured at 5σ and 15σ and depicted in green and blue mesh, respectively) showing no significant density peaks in the extracellular vestibule.
We suggest that Trp602, located ∼4 Å above Trp601, represents a low affinity, transiently occupied site for amino acids as they move from the extracellular vestibule to the substrate binding pocket, perhaps with R30 and D404 serving to dehydrate the incoming amino acid. Although molecular dynamics studies (26, 27) and binding assays (27) suggest that the substrate leucine can bind to a site similar to the Trp602 site when LeuT is in an occluded-like state, the LeuT-Trp crystal structure demonstrates that occupancy of the Trp602 site requires an open-to-out state, with the guanidium group of R30 and the side chain carboxylate of D404 separated by ∼7-8 Å. To determine if substrates can bind to the Trp602 site in the occluded state, we solved the crystal structure of LeuT with 30 mM leucine and did not observe any amino acid density in the extracellular vestibule (fig. S7). We also measured diffraction data on a LeuT-selenomethionine complex. Anomalous difference Fourier maps show a large anomalous peak (25σ) in the primary binding site but no significant peaks elsewhere (Fig. 4B).
In addition to Trp601 and 602, we observed two additional tryptophans (603 and 604) at the cytoplasmic and extracellular faces of LeuT, respectively (Fig. 3C). Trp603 is located in the cleft between EL2 and EL4, while Trp604 is situated at the cytoplasmic-face of LeuT and forms a salt bridge with R11. At present we believe these tryptophan molecules are not relevant to the function of LeuT, in large part because they are removed from mechanistically crucial regions of the transporter.
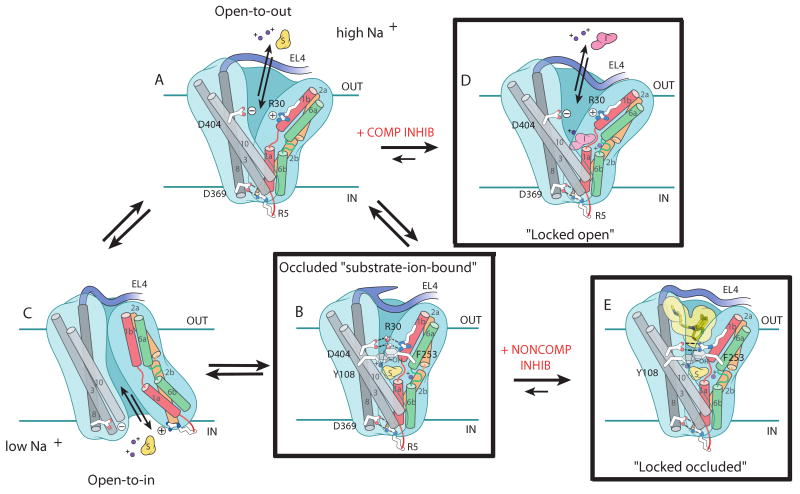
A Model for Transport and Inhibition
How do inhibitors prevent substrate translocation? For LeuT we postulate that inhibition of transport occurs by preventing distinct steps of the transport cycle (Fig. 5, D and E). Here we show that a competitive inhibitor displaces substrate and traps the transporter in an open-to-out conformation, thereby preventing progression to the occluded state (Fig. 5, A and D). The LeuT-Trp complex demonstrates how the extracellular-facing TMs 1b, 2a (residues 40-54), and 6a are involved in the binding of a competitive inhibitor and in the ensuing conformational changes, showing that TMs 1b, 2a and 6a move independently of their respective intracellular-facing counterparts, TMs 1a, 2b (residues 55-70), and 6b and that the TM1, 2, 6 and 7 helix bundle does not move as rigid body, in contrast to a recent proposal by Forrest and colleagues (28). The notion that TMs 1b and 6a undergo conformational changes upon substrate or inhibitor binding is further supported by chemical modification experiments on single cysteine mutants of the eukaryotic GABA and serotonin transporters (29-31).
Fig. 5.
Schematic of transport and inhibition in LeuT. Postulated conformational changes associated with isomerization from the open-to-out (A) to the outward facing occluded state (B) upon binding of substrate and ions, from the occluded (B) to open-to-in state (C) and dissociation of transported substrate and ions, and from the open-to-in (C) back to the open-to-out state (A). (D) Effect of a competitive inhibitor on transport: stabilizing the open-to-out conformation. (E) TCAs are noncompetitive inhibitors that stabilize the occluded state. The boxed conformations represent actual crystal structures, while the unboxed conformations are hypothetical.
What are the molecular principles associated with binding, i.e. formation of the occluded state? We suggest that substrates permeate from extracellular solution to the primary substrate site, located halfway across the membrane, by transiently binding to the extracellular gate residues R30 and D404. We argue that this binding event is only possible when the transporter is in the open-to-out conformation, typified by the LeuT-Trp complex. The substrate then moves to the primary binding site and the open-to-out state ‘collapses’ to the occluded state (Fig. 5, A and B) before isomerizing to the open-to-in state and permitting release of substrate to the cytoplasm (Fig. 5, B and C). The transporter can then cycle to the open-to-out state, perhaps through an apo yet occluded-like conformation. Optimal substrate binding and formation of the occluded state requires complementary shape and charge and is best satisfied by leucine and methionine. The apparent paradox posed by these two amino acids, which exhibit the highest binding affinities but the lowest turnover rates and catalytic efficiencies, is reconciled by the notion that transport is a balance between affinities for different intermediates in the transport cycle. Accordingly, the slow turnover rates of leucine and methionine are due to the fact that their occluded state complexes are very stable and the energy barriers associated with isomerization to open-to-in (Fig. 5, B and C) or -out conformations are relatively high. By contrast, the reduced affinity but higher turnover rate and catalytic efficiency of the smaller alanine is likely a reflection of the limited degree to which it can stabilize the occluded state compared to leucine or methionine.
The distinction between a substrate and a competitive inhibitor is provided by the ability of the ligand to promote formation of the occluded state. For LeuT, this distinction is highlighted by the differences between tyrosine and tryptophan. Because tyrosine is a substrate and tryptophan is not, there is an apparent size “boundary” for transport between 197 and 228 Å3, the volumes of tyrosine and tryptophan, respectively. In GAT1, the existence of a size boundary between transport and inhibition has been shown, where the addition of an aromatic moiety transforms the substrate nipecotic acid into the non-transportable competitive inhibitor SKF89976A (32). In the SLC5 family, this size boundary has also been demonstrated with a series of glycoside derivatives in experiments on the human glucose transporter (hSGLT1) (32). Whereas galactose is a substrate, 1-naphtylgalactose is a non-transportable, competitive inhibitor. But the boundary between a substrate and competitive inhibitor does not solely reside with the ligand. More generally, the definition of this boundary depends on the size, shape, and rigidity of the ligand relative to the constraints imposed by the binding pocket. Thus, amino acid substitutions in the binding pocket that alter these constraints, i.e. increase volume, should also alter this boundary. An example of this second case is TnaT, a prokaryotic SLC6 tryptophan transporter (33). Comparison of the amino acids lining the substrate-binding pocket in LeuT and TnaT (34) reveals that a prominent difference is substitution of the larger Phe at position 259 of LeuT with the smaller Val in TnaT. These substitutions would increase the volume of the binding pocket in TnaT to permit accommodation of Trp in an occluded state. These molecular principles are not only relevant to our understanding of LeuT and its SLC6-orthologs but are also germane to the structurally related glucose and nucleobase transporters (25, 35).
Supplementary Material
Acknowledgments
We are grateful to the staff of National Synchrotron Light Source beamline X29A and Advanced Light Source beamlines 8.2.1 and 8.2.2 for assistance with data collection. S.K.S. was supported by an individual NIH/NINDS NRSA postdoctoral fellowship and a NIH/NIMH K99/R00 Pathway to Independence Award. C.L.P. was supported by an institutional NIH Multidisciplinary Training in Neuroendocrinology grant. A.Y. was on leave from the Laboratory for Structural Biochemistry, RIKEN Harima Institute at SPring-8, Japan. This work was supported by the NIH (E.G.). E.G. is an investigator with the Howard Hughes Medical Institute. Coordinates and structure factors of the LeuT complexes have been deposited in the Protein Data Bank with the following codes: glycine (3F4J), alanine (3F48), 30 mM leucine (3F3E), methionine (3F3D), selenomethionine (3F4I), 4-fluorophenylalanine (3F3C) and tryptophan (3F3A).
Footnotes
Supplementary materials include Supplemental Experimental Methods, 6 figures, 2 tables, and a movie.
References and Notes
- 1.Parsons SM. FASEB J. 2000;14:2423. doi: 10.1096/fj.00-0203rev. [DOI] [PubMed] [Google Scholar]
- 2.Roux MJ, Supplisson S. Neuron. 2000;25:373. doi: 10.1016/s0896-6273(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 3.Zerangue N, Kavanaugh MP. Nature. 1996;383:634. doi: 10.1038/383634a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen NH, Reith ME, Quick MW. Pflugers Arch. 2004;447:519. doi: 10.1007/s00424-003-1064-5. [DOI] [PubMed] [Google Scholar]
- 5.Kanner BI. J Membr Biol. 2006;213:89. doi: 10.1007/s00232-006-0877-5. [DOI] [PubMed] [Google Scholar]
- 6.Warskulat U, Reinen A, Grether-Beck S, Krutmann J, Haussinger D. J Invest Dermatol. 2004;123:516. doi: 10.1111/j.0022-202X.2004.23313.x. [DOI] [PubMed] [Google Scholar]
- 7.Masson J, Sagne C, Hamon M, Mestikawy SE. Pharm Rev. 1999;51:439. [PubMed] [Google Scholar]
- 8.Hahn MK, Blakely RD. Pharmacogenomics J. 2002;2:217. doi: 10.1038/sj.tpj.6500106. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki N, et al. Mol Psychiatry. 2003;8:933. doi: 10.1038/sj.mp.4001365. [DOI] [PubMed] [Google Scholar]
- 10.Richerson GB, Wu Y. Adv Exp Med Biol. 2004;548:76. doi: 10.1007/978-1-4757-6376-8_6. [DOI] [PubMed] [Google Scholar]
- 11.Sutcliffe JS, et al. Am J Hum Genet. 2005;77:265. doi: 10.1086/432648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon JR, et al. N Engl J Med. 2000;342:541. doi: 10.1056/NEJM200002243420803. [DOI] [PubMed] [Google Scholar]
- 13.Salomons GS, et al. Am J Hum Genet. 2001;68:1497. doi: 10.1086/320595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller-Stilb B, et al. FASEB J. 2002;16:231. doi: 10.1096/fj.01-0691fje. [DOI] [PubMed] [Google Scholar]
- 15.Barker EL, Blakely RD. In: Psychopharmacology - the Fourth Generation of Progress. Bloom FE, Kupfer DJ, editors. Raven Press; New York: 2000. [Google Scholar]
- 16.Krogsgaard-Larsen P, Frolund B, Frydenvang K. Curr Pharm Des. 2000;6:1193. doi: 10.2174/1381612003399608. [DOI] [PubMed] [Google Scholar]
- 17.Amara SG, Sonders MS. Drug Alcohol Depend. 1998;51:87. doi: 10.1016/s0376-8716(98)00068-4. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Nature. 2005;437:215. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 19.Singh SK, Yamashita A, Gouaux E. Nature. 2007;448:952. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, et al. Science. 2007;317:1390. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talvenheimo J, Nelson PJ, Rudnick G. J Biol Chem. 1979;254:4631. [PubMed] [Google Scholar]
- 22.Apparsundaram S, Stockdale DJ, Henningsen RA, Milla ME, Martin RS. J Pharmacol Exp Ther. 2008 doi: 10.1124/jpet.108.142315. DOI:10.1124. [DOI] [PubMed] [Google Scholar]
- 23.Kimelberg HK, Pelton EW., 2nd J Neurochem. 1983;40:1265. doi: 10.1111/j.1471-4159.1983.tb13565.x. [DOI] [PubMed] [Google Scholar]
- 24.Henry LK, et al. J Biol Chem. 2006;281:2012. doi: 10.1074/jbc.M505055200. [DOI] [PubMed] [Google Scholar]
- 25.Faham S, et al. Science. 2008;321:810. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Celik L, Schiott B, Tajkhorshid E. Biophys J. 2008;94:1600. doi: 10.1529/biophysj.107.117580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. Mol Cell. 2008;30:667. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forrest LR, et al. Proc Natl Acad Sci U S A. 2008;105:10338. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Bennett ER, Kanner BI. J Biol Chem. 2004;279:13800. doi: 10.1074/jbc.M311579200. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg A, Kanner BI. J Biol Chem. 2008;283:14376. doi: 10.1074/jbc.M801093200. [DOI] [PubMed] [Google Scholar]
- 31.Henry LK, Adkins EM, Han Q, Blakely RD. J Biol Chem. 2003;278:37052. doi: 10.1074/jbc.M305514200. [DOI] [PubMed] [Google Scholar]
- 32.Hirayama BA, Diez-Sampedro A, Wright EM. Br J Pharmacol. 2001;134:484. doi: 10.1038/sj.bjp.0704274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Androutsellis-Theotokis A, et al. J Biol Chem. 2003;278:12703. doi: 10.1074/jbc.M206563200. [DOI] [PubMed] [Google Scholar]
- 34.Beuming T, Shi L, Javitch JA, Weinstein H. Mol Pharmacol. 2006;70:1630. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 35.Weyand S, et al. Science. 2008 DOI:10.1126. [Google Scholar]
- 36.Tsai J, Taylor R, Chothia C, Gerstein M. J Mol Biol. 1999;290:253. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.