Abstract
Rationale and Objectives
Dynamic contrast enhanced MRI (DCE-MRI) is a clinical imaging modality for detection and diagnosis of breast lesions. Analytical methods were compared for diagnostic feature selection and performance of lesion classification to differentiate between malignant and benign lesions in patients.
Materials and Methods
The study included 43 malignant and 28 benign histologically-proven lesions. Eight morphological parameters, ten gray level co-occurrence matrices (GLCM) texture features, and fourteen Laws’ texture features were obtained using automated lesion segmentation and quantitative feature extraction. Artificial neural network (ANN) and logistic regression analysis were compared for selection of the best predictors of malignant lesions among the normalized features.
Results
Using ANN, the final four selected features were compactness, energy, homogeneity, and Law_LS, with area under the receiver operating characteristic curve (AUC) = 0.82, and accuracy = 0.76. The diagnostic performance of these 4-features computed on the basis of logistic regression yielded AUC = 0.80 (95% CI, 0.688 to 0.905), similar to that of ANN. The analysis also shows that the odds of a malignant lesion decreased by 48% (95% CI, 25% to 92%) for every increase of 1 SD in the Law_LS feature, adjusted for differences in compactness, energy, and homogeneity. Using logistic regression with z-score transformation, a model comprised of compactness, NRL entropy, and gray level sum average was selected, and it had the highest overall accuracy of 0.75 among all models, with AUC = 0.77 (95% CI, 0.660 to 0.880). When logistic modeling of transformations using the Box-Cox method was performed, the most parsimonious model with predictors, compactness and Law_LS, had an AUC of 0.79 (95% CI, 0.672 to 0.898).
Conclusion
The diagnostic performance of models selected by ANN and logistic regression was similar. The analytic methods were found to be roughly equivalent in terms of predictive ability when a small number of variables were chosen. The robust ANN methodology utilizes a sophisticated non-linear model, while logistic regression analysis provides insightful information to enhance interpretation of the model features.
INTRODUCTION
Dynamic contrast enhanced MRI (DCE-MRI) is a clinical imaging modality for detection and diagnosis of breast lesions. Computer algorithms have been employed for automated lesion segmentation (1–4) with statistical analysis techniques applied to select an optimal set of features to achieve the highest diagnostic accuracy (5). Breast MRI has demonstrated a high sensitivity (>95%) with specificity of approximately 67% reported by meta analysis (6–11). To address the challenge of accuracy and efficiency in interpretation of breast MRI, a computer-aided diagnosis (CAD) system that can automatically analyze lesion features to differentiate between malignant and benign lesions would be very useful.
The general approach of breast CAD to predict a dichotomous outcome, malignant versus benign, involves applying computer algorithms for tumor characterization and then developing models using techniques including linear discriminate analysis (12), logistic regression analysis (13), and artificial neural networks (ANN) (14) for classifying a lesion as either malignant or benign. Because of inherent differences in these techniques, it is of interest to compare the diagnostic performance of analysis methods. In a review of 28 studies comparing ANN with other statistical approaches, Sargent found that ANN outperformed regression analysis in 36% of cases, ANN was outperformed by regression analysis in 14% of cases, and that similar results were obtained in 50% of cases (15). The advantages of ANN for predicting a dichotomous outcome include a requirement of less formal statistical training, an ability to implicitly detect complex nonlinear relationships between dependent and predictor variables, consideration of all possible interactions between predictor variables, and the availability of multiple training algorithms. Disadvantages include the “black box” nature of the ANN procedure, a greater computational burden, the tendency for overfitting, and the empirical nature of model development (16). Logistic regression is superior for examining possible causal relationships between independent and dependent variables, and understanding the effect of predictors on outcome variables (16). In summary, there is no universal approach to select models for data classification; evaluation of tasks on a case-by-case basis is necessary.
To our knowledge, only one study has compared ANN and logistic regression for breast cancer diagnosis using sonograms by Song et al. to differentiate between 24 malignant and 30 benign lesions (17). No difference was found in the diagnostic performance of ANN and logistic regression when measured by the area under the ROC curve. We have developed a CAD that incorporates a clustering-based algorithm for lesion segmentation and derives a full panel of quantitative morphological and texture descriptors for lesion characterization (18). In that study the ANN was used to select diagnostic features. In the present work we investigated and compared the diagnostic performance using the ANN and logistic regression.
Two aims were studied. Firstly we applied ANN to select features and investigated the diagnostic performance of different feature sets (models) selected based on the morphology and texture of the lesion. While ANN can select features robustly, the non-linear diagnostic model makes the weighting of each individual feature not interpretable. The logistic regression was applied to gain more insightful understanding for how each selected feature can be interpreted. Secondly, we illustrated the use of logistic regression for feature selection. The following statistical procedures were applied sequentially: 1) transformation of feature values to induce normality, 2) consideration of strategies for model selection and validation, 3) assessment of correlation between features to reduce collinearity, 4) reduction of the number of variables in models to avoid overfitting, and 5) evaluation and comparison of the diagnostic performance of classifiers. The selected features were compared to those selected by ANN. Also the diagnostic performance achieved by each selected model was evaluated and compared to that achieved by ANN.
MATERIALS AND METHODS
Malignant and Benign Lesion Database
The database analyzed in this study included 43 malignant and 28 benign lesions, the same database as reported in a previous study (18). The 43 patients with malignant lesions were from 29 to 76 years old with mean (± SD) 48±9 yr and median 48 yr. The 28 patients with benign lesions were from 21 to 74 yr, with mean 45±7 yr. and median 45 yr. The MRI studies were performed on a Philips Eclipse 1.5T scanner (Cleveland, OH). The images were acquired using a T1-weighted 3D SPGR (RF-FAST) pulse sequence, with TR=8.1 ms, TE=4.0 ms, flip angle=20°, matrix size=256×128, FOV (Field of View) varying between 32 and 38 cm. The lesion was identified based on contrast enhanced images at 1-min after injection of the MR contrast medium Omniscan® (0.1 mmol/kg body weight).
The lesion was segmented on contrast-enhanced images. The detailed procedures have been reported previously (18). The operator inspected the lesion presentations on MRI, and determined the beginning and the ending image slices that contain the lesion, then one square box was placed on the lesion on one image slice to indicate the lesion location. With these inputs the 3-dimensional boundary of the lesion (or, lesion ROI- Region Of Interest) was obtained using an automated computer program based on the fuzzy c-means (FCM) clustering algorithm (1). After the 3-dimensional ROI for one lesion was defined, computer algorithms were used to extract quantitative features representing the morphological and texture properties of this lesion.
Eight morphological parameters were obtained for each tumor, including: volume, surface area, compactness, normalized radial length (NRL) mean, sphericity, NRL entropy, NRL ratio, and roughness, were obtained. Texture is a repeating pattern of local variations in image intensity, and is characterized by the spatial distribution of intensity levels in a neighborhood. Therefore, the texture features analyzed within the contrast enhanced lesion represent the distribution of enhancements. Ten GLCM texture features (energy, maximum probability, contrast, homogeneity, entropy, correlation, sum average, sum variance, difference average, and difference variance), as defined by Haralick and colleagues, were obtained for each lesion (19).
In addition, commonly used Laws’ texture features were also obtained. The Laws’ features were computed by first applying small convolution kernels to a digital image and then performing a nonlinear windowing operation. The 2D convolution kernels were generated from a set of 1D convolution kernels: L5=[1 4 6 4 1], E5=[−1 −2 0 2 1], S5=[−1 0 2 0 −1], R5=[1 −4 6 −4 1], W5=[−1 2 0 −2 −1]. These mnemonics stood for Level, Edge, Spot, Ripple and Wave. 2D convolution kernels such as Law_LE were obtained by convolving a vertical L5 kernel with a horizontal E5 kernel. In total, a set of 14 texture features that were rotationally invariant were obtained (Law_LE, Law_LS, Law_LW, Law_LR, Law_EE, Law_ES, Law_EW, Law_ER, Law_SS, Law_SW, Law_SR, Law_WW, Law_WR, and Law_RR) (20).
Artificial Neural Network (ANN) for diagnostic feature selection
A three-layer back-propagation neural network, known as multi-layer perceptrons (MLP) artificial neural network (ANN) was utilized to obtain optimal classifiers. The three-layer topology has an input layer, one hidden layer, and an output layer. The number of nodes in the input corresponds to the number of input variables. The output layer contains one node with values from 0 to 1 indicating level of malignancy, where 0 means absolutely benign and 1 means absolutely malignant. The number of hidden nodes is usually determined by a number of trial-and-error runs. Different neural network architectures with hidden nodes from 2 to 31 were tested. A stochastic gradient decent with the mean squared error function was used as the learning algorithm. The optimal architecture was chosen as the one for which the validation error was the lowest. With the determined number of hidden nodes, both the learning rate (0.1–0.7) and the momentum coefficient (0.2–0.8) were varied during network training to ensure a high probability of global network convergence. For training, criteria for convergence was met with a root mean squared error less than or equal to 0.0001 or a maximum of 1000 iterations. With the determined number of hidden nodes, both the learning rate, (0.1–0.7), and the momentum coefficient, (0.2–0.8), were varied during network training to ensure a high probability of global network convergence.
After the topology was chosen, feature selection was performed to find potential predictors within each set of morphology, GLCM texture, and Laws’ texture features. The features were selected using the LNKnet (http://www.ll.mit.edu/IST/lnknet/) package in order to identify those yielding maximum discrimination capability thus achieving the optimal diagnostic performance. Each feature from 71 lesions was transformed to have zero mean and unit variance before training; after determination of the mean and standard deviation (STD) for a particular feature with n=71, the transformation for the ith value of a feature is zi= (feature value – mean)/STD. This transformation is known as a z-score.
A forward search strategy was applied to find the optimal feature subset, which was obtained when the trained classifier produced the least error rate. The k-fold stratified cross-validation technique with k=4 was applied to test the performance of the generated classifiers. Weights and bias of the neural network were determined by a two-phase training procedure. The first phase had 30 iterations of back propagation, and the second phase had a longer run of conjugated gradient descent to ensure full convergence. The logistic sigmoid function was used to interpret the output variation in terms of probability of class membership within the range (0 to 1). To control for overfitting, the potential feature set was limited to no more than 4 in each category which had the least error. After the features from each of three categories (morphology, GLCM, and Laws’) were individually obtained (Models A, B, and C respectively), all selected predictors were considered in a combined model (Model D), and a final model, restricted to contain no more than four features (Model E), was selected using ANN. Specifically, these models consisted of Model A with three features selected from eight morphology features, Model B with three features selected from 10 GMLM features, and model C with one selected from 14 Laws’ features, Model D consisting of the combined seven selected features, and Model E consisting of four features selected from the combined seven features.
Logistic Regression Model
For this study the binary response variable of interest is the presence of a malignant breast lesion, coded 1=yes and 0=no. Note that the mean of the response values for a sample of lesions is the same as the proportion of lesions that are malignant. A logistic regression model that predicts a transformation of the response variable is used, logit(p). Let p = probability that a lesion is malignant and 1-p = probability that a lesion is benign. Then the log odds is defined as
| (1) |
The logistic regression model enables prediction of the probability of a malignant breast lesion in relation to m lesion features, x1, x2, x3,⋯, xm from an equation of the form
| (2) |
where β0 is the intercept term, and β1, β2, β3,…βm, are the coefficients in the model associated with the m lesion features. It is assumed that the lesion features are linearly related to the log odds of the response. For a continuous lesion feature, the odds ratio for malignant versus benign lesions can be estimated as the exponentiation of the associated coefficient in the model (21).
Evaluation of ANN models using Logistic Regression
To address the first aim of the study, the selected diagnostic features in the five final models (A-E) by ANN were analyzed using logistic regression. Lesions were classified as benign or malignant on the basis of increasing threshold probabilities of malignancy. The ROC curve was constructed from the full range of probability thresholds with corresponding data points (sensitivity, 1-specificity) and the AUC was calculated. A nonparametric approach was used to compute the 95% confidence interval for the AUC (22, 23). The classification of lesions on the basis of probability of being malignant > 0.5 was used to compare results for logistic regression with ANN for accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
Logistic Regression Analysis for Diagnostic Feature Selection
To address the second aim of the study, logistic regression was used for initial selection of predictive models from feature z-scores. Descriptive statistics were calculated for features obtained from 71 lesions. The k-fold stratified cross-validation was applied with k=4. Forty-three malignant and twenty-eight benign lesions were separately and randomly assigned into four sub-cohorts so that each sub-cohort contained approximately the same proportion of malignant cases as the original cohort. To build predictive models, four sets of any three sub-cohorts were combined and used as a training cohort, with the remaining sub-cohort used as the corresponding validating cohort. These were labeled training cohorts 1–4 with corresponding validating cohorts 1–4.
The analysis method is now summarized. For each training set, logistic regression analysis was applied to examine the association between each individual feature and the dichotomous outcome, malignant lesion versus benign lesion. For example, univariate logistic regression models were formed, one for each of the eight morphological features. For each model, the likelihood ratio statistic was computed and the Bonferroni-Holm multiple comparisons method was applied to indicate statistically significant features at a 5% experimentwise significance level. These procedures were repeated for the 10 GLCM texture features and for the 14 Laws’ texture features and the statistically significant features for each group were retained.
Next, for each training set, stepwise logistic model selection was applied to each set of features. For example, stepwise logistic regression was applied to the eight morphological features as predictors. The Score chi-square statistics were calculated and used to determine addition and removal of variables at the 0.05 significance level. The stepwise procedure was repeated for the 10 GLCM texture features and for the 14 Laws’ texture features. Due to the small sample size of the cohorts, at most two predictors were retained from each set of features.
Pearson’s correlation coefficient was computed for the pairs of predictors retained on the basis of univariate and stepwise modeling. Multivariate models were then built considering the combined morphology, GLCM texture and Laws’ texture features that did not have statistically significant pairwise correlation. For each multivariate model, the variance inflation factor (24) for features in a model were examined with the objective of minimizing multicollinearity. No effect of multicollinearity is indicated by a VIF of zero. A common rule of thumb is that if the variance inflation is >5, then multicollinearity is high.
For each training set, a list of models was generated in ascending order of estimated AUC. The c index of concordance statistic was calculated to compare training datasets with regard to the receiver operating characteristic area (25, 26) and the 95% confidence interval of the average of concordance statistics was obtained for each model. Moreover, the Hosmer-Lemeshow goodness-of-fit test was performed for each model (26, 27). The models selected from each training cohort were applied to the corresponding validating sub-cohort.
Finally, the models with the greatest AUC for each validating sub-cohort were applied to the entire cohort. For each of these models, the accuracy, sensitivity, specificity, PPV, and NPV were calculated.
The odds of a lesion to be malignant was computed for every increase of 1 SD in a feature, adjusted for differences in the other features in the model. For the comparison, the same analysis method was used after Box-Cox transformations were applied to induce normality, as needed.
RESULTS
Artificial Neural Network Diagnostic Feature Selection and Evaluation
Before ANN training, features were transformed to z-scores based on the mean and standard deviation of the entire set of 71 lesions. A three layer ANN with 5 hidden nodes in the hidden layer was chosen after a number of trial-and-error runs. The diagnostic measures used to assess differentiation between 43 malignant and 28 benign lesions are summarized in Table 1. Considering the eight morphology features, the classifier selected by ANN included three parameters: lesion volume, NRL entropy, and compactness (Model A). Using these three features for ROC analysis of the entire cohort of 71 lesions, an AUC of 0.80 and accuracy of 0.77 was achieved. Considering the ten GLCM texture features, the selected parameters were: gray level energy, gray level sum average, and homogeneity (Model B), with an AUC of 0.81 and accuracy of 0.73. From fourteen Laws’ energy features, the best classifier contained only one parameter, Law_LS (Model C), with an AUC of 0.70 in the ROC analysis and accuracy of 0.65. Applying ANN to the combined set of seven features, including compactness, lesion volume, and NRL entropy, energy, gray level sum average, homogeneity, and the Law_LS parameter (Model D), the resulting AUC and accuracy were improved to 0.87 and 0.79, respectively, with positive and negative predictive values of 0.83 and 0.72. When these seven features were considered by a further ANN selection process, the final four selected features were: compactness, energy, homogeneity, and Law_LS, (Model E) and achieved an AUC of 0.82, accuracy of 0.76, PPV of 0.78, and NPV of 0.72.
Table 1.
Diagnostic evaluation of models selected using the artificial neural network (ANN) technique. For each model the corresponding logistic regression equation also was applied to data from the full cohort (N = 71: Malignant = 43; Benign = 28).
| Model | Imaging Descriptorsa | Method | Accuracy1 (%) |
Sensitivity2 (%) |
Specificity3 (%) |
PPV4 | NPV5 | Estimated Area under ROC |
||
|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | |||||||||
| A | Morphology (3 selected from 8) | ANN | 77 | 88 | 61 | 0.78 | 0.77 | 0.80 | ||
| Volume, NRL Entropy, Compactness |
Logistic Regression |
76 | 93 | 50 | 0.74 | 0.82 | 0.80 | 0.686 | 0.912 | |
| B | GLCM (3 selected from 10) | ANN | 73 | 84 | 57 | 0.75 | 0.70 | 0.81 | ||
| Energy, Gray Level Sum Average, Homogeneity |
Logistic Regression |
68 | 86 | 39 | 0.69 | 0.65 | 0.77 | 0.660 | 0.880 | |
| C |
LAWs (only 1 selected from 14) |
ANN | 65 | 84 | 36 | 0.67 | 0.59 | 0.70 | ||
| Law_LS | Logistic Regression |
65 | 86 | 32 | 0.66 | 0.60 | 0.70 | 0.573 | 0.819 | |
| D | Combining all 7 selected features | ANN | 79 | 81 | 75 | 0.83 | 0.72 | 0.87 | ||
| Volume, NRL Entropy, Compactness, Energy, Gray Level Sum Average, Homogeneity, Law_LS |
Logistic Regression |
80 | 86 | 71 | 0.82 | 0.77 | 0.86 | 0.772 | 0.949 | |
| E | Final (4 selected from 7) | ANN | 76 | 84 | 64 | 0.78 | 0.72 | 0.82 | ||
| Compactness, Energy, Homogeneity, Law_LS |
Logistic Regression |
72 | 86 | 50 | 0.73 | 0.70 | 0.80 | 0.688 | 0.905 | |
Accuracy = (number of correctly identified cases / 71) × 100%
Sensitivity = (number of correctly identified as malignant / 43) × 100%
Specificity = (number of correctly identified as benign / 28) × 100%
Positive Predictive Value (PPV)= number of lesions correctly identified as malignant/ number of lesions identified as malignant
Negative Predictive Value (NPV)= number of lesions correctly identified as benign/ number of lesions identified as benign.
Each variable in the model was standardized by subtracting the mean and dividing by the standard deviation of data from 71 subjects.
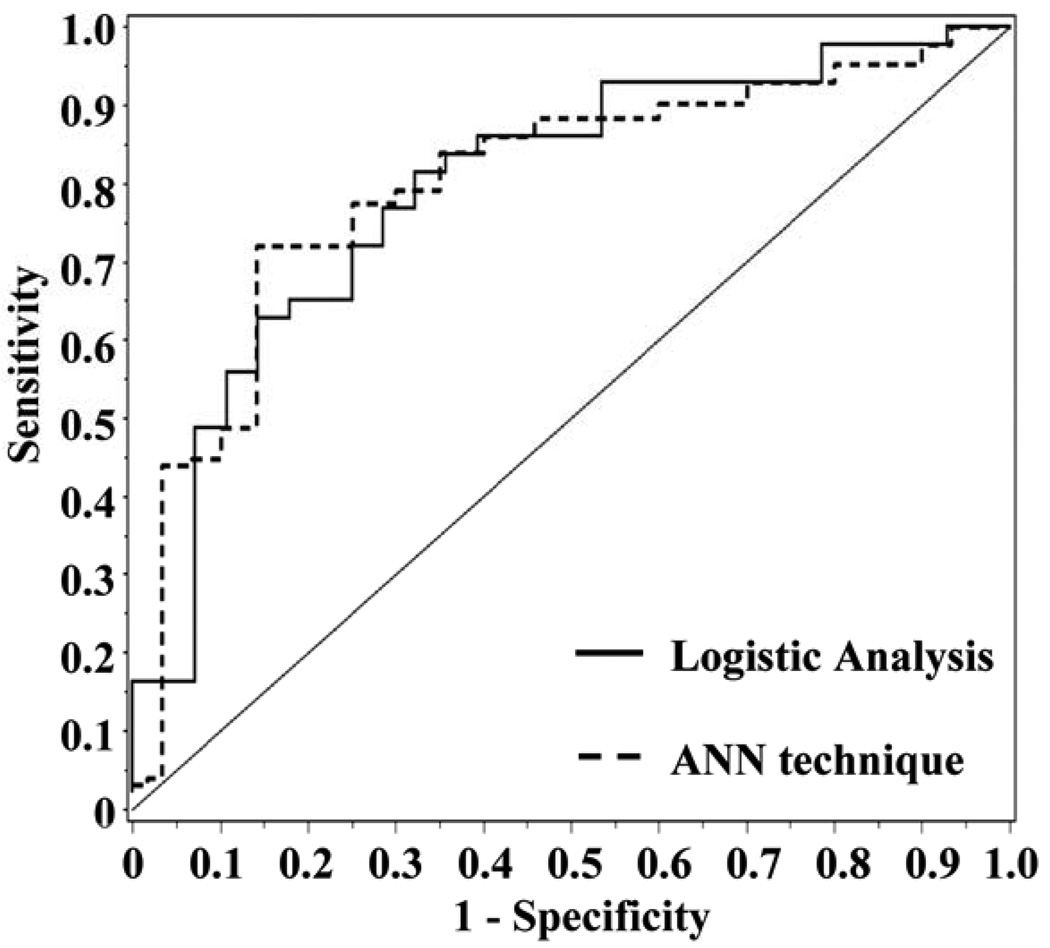
Logistic regression was applied to each of the models selected by ANN for the cohort of 71 lesions, classifying a lesion as malignant if the threshold probability of malignancy was >0.5. The diagnostic measures are also displayed in Table 1 for comparison with the ANN results. The values for diagnostic measures are similar to those recorded from the ANN technique. For example, the 3-feature morphology model of volume, NRL entropy, and compactness (Table 1, model A) has an AUC of 0.80 recorded by the ANN technique, and the same AUC of 0.80 (95% CI, 0.686 to 0.912) computed using logistic regression. The 95% confidence interval for the AUC from logistic regression contains the 0.80 value recorded from ANN. Diagnostic accuracy of 0.77, recorded by ANN, was higher than the value of 0.76 computed from logistic regression. Similarly, the final 4-feature model of compactness, energy, homogeneity, and Law_LS, (Table 1, Model E), had an AUC of 0.82 recorded by the ANN technique, as compared to 0.80 (95% CI, 0.688 to 0.905) computed on the basis of logistic regression. The 95% confidence interval for AUC contains the computed value of 0.82 from ANN. Figure 1 displays the ROC curves for the 4-feature model (E) analyzed using the ANN and logistic regression. The diagnostic accuracy of 0.76 recorded by ANN was slightly higher than 0.72 computed by logistic regression.
Figure 1.
The dashed line represents the ROC curve for ANN modeling of z-scores (Table 1, Model E; Compactness, Energy, Homogeneity and Law_LS; AUC 0.82). The solid line represents the ROC curve for the four features as assessed by logistic regression (Table 1, Model E, AUC 0.80).
For the outcome of presence of a malignant lesion and modeling of z-scores from feature predictors with n=71, the logistic regression equation was
| (3) |
Law_LS was the most predictive variable in the model (Wald statistic, P=0.028). The P-value of the Hosmer-Lemeshow test was 0.76 indicating a reasonably good fit of the model to the data. However, the adjusted R2 for the model was 0.30 which indicates that only 30% of the total variation in values could be explained by the predictor features in the model (28). The estimated odds ratio for Law_LS was calculated as the exponentiation of the associated estimated coefficient, exp(−0.73) = 0.48. Thus the odds of a malignant lesion decreased by 48% (95% CI, 25% to 92%) for every increase of 1 SD in the Law_LS feature, adjusted for differences in compactness, energy, and homogeneity. For example, comparing a lesion with Law_LS of 0.19, one standard deviation above the mean of 0.13 for the set of 71 lesions, to a lesion Law_LS of 0.25, two standard deviations above the mean, the odds of the second lesion being malignant are about one-half of that of the first lesion, assuming that they have similar values for compactness, energy, and homogeneity.
Diagnostic Feature Selection using Logistic Regression Analysis
To address the second aim of the study, we applied logistic regression to z-scores for morphology, GLCM texture, and Laws’ texture features and followed the modeling strategy described in Materials and Methods. Within training sets, features preselected for consideration by multivariate models had no statistically significant correlation between pairs of features (P>0.13 for all). Table 2 displays the best single model selected from each training cohort. For each training cohort, the model chosen had the highest AUC from among those applied to the corresponding validation cohort. For a particular model, the diagnostic values presented in the table were calculated when the model was used to reclassify the full data set of 71 lesions on the basis of probability of being benign or malignant. Each of the four models (Table 2, training cohort 1–4) contained two morphological parameters, compactness and NRL Entropy. In addition, models chosen from training cohort 1 and 2 contained a GLCM texture feature. The model selected from training cohort 1 included energy; while the model from training cohort 2 included gray level sum average. Identical models were chosen from training cohorts 3 and 4 and contained the Laws’ texture feature, Law_LW. When the chosen models were applied to the full data set of 71 lesions, all models had P-values for the likelihood ratio test less than 0.004 indicating the joint significance of all predictor features with respect to the outcome. Values for AUC varied from 0.75 for the model from training cohort 1 (compactness, NRL entropy, energy) to 0.80 for the model selected form training cohorts 3 and 4 (compactness, NRL entropy, Law_LW). However, the 95% confidence intervals for the AUC overlap, indicating no significant difference between AUC values. Estimated accuracy of models varied from 72% to 75%. The model chosen from training cohort 2 had the highest overall accuracy, correctly classifying 75% of lesions, and an AUC of 0.77 (95% CI, 0.660 to 0.880). This model was comprised of three features including compactness, NRL entropy, and gray level sum average. The model had high sensitivity, correctly identifying 91% of malignant lesions and moderate specificity, correctly identifying 50% of benign lesions. While the model selected from training cohort 1 (compactness, NRL entropy, and energy) had the highest sensitivity and correctly identified 95% of malignant cases, the specificity was only 39%.
Table 2.
Diagnostic evaluation of models selected using logistic regression of feature z-scores with 4-fold cross validation and applied to data from the full cohort (N = 71: Malignant = 43; Benign = 28).
| Training Cohort |
Model with highest AUC for corresponding validation cohorta |
Likelihood ratio ▯2 |
P- value |
Accuracy1 (%) |
Sensitivity2 (%) |
Specificity3 (%) |
PPV4 | NPV5 | Estimated Area under ROC6 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | ||||||||||
| 1 | Compactness, NRL Entropy, Energy |
13.54 | 0.0036 | 73 | 95 | 39 | 0.71 | 0.85 | 0.75 | 0.626 | 0.879 |
| 2 | Compactness, NRL Entropy, Gray Level Sum Average |
17.86 | 0.0005 | 75 | 91 | 50 | 0.74 | 0.78 | 0.77 | 0.660 | 0.880 |
| 3,4 | Compactness, NRL Entropy, Law_LW |
19.16 | 0.0003 | 72 | 86 | 50 | 0.73 | 0.70 | 0.80 | 0.690 | 0.908 |
Accuracy = (number of correctly identified cases / 71) × 100%;
Sensitivity = (number of correctly identified as malignant / 43) × 100%
Specificity = (number of correctly identified as benign / 28) × 100%;
Positive Predictive Value (PPV)= number of lesions correctly identified as malignant/ number of lesions identified as malignant
Negative Predictive Value (NPV)= number of lesions correctly identified as benign/ number of lesions identified as benign.
Classifiction criteria: If the predicted value > 0.5, then the case was classified as malignant case by the model. If the predicted value < 0.5, then the case was classified as benign case by the model.
Each variable in the model was standardized by subtracting the mean and dividing by standard deviation of values from 71 subjects.
Figure 2 displays the ROC curve, illustrated with a solid line, for the 3-feature model selected from training cohort 2 with the highest overall accuracy among validating cohorts, correctly classifying 75% of the 71 lesions. The model was comprised of two morphological parameters, compactness and NRL entropy, and one GLCM feature, gray level sum average (Table 2, Figure 2). Applied to the entire cohort of 71 lesions, there was no statistically significant correlation between pairs of variables (P>0.10 for all). A good fit of the model to the data was demonstrated (Hosmer and Lemeshow goodness-of-fit statistic, P=0.88) and a high concordance statistic (0.78; 95% CI, 0.755 to 0.813) implied a good ability to predict the lesion type. As expected, the average variance inflation factor (VIF) for the three features in the model was 1.02 indicating low effect of multicollinearity on the variance of model coefficients. The strongest predictor in the model was gray level sum average. The odds of a malignant lesion increased by 2.0 (95% CI, 1.14 to 3.68) for every increase of 1 SD in the texture feature, gray level sum average, adjusted for differences in morphology features, compactness and NRL entropy. The logistic regression equation was
| (4) |
Gray level sum average was the most predictive variable in the model (Wald statistic, P=0.016). For this variable, the estimated odd ratio was exp(0.72) = 2.1 (95% CI, 1.14 to 3.68), thus the odds of a malignant lesion increased by 2.1 for every increase of 1 SD in the gray level sum average feature, adjusted for differences in compactness and NRL entropy. For example, comparing a lesion with gray level sum average of 27.6, the mean for the 71 lesions, to a lesion with feature value of 34.2, one standard deviation above the mean, the odds of the second lesion being malignant are over twice that of the first lesion, assuming that they have similar values for compactness and NRL entropy.
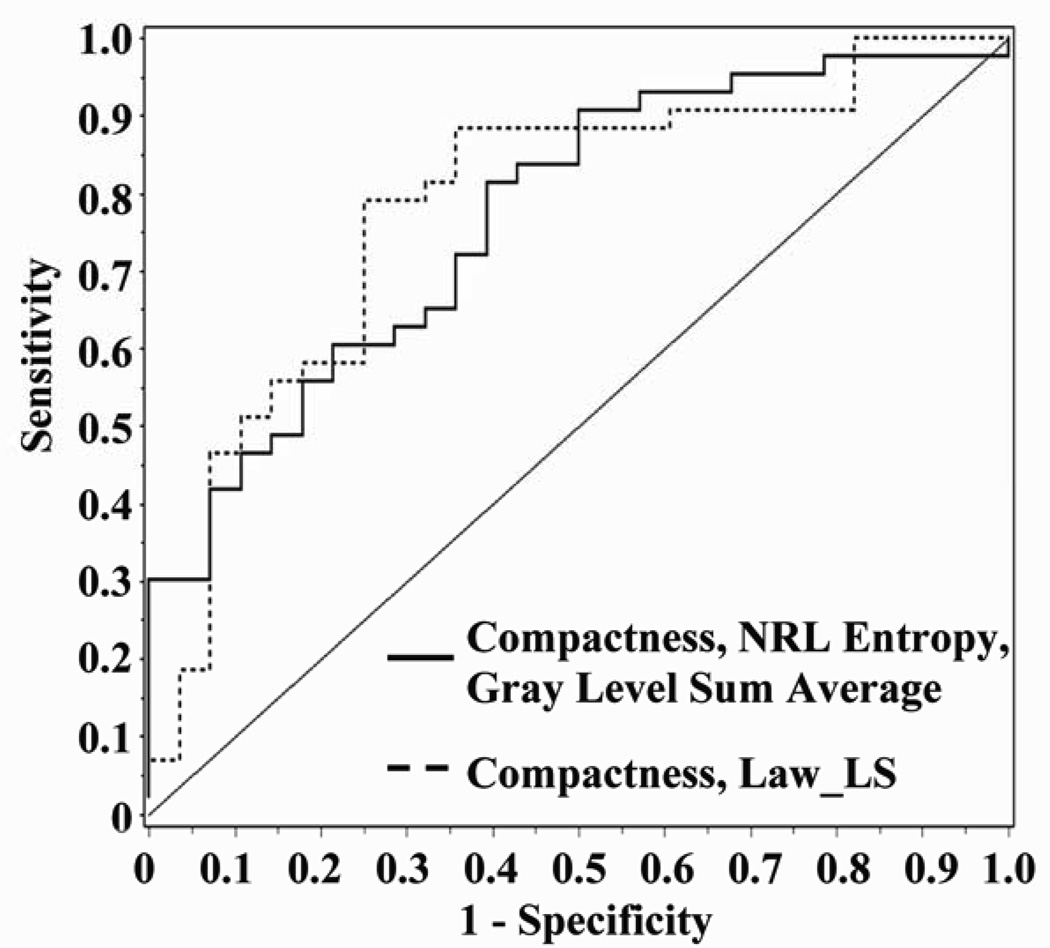
Figure 2.
The solid line represents the ROC curve for logistic regression modeling of z-scores for compactness, NRL entropy and gray level sum average (AUC 0.77; 95% CI, 0.660 to 0.880). The dashed line represents logistic regression model of Box-Cox transformed values for compactness and Law_LS (AUC 0.79; 95% CI, 0.672 to 0.898).
We compared results of modeling the feature z-scores to those obtained after applying the Box-Cox method of selecting the most appropriate transformation to induce normality. A two-feature model of ln compactness and Box-Cox transformed Law_LS had the highest accuracy of 0.79, with estimated AUC of 0.79 (95% CI, 0.672 to 0.898), sensitivity 0.88 and specificity 0.64. Figure 2 displays the ROC curves for the 2-feature model. The likelihood ratio chi-square for the model was statistically significant (P<0.0001). As expected, the correlation between ln compactness and transformed Law_LS was not statistically significant (r=0.196, P=0.10) and a good fit of the model to the data was demonstrated (Hosmer and Lemeshow Goodness-of-Fit, P=0.66) with a concordance statistic 0.79 (95% C.I., 0.724 to 0.855). The average variance inflation factor was 1.04, indicating a low effect of multicollinearity on the variance of model coefficients. In the model, both features added information to the model (Wald statistic, P<0.007 for each). For the outcome of presence of a malignant lesion, the logistic regression equation was
| (5) |
For ln compactness, the estimated odds ratio was exp(0.666) = 1.9. This indicates that the odds of a malignant lesion increased by 1.9 (95% CI, 1.20 to 3.56) for every increase of 1 unit in the natural log of the morphology feature, compactness, adjusted for differences in the Laws’ feature, Law_LS. For example, comparing a lesion with ln compactness of 3.0, one standard deviation above the mean of 1.6 for the set of 71 lesions, to that of a lesion with ln compactness of 4.5, two standard deviations above the mean, the second lesion would be nearly twice as likely to be malignant as first lesion, assuming equal values for Law_LS.
DISCUSSION
The primary aim of the study was to compare ANN and logistic regression analysis for lesion classification to differentiate between malignant and benign breast lesions in patients. Using our dataset of 71 lesions, the ANN procedure was applied to select the best classifiers for morphology and texture (GLCM and Laws’) category features. The three selected morphology features (volume, NRL entropy, compactness) achieved a moderate AUC of 0.80 and estimated accuracy of 0.77. The three selected GLCM features (energy, gray level sum average, and homogeneity) achieved higher AUC (0.81) and estimated accuracy (0.73). Only one Laws’ feature (Law_LS) was selected, and achieved lower AUC (0.70) and lower accuracy (0.65). When all seven features were combined the model achieved an improved AUC of 0.87 and estimated accuracy of 0.79. Submitting these seven features into another ANN selection, resulted in selection of four features, one morphology feature (compactness), two GLCM features (energy, homogeneity), and one Laws’ texture feature (Law_LS) with an AUC of 0.82 and accuracy of 0.76. These results demonstrate that it is possible to use ANN to select the best combined indicators to predict tumor malignancy.
For the 4-feature model (Table 1, Model E), logistic regression analysis revealed that Law_LS was the most predictive variable in the model (Wald statistic, P=0.028). The estimated odds ratio for Law_LS was calculated as exp(-0.73) = 0.48. Thus the odds of a malignant lesion decreased by 48% (95% CI, 25% to 92%) for every increase of 1 SD in the Law_LS feature, adjusted for differences in compactness, energy, and homogeneity. This result provides an illustration of the complementary manner in which ANN and logistic regression can be used (15). While ANN is more robust in that features are selected with minimal intellectual judgment of the operator, logistic regression can provide more insight and understanding into the relationship between selected features and the outcome.
Artificial neural networks have been used elsewhere in clinical data modeling, and similar results with that of regression modeling techniques were demonstrated (29–33). Compared to logistic regression modeling, ANN was found to have higher prediction rates in complex and non-linear relationships among a large number of variables; however even when the difference was significant (due to very large sample size) the improved performance was only marginal. Nilsson et al. used data from 18,362 patients undergoing cardiac surgery to predict the operative mortality. ANN selected 34 of the total 72 risk variables as relevant for mortality prediction. The area under ROC curve for ANN (0.81) was larger than that of the logistic regression model (0.79, P=0.0001) with the same 34 top-ranked risk variables (29). Delen and colleagues acquired a large dataset (202,932 cases with 17 variables) to predict five-year breast cancer survival using 10-fold cross-validation. The results indicated that when all variables were used, the ANN estimated area under the ROC curve with 0.91, compared to 0.89 computed by logistic regression (31). Lundin et al. tried to predict five-year breast cancer survival using data from 951 breast cancer patients. Using eight input variables, the ANN and logistic regression models achieved similar values for AUC of 0.901 and 0.897 respectively (33). Jaimes et al. (32) and Clermont et al. (30) also found that both ANN and logistic regression have similar performance when considering a small number of variables.
Advantages of neural network analysis are that few prior assumptions or knowledge about data distributions are required, so knowledge about complex variable transformations is not needed before training, and the search for the optimal diagnostic classifier involves minimal user input. Another advantage is that ANN has the capacity to model complex nonlinear relationships between independent and predictor variables, allowing the inclusion of a large number of variables. A disadvantage of ANN is the long training process and requirement of an experienced operator to determine the optimal network topology. The major factor that needs to be experimentally determined is the number of hidden layer nodes. If too few hidden nodes are used, proper training is impeded. If too many are used, the neural network is over-trained. In our study the number of hidden nodes was determined by a number of trial-and-error runs. Another limitation of the ANN technique is the poor interpretability of selected models. Neither standardized coefficients nor odd ratios corresponding to selected variable can be calculated and presented as in regression models. Logistic regression can be applied in a complementary manner to provide this information, thus overcoming the problem. Furthermore, the technique can be used for hypothesis testing regarding univariate and multivariate associations between predictor variables and the outcome of interest (15) and to enhances understanding and interpretation of the effect of predictor variables on the response (16). The logistic regression analysis may be preferred to ANN due to improved interpretation of individual predictors.
Both ANN and logistic regression are subject to issues of overfitting, assessing model convergence, and collinearity that affect the generalization of results (15). Methods to avoid overfitting include cross-validation as applied in our study (34). Cross-validation serves to check internal validity (reproducibility) (35). Leave-one-out cross validation, although almost unbiased, may have high variance leading to unreliable estimates (36). Kohavi studied cross-validation for accuracy of estimation and model selection and recommends k-fold stratified cross validation for model selection on the basis of stability of predictions and accuracy, when compared to leave-one-out cross validation (34, 37). As recommended, in this study we use 4-fold cross validation for ANN and logistic regression modeling. Finally, it is important to note that ANN does not strictly check collinearity among features during the selection process and collinearity can affect the variance of model estimates. Since the potential for collinearity among the features is not specifically taken into account, this can lead to stability problems (38–40).
The second aim of the study was to illustrate the use of logistic regression for feature selection. We examined differences between methods of standardizing features and addressed the issue of potential collinearity by incorporating statistical testing for correlation between variables to pre-select variables for further modeling. As illustrated in Table 2, the logistic regression model of feature z-scores with the highest estimated accuracy of 0.75 and AUC (0.77; 95% CI, 0.660 to 0.880) included compactness, NRL entropy, and gray level sum average (Figure 2). The model of Box-Cox transformed values with the highest accuracy of 0.79 contained two features, ln compactness and Box-Cox transformed Law_LS, with estimated AUC of 0.79 (95% CI, 0.672 to 0.898), sensitivity 0.88 and specificity 0.64 (Figure 2). The results suggest that the diagnostic performance of the models selected by logistic regression was comparable to that of ANN; also that the method of standardization may improve on model selection.
Regarding the generalizability of our results, although we analyzed a relatively small dataset, the selected features have been well accepted and commonly used in the development of breast CAD systems. Using this same dataset, we previously attempted to establish the association between the extracted quantitative features and the lesion phenotype appearance on MRI as described in the BI-RADS breast MRI lexicon (18). For example, the compactness morphological parameter is strongly linked to the shape and margin of the lesion, and the GLCM texture features are associated with the degree of the enhancement and the heterogeneous enhancement patterns within the lesion. These BI-RADS descriptors are well-established diagnostic features. Therefore, although the generalization of the selected quantitative features need validation using independent datasets, they are generlizable in the sense that they are closely related to visual diagnostic features commonly used by radiologists.
In summary, we have shown that the diagnostic performance of models selected by ANN and logistic regression was similar and the analytic methods were found to be roughly equivalent in terms of predictive ability when a small number of variables were chosen. We have emphasized interpretation of the predictors in the model and illustrated comparison of lesion features in terms of the odds of a lesion being malignant, enhancing the usefulness of the logistic regression modeling. The ANN methodology is more robust (i.e. it does not require a high level of operator judgment), and it utilizes a sophisticated non-linear model to achieve a high diagnostic performance. On the other hand, logistic regression may generate many sets of models that yield similar diagnostic performance, and the operator will need to make intellectual judgments to select the best model(s). The modeling strategy provided in the present work requires statistical judgment and thus may be more difficult to implement in a large dataset that has a many variables compared to the “black box” ANN approach. Nonetheless, logistic regression analysis provides insightful information to enhance interpretation of the model features. Finally, many diagnostic models (feature sets) could be selected using ANN and logistic regression based on cross-validation within one dataset; and the ultimate diagnostic value of these models will have to be determined in an independent validation dataset.
Acknowledgements
This work was supported in part by NIH/NCI R01 CA90437 (O. Nalcioglu), CA121568 (M-Y Su), the California Breast Cancer Program grant #9WB-002 (M-Y Su), and the UC Irvine Cancer Center Support Grant No. 2P30CA062203-13S (F.L. Meyskens, Jr.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chen W, Giger ML, Bick U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad Radiol. 2006;13:63–72. doi: 10.1016/j.acra.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Giger ML, Bick U, et al. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys. 2006;33:2878–2887. doi: 10.1118/1.2210568. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Giger ML, Lan L, et al. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys. 2004;31:1076–1082. doi: 10.1118/1.1695652. [DOI] [PubMed] [Google Scholar]
- 4.Liney GP, Sreenivas M, Gibbs P, et al. Breast lesion analysis of shape technique: semiautomated vs. manual morphological description. J Magn Reson Imaging. 2006;23:493–498. doi: 10.1002/jmri.20541. [DOI] [PubMed] [Google Scholar]
- 5.Meinel LA, Stolpen AH, Berbaum KS, et al. Breast MRI lesion classification: improved performance of human readers with a backpropagation neural network computer-aided diagnosis (CAD) system. J Magn Reson Imaging. 2007;25:89–95. doi: 10.1002/jmri.20794. [DOI] [PubMed] [Google Scholar]
- 6.Esserman L, Hylton N, Yassa L, et al. Utility of magnetic resonance imaging in the management of breast cancer: evidence for improved preoperative staging. J Clin Oncol. 1999;17:110–119. doi: 10.1200/JCO.1999.17.1.110. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Kopka L, Grabbe E. Breast carcinoma: effect of preoperative contrast-enhanced MR imaging on the therapeutic approach. Radiology. 1999;213:881–888. doi: 10.1148/radiology.213.3.r99dc01881. [DOI] [PubMed] [Google Scholar]
- 8.Mumtaz H, Hall-Craggs MA, Davidson T, et al. Staging of symptomatic primary breast cancer with MR imaging. AJR Am J Roentgenol. 1997;169:417–424. doi: 10.2214/ajr.169.2.9242745. [DOI] [PubMed] [Google Scholar]
- 9.Rieber A, Schirrmeister H, Gabelmann A, et al. Pre-operative staging of invasive breast cancer with MR mammography and/or PET: boon or bunk? Br J Radiol. 2002;75:789–798. doi: 10.1259/bjr.75.898.750789. [DOI] [PubMed] [Google Scholar]
- 10.Schelfout K, Van Goethem M, Kersschot E, et al. Contrast-enhanced MR imaging of breast lesions and effect on treatment. Eur J Surg Oncol. 2004;30:501–507. doi: 10.1016/j.ejso.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Fukatsu H, Naganawa S, et al. The role of contrast-enhanced MR mammography for determining candidates for breast conservation surgery. Breast Cancer. 2002;9:231–239. doi: 10.1007/BF02967595. [DOI] [PubMed] [Google Scholar]
- 12.Gilhuijs KG, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys. 1998;25:1647–1654. doi: 10.1118/1.598345. [DOI] [PubMed] [Google Scholar]
- 13.Chou YH, Tiu CM, Hung GS, et al. Stepwise logistic regression analysis of tumor contour features for breast ultrasound diagnosis. Ultrasound Med Biol. 2001;27:1493–1498. doi: 10.1016/s0301-5629(01)00466-5. [DOI] [PubMed] [Google Scholar]
- 14.Chan HP, Sahiner B, Petrick N, et al. Computerized classification of malignant and benign microcalcifications on mammograms: texture analysis using an artificial neural network. Phys Med Biol. 1997;42:549–567. doi: 10.1088/0031-9155/42/3/008. [DOI] [PubMed] [Google Scholar]
- 15.Sargent DJ. Comparison of artificial neural networks with other statistical approaches. Cancer. 2001;91:1636–1642. doi: 10.1002/1097-0142(20010415)91:8+<1636::aid-cncr1176>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 16.Tu JV. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol. 1996;49:1225–1231. doi: 10.1016/s0895-4356(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 17.Song JH, Venkatesh SS, Conant EA, et al. Comparative analysis of logistic regression and artificial neural network for computer-aided diagnosis of beast masses. Acad Radiol. 2005;12:487–495. doi: 10.1016/j.acra.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Nie K, Chen J-H, Yu HJ, et al. Quantitative Analysis of Lesion Morphology and Texture Features for Diagnostic Prediction in Breast MRI. Acad Radiol. 2008 doi: 10.1016/j.acra.2008.06.005. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973;SMC-3:610–621. [Google Scholar]
- 20.Laws KI. Society of Photo-Optical Instrumentation Engineers. San Diego, CA: Image processing for missile guidance; 1980. Rapid texture identification; pp. 376–380. [Google Scholar]
- 21.Altman DG. London: Chapman & Hall; 1991. Practical statistics for medical research. [Google Scholar]
- 22.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 23.Puri ML, Sen PK. New York: Wiley; 1971. Nonparametric Methods in Multivariate Analysis. [Google Scholar]
- 24.Neter J, Kutner M, Nachtsheim C, et al. New York: WCB McGraw-Hill; 1996. Applied linear statistical models. [Google Scholar]
- 25.Harrell FE, Cadiff RM, Pryor DB, et al. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 26.Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst. 2007;99:715–726. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 27.Hosmer DW, Lemeshow S. New York: John Wiley & Sons; 1989. Applied Logistic Regression. [Google Scholar]
- 28.Nagelkerke NJD. A Note on a General Definition of the Coefficient of Determination. Biometrika. 1991;78:691–692. [Google Scholar]
- 29.Nilsson J, Ohlsson M, Thulin L, et al. Risk factor identification and mortality prediction in cardiac surgery using artificial neural networks. J Thorac Cardiovasc Surg. 2006;132:12–19. doi: 10.1016/j.jtcvs.2005.12.055. [DOI] [PubMed] [Google Scholar]
- 30.Clermont G, Angus DC, DiRusso SM, et al. Predicting hospital mortality for patients in the intensive care unit: a comparison of artificial neural networks with logistic regression models. Crit Care Med. 2001;29:291–296. doi: 10.1097/00003246-200102000-00012. [DOI] [PubMed] [Google Scholar]
- 31.Delen D, Walker G, Kadam A. Predicting breast cancer survivability: a comparison of three data mining methods. Artif Intell Med. 2005;34:113–127. doi: 10.1016/j.artmed.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Jaimes F, Farbiarz J, Alvarez D, et al. Comparison between logistic regression and neural networks to predict death in patients with suspected sepsis in the emergency room. Crit Care. 2005;9:R150–R156. doi: 10.1186/cc3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundin M, Lundin J, Burke HB, et al. Artificial neural networks applied to survival prediction in breast cancer. Oncology. 1999;57:281–286. doi: 10.1159/000012061. [DOI] [PubMed] [Google Scholar]
- 34.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. The Fourteenth International Joint Conference on Artificial Intelligence Morgan Kaufmann; San Mateo. 1995. pp. 1137–1143. [Google Scholar]
- 35.Terrin N, Schmid CH, Griffith JL, et al. External validity of predictive models: a comparison of logistic regression, classification trees, and neural networks. J Clin Epidemiol. 2003;56:721–729. doi: 10.1016/s0895-4356(03)00120-3. [DOI] [PubMed] [Google Scholar]
- 36.Efron B. Estimating the Error Rate of a Prediction Rule: Improvement on Cross-Validation. J Am Stat Assoc. 1983;78:316–331. [Google Scholar]
- 37.Breiman L, Spector P. Submodel Selection and Evaluation in Regression. The X-Random Case. Int Stat Rev. 1992;60:291–319. [Google Scholar]
- 38.Martens H, Næs T. Chichester: Wiley; 1989. Multivariate Calibration. [Google Scholar]
- 39.Næs T, Mevik B-H. Understanding the collinearity problem in regression and discriminant analysis. Journal of Chemometrics. 2001;15:413–426. [Google Scholar]
- 40.Weisberg S. New York: Wiley; 1985. Applied Linear Regression. [Google Scholar]