Abstract
Dendritic cells (DC) are important adjuvants for cancer vaccines. Immature dendritic cells (iDCs) are often produced by the stimulation of peripheral blood monocytes with IL-4 and GM-CSF. For many applications iDCs are treatment with cytokines or inflammatory signals to produce mature DCs (mDCs). Immature DCs are often treated ex-vivo with LPS and IFN-γ to produce mature DCs for clinical therapy. The purpose of this study was to determine if the DC maturation cocktail LPS plus IFN-γ could be improved by the addition of two other DC maturation agents IL-1β and TNF-α. Peripheral blood mononuclear cells (PBMCs) were collected from 6 healthy subjects. Monocytes were isolated from the PBMC concentrates by elutriation and were incubated for 3 days with GM-CSF and IL-4 to produce iDCs. Immature DCs from each subject were divided into 3 and were incubated for 24 hours with LPS plus IFN-γ; LPS, IFN-γ plus IL-1β; or LPS, IFN-γ, IL-1β plus TNF-α to produce mDCs. The DCs were compared by measuring the expression of co-stimulator and antigen presenting molecules (CD80, CD83, CD86, and HLA-DR) by flow cytometry, cytokine production (IL-12p70 and IL-10) by ELISA and global gene expression using an oligonucleotide microarray. There were no differences in the expression of co-stimulatory molecules, HLA-DR and CCR7 and production of IL-12p70 among the mDCs produced with the 3 cocktails. Global gene expression analysis found that the expression of 9,576 genes differed between the iDCs and mDCs, but the expression of only 13 differed among the 3 different groups of mDCs. There was no benefit of adding IL-1β and TNF-α to LPS and IFN-γ to order to produce mDCs.
INTRODUCTION
Adoptive immune therapy is being used to treat many types of cancer. Adaptive immune responses are induced, coordinated and regulated by dendritic cells (DCs).1 DCs are increasingly being used as adjuvants for cancer vaccination. They are potent antigen presenting cells that recognize, process, and present foreign antigens to T-cells.2;3 Adoptive immune cancer therapy protocols often use antigen-loaded DCs alone or in combination with other leukocytes in clinical trials.4 Clinical vaccine protocols that make use of DCs as adjuvants require the administration of several million DCs. DCs are present in the peripheral blood, however, the concentration of DCs in the blood is too low to make their collection practical for clinical applications.5 Instead, DCs are manufactured from either hematopoietic stem cells or peripheral blood monocytes.5;6
DCs are most frequently produced from peripheral blood mononuclear cells (PBMCs). Large quantities of PBMCs can be collected by leukapheresis and monocytes can be isolated with high purity from the PBMC concentrates by adherence, elutriation, or selection using immunomagnetic beads.7;8 The monocytes are then incubated with GM-CSF and IL-4 to produce DCs. The DCs produced by incubation of monocytes with GM-CSF and IL-4 are quiescent or immature. These immature DCs (iDCs) can be matured using various exogenous stimuli including cytokines, growth factors, costimulatory molecules and inflammatory signals.4;9
Some adoptive immune therapy studies have used iDCs. However, mature DCs (mDCs) are increasingly being used because they express increased levels of co-stimulatory molecules, produce greater quantities of cytokines and are superior in the stimulation of cytotoxic T-cell response compared to iDCs.1 mDCs with effective anti-tumor responses have the following features: high levels of expression of co-stimulatory molecules CD80, CD83, and CD86,10 high migratory activity in response to the lymphoid organ chemokines CCL19 and CCL21,11 and high capacity to produce interleukin-12p70 (IL-12p70) which is the major factor driving Th1 reactions.9;12
To produce mDCs, imDCs are treated with various stimuli known to induce DCs maturation.12–14 DC maturation can be induced using inflammatory signals such as tumor necrosis factor (TNF), interleukin-1beta (IL-1β) or bacterial derivatives such as lipopolysaccharide (LPS). Double stranded RNA, interferons and postaglandins can also induce DC maturation.15;16 The “conventional” DC maturation cocktail has been IL-1β, IL-6, TNF-α, and prostaglandin E2 (PGE2).17 This cocktail generates mDCs with high co-stimulatory and migratory functions but they produce relatively low levels of IL-12.4 As a result, a number of an ex vivo maturation cocktails have been developed for clinical use that generate mDC that produce higher levels of IL-12p70 than IL-1β, IL-6, TNF-α and PGE2.12
LPS is a toll-like receptor (TLR) ligand and a microbial compound originating from the outer membrane of the gram-negative bacterial cell wall. It is being used as a DC maturation stimulant since it is a good inducer of IL-12p70 production.13;14 LPS stimulates Toll-like receptor 4 (TLR4), mediates the activation of nuclear factor-kappa B (NF-kB) and mitogen activated protein kinases (MAPKs), and finally initiates DC maturation.18;19 Another important DC maturation stimulant is IFN-γ which enhances the TLR signaling pathway and up-regulate DC inflammatory cytokines. It has been also reported that IFN-γ can significantly augment IL-12p70 production.20
LPS and IFN-γ is being used to produce mDCs for clinical cancer vaccines to treat patients with childhood solid tumors and breast cancer.21;22 HER2/neu-positive breast cancer patients have been vaccinated with peptide-loaded mDC that were matured with LPS and IFN-γ. This vaccine induced HER2/neu-specific T cell responses and measurable decreases in tumor volume.22 In addition, GMP grade LPS and IFN-γ is being used to produce mDC from peripheral blood moncytes treated with GM-CSF plus IL-4 for two clinical protocols at our institution.
In order to determine if the maturation of DCs by the combination of LPS plus IFN-γ can be improved by the addition of IL-1β or IL-1β plus TNF-α, we compared mDCs produced from peripheral blood monocytes with these agents. Monocyte derived DCs were prepared by the elutriation of monocytes from PBMCs followed by incubation in GM-CSF and IL-4 to produce iDCs. To produce mDCs the iDCs were stimulated with LPS plus IFN-γ; LPS, IFN-γ, plus IL-1b; and LPS, IFN-γ, IL-1β plus TNF-α. To evaluate the effectiveness of these DC maturation cocktails the three types of mDCs were compared by measuring IL-12p70 and IL-10 production by ELISA and the expression of HLA-DR and co-stimulatory molecules CD80, CD83, and CD86 by flow cytometry. We also compared the mDCs using global gene expression analysis and confirmed the expression levels of important genes using quantitative PCR (qPCR). We found no differences in the three types of mDCs and demonstrated that the addition of inflammatory cytokines IL-1β and TNF-α was of no benefit to the LPS and IFN-γ maturation cocktail.
Materials and Methods
PBMC collection and sample preparation
PBMC concentrates were collected using a CS3000 Plus blood cell separator (Baxter Healthcare Corp., Fenwal Division, Deerfield, IL) from 6 healthy donors in the Department of Transfusion Medicine (DTM), Clinical Center, National Institutes of Health (NIH). All donors signed an informed consent approved by a NIH Institutional Review Board (IRB). Monocytes are isolated from the PBMC concentrate by elutriation on the same day of collection.
Elutriation
Continuous counter-flow elutriation was performed using the Elutra® (Gambro BCT, Lakewood, CO) using the automatic mode according to the manufacturer’s recommendations. The monocyte fraction was suspended in HBSS without magnesium, calcium, and phenol red (Gambro BCT, Lakewood, CO).
DC preparation, maturation, and harvest
The elutriated monocytes from each donor were suspended at 6.7 × 106/mL with RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Invitrogen), 2 mM L-glutamine (Invitrogen), 1% nonessential aminoacid (Invitrogen), 1% pyruvate (Invitrogen), 100 units/mL penicillin/streptomycin (Invitrogen), and 50 µM 2-mercaptoethanol (Sigma, St Louis, MO). A total of 10 mL monocytes suspensions were cultured in T25 culture flasks (Nalge Nunc International, Rochester, NY) overnight in humidified incubator containing 5% CO2 at 37°C. On Day 1, 2000 IU/mL human IL-4 (R&D Systems, Minneapolis, MN) and 2000 IU/mL GM-CSF (R&D Systems) were added. On Day 3, an additional 2000 IU/mL IL-4 and GM-CSF were added. On day 4, to induce DC maturation, 3 kinds of cocktails were added. Cocktail 1 was a mixture of 100 mg/mL LPS (Sigma) and 1000 IU/mL IFN-γ (R&D Systems). Cocktail 2 contained 100 mg/mL LPS, 1000 IU/mL IFN-γ, and 2000 IU/mL IL-1β (R&D Systems). Cocktail 3 contained 100 mg/mL LPS, 1000 IU/mL IFN-γ 2000 IU/mL IL-1β, and 1000 IU/mL TNF-α (R&D Systems). After addition of maturation cocktails, DCs were harvested at 0 hours (h) and 24 h. To retrieve adherent cells, 2mM EDTA-PBS was added to each flask on ice. Harvested cells were pelleted, washed twice with HBSS, and resuspended in the RPMI 1640. After harvesting, total cell number and viability were measured microscopically by the trypan blue test.
Flow cytometeric analysis
For evaluation of the purity of the elutriated monocytes they were tested with CD14-PE, CD19-FITC, CD3-PE-Cy5, and CD56-APC (all Becton Dickinson, San Jose, CA) and isotype controls (Becton Dickinson Mountain View, CA). To confirm the maturation of the DCs, the harvested DCs were labeled with CD80-FITC, CD83-PE, CD86-FITC, HLA-DR-PE-Cy5, and CD14-APC (all Becton Dickinson) and isotype controls (all Becton Dickson). The flow cytometry acquisition and analysis were performed with a FACScan with CellQuest software (Becton Dickinson, Mountain View, CA)
Dendritic cell cytokine generation
To measure DC cytokine production after restimulation, iDC and mDCs from each of three cocktails (100,000 cells/ml) were co-incubated with 50,000 cells/ml of adherent mouse fibroblasts transfected to express human CD40-Ligand (CD40L-LTK) in 48 well plates. This cell line was kindly provided by Dr. Kurlander (Department of Laboratory Medicine, Clinical Center, National Institutes of Health, Bethesda, MD). As controls, imDC and mDCs were cultured without CD40L-LTK cells. At 0 h and after 24 h, the supernatant was collected for analysis of human IL-12p70 (eBioscience, San Diego, CA) and IL-10 (eBioscience) by ELISA.
RNA preparation, RNA amplification and labeling for oligonucleotide microarray
Total RNA was extracted from the harvested DCs using the Trizol (Invitrogen, Carlsbad, CA). RNA integrity was assessed using the Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany). Total RNAs (3 µg) from DCs were amplified into anti-sense RNA (aRNA). Also, total RNA from PBMCs pooled from six normal donors was extracted and amplified into aRNA to serve as the reference.23 Pooled reference and test aRNA were isolated and amplified in identical conditions during the same amplification/hybridization procedure to avoid possible interexperimental biases. Both reference and test aRNA were directly labeled using ULS aRNA Fluorescent Labeling kit (Kreatech, Amsterdam, The Netherlands) with Cy3 for reference and Cy5 for test samples.
Whole-genome human 36K oligonucleotide arrays were printed in the Infectious Disease and Immunogenetics Section, DTM, Clinical Center, NIH (Bethesda, MD) using a commercial probe set which contains 35,035 oligonucleotide probes, representing approximately 25,100 unique genes and 39,600 transcripts excluding control oligonucleotides (Operon Human Genome Array-Ready Oligo Set version 4.0, Huntsville, AL, USA). The design is based on the Ensemble Human Database build (NCBI-35c), with a full coverage on NCBI human Refseq dataset (04/04/2005). The microarray was composed of 48 blocks and one spot was printed per probe per slide. Hybridization was carried out in a water bath at 42°C for 18 to 24 hours and the arrays were then washed and scanned on a GenePix scanner Pro 4.0 (Axon, Sunnyvale, CA) at variable photomultiplier tube to obtain optimized signal intensities with minimum (<1% spots) intensity saturation.
Data processing and statistical analyses
The raw data set was filtered according to standard procedure to exclude spots with minimum intensity that arbitrarily was set to an intensity parameter of 200 in both fluorescence channels. If the fluorescence intensity of one channel was great than 200, but the other was below 200, the fluorescence of the low intensity channel was arbitrarily set to 200. Spots with diameters < 20µm and flagged spots were also excluded from the analyses. Then, the filtered data were normalized using Lowess Smoother and retrieved by the BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html) developed at the National Cancer Institute (NCI), Biometric Research Branch, Division of Cancer Treatment and Diagnosis. Hierarchical cluster analysis and TreeView software were used for visualization.24;25 For annotation of genes and functional pathways, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) 2007 software 26 and GeneCards website (http://www.genecards.org/index.shtml).
Gene expression Quantitative PCR
To validate the microarray analysis, 4 genes were selected for Quantitative PCR. Gene expressions for HLA-DRA (Assay ID Hs00219578_m1), HLA-DRB1 (Assay ID Hs99999917_m1), CCR7 (Assay ID Hs99999080_m1), and CD86 (Assay ID Hs00199349_m1) were quantified by TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA) according to manufacturers’ protocol and normalized by GAPDH (Assay ID Hs99999905_m1). PCR amplification of target genes and quantification of the amount of PCR products were performed by ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Differences in expression were determined by the relative quantification method; the Ct values of the test genes were normalized to the Ct values of endogenous control GAPDH. The fold change was calculated using the equation 2 −ΔΔCt.
Results
Donors and Products
The age of the 6 healthy donors ranged from 19 to 49 years, all were male, three were Caucasian and three were African American (Table 1). A PBMC concentrate was collected from each donor and monocytes were isolated from the PBMC concentrates by elutriation. The elutriated monocyte products contained 1.64±0.59×109 cells (range 1.38−2.66×109) that were 91±4% (range 86% to 96%) in purity (Table 1).
Table 1.
Characteristics of the peripheral blood mononuclear cell donors and the elutriated monocyte product
| Donor Number |
Age (years) |
Gender | Race | Elutriated monocyte product total and differential WBC counts |
||
|---|---|---|---|---|---|---|
| WBC Count (total × 109) |
Lymphocytes (%) |
Monocytes (%) |
||||
| 1 | 30 | M | Black | 1.92 | 3 | 94 |
| 2 | 30 | M | Caucasian | 2.10 | 6 | 87 |
| 3 | 49 | M | Black | 1.60 | 14 | 86 |
| 4 | 34 | M | Caucasian | 2.95 | 7 | 90 |
| 5 | 22 | M | Caucasian | 2.09 | 3 | 96 |
| 6 | 19 | M | Black | 1.00 | 4 | 92 |
Yield, viability, and maturation of DCs
The monocytes were cultured in GM-CSF and IL-4 for three days to produce imDCs. The yield of iDCs represented 60.0±0.3% (range 49.5 to 85.0%) of the original monocytes. The viability of the iDC was 97.6 ± 1.9% (range 90.4 to100%).
iDCs from each donor were divided into three and incubated with LPS plus IFN-γ; LPS, IFN-γ plus IL-1β; or LPS, IFN-γ, IL-1β plus TNF-α to produce mDCs. There was no difference in yield and viability among the mDCs produced with the three maturation cocktails. The yield of harvested mDCs were 55.6±6.4% of the monocytes (range 51.0 to 80.0%) for cocktail 1, 54.4±7.4% (range 49.5 to 75.0%) for cocktail 2, and 55.0±7.0% (range 55.0 to 85.0%) for cocktail 3. The viabilities of the mDCs were 94.5±2.0% (range 92.3 to 100%), 95.4±1.6% (range 95.1 to 100%), and 95.5 ± 2.2% (range 90.4 to 100%) for cocktail 1, 2, and 3 respectively.
The expression of DC maturation markers CD80, CD83, CD86, and HLA-DR as assessed by flow cytometry was greater on the mDCs than on iDCs (Table 2). However, there was no difference in the expression of these antigens among mDCs produced with the 3 maturation cocktails (Table 2).
Table 2.
Comparison of dendritic cell (DC) expression of CD16, CD80, CD83, CD86, and HLA-DR antigens by DC maturation cocktail and duration of exposure to each cocktail
| Antigen Expression (Percent of all cells) | |||||||
|---|---|---|---|---|---|---|---|
| Cocktail | Time | CD80 | CD83 | CD86 | CD83 & CD86 | HLA-DR | CD14 |
| LPS+IFNγ | 0h (iDCs) | 29 ± 10 | 37 ± 12 | 26. ± 13 | 21 ± 15 | 81 ± 10 | 0.2 ± 0.1 |
| 24h (mDCs) | 90 ± 8 | 94 ± 6 | 97 ± 2 | 98 ± 1 | 98 ± 1 | 0.1 ± 0.1 | |
| LPS+IFNγ+IL-1β | 0h (iDCs) | 30 ± 10 | 38 ± 13 | 29 ± 13 | 22 ± 16 | 82 ± 10 | 0.2 ± 0.1 |
| 24h (mDCs) | 90 ± 8 | 93 ± 7 | 97 ± 2 | 98 ± 1 | 99 ± 2 | 0.2 ± 0.3 | |
| LPS+IFNγ+IL-1β+TNF-α | 0h (iDCs) | 30 ± 11 | 36 ± 12 | 28 ± 14 | 20 ± 16 | 80 ± 12 | 0.1 ± 0.1 |
| 24h (mDCs) | 87 ± 10 | 93 ± 6 | 96 ± 1 | 97 ± 3 | 97 ± 1 | 0.2 ± 0.2 | |
Expression was measured by flow cytometry.
The results are expressed as a percentage of all cells.
iDCs = immature dendritic cells
mDCs = mature dendritic cells
h = hours
Dendritic cell cytokine generation
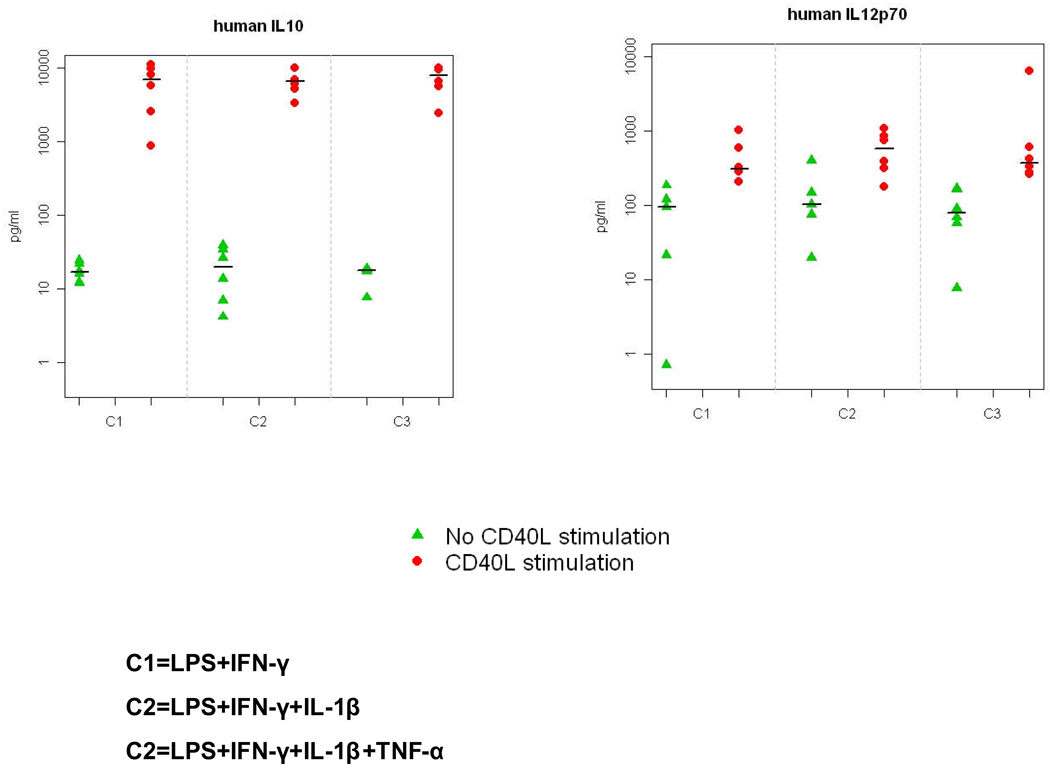
mDCs produced IL-12p70 in response to CTK-CD40L stimulation, but there was no significant difference at IL-12p production among the three mDCs (Figure 1, ANOVA p=0.540). mDCs stimulated with CTK-CD40L also produced IL-10 but there was no significant difference in IL-10 secretion among the three types of mDCs (Figure 1, ANOVA p=0.429).
Figure 1.
IL-12p70 and IL-10 secretion by mature dendritic cells (mDCs). DC production IL-10 (left panel) and IL-12p70 (right panel) was measured by ELISA before and in response to stimulation by mouse fibroblasts transfected to express human CD40-ligand (CD40L-LTK). The production of IL-10 and IL-12p70 by all three types of mDCs increased with CD40-ligand stimulation. There were no significant differences in the quantities of IL-10 and IL-12p70 produced among all types of DCs. Abbreviations: C1 = mDCs produced with LPS plus IFN-γ; C2 = mDC produced with LPS, IFN-γ, plus IL-1β; C3 = mDCs produced with LPS, IFN-γ, IL-1β, plus TNF-α.
Gene expression profiling of DCs by unsupervised hierarchical clustering
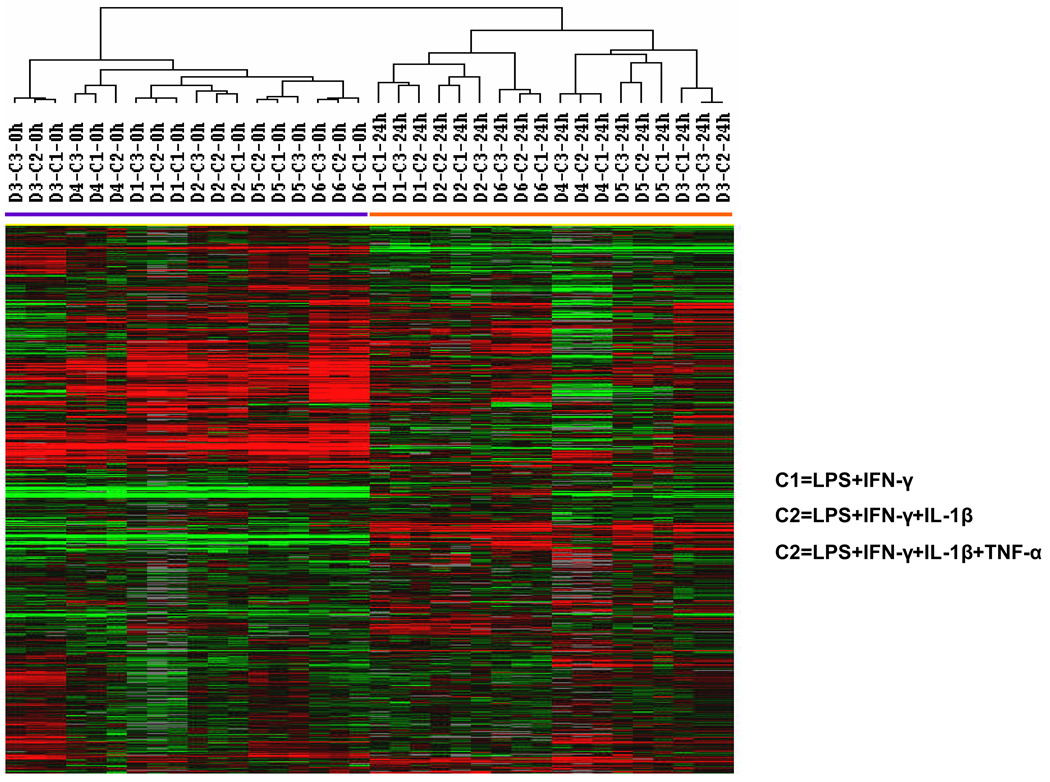
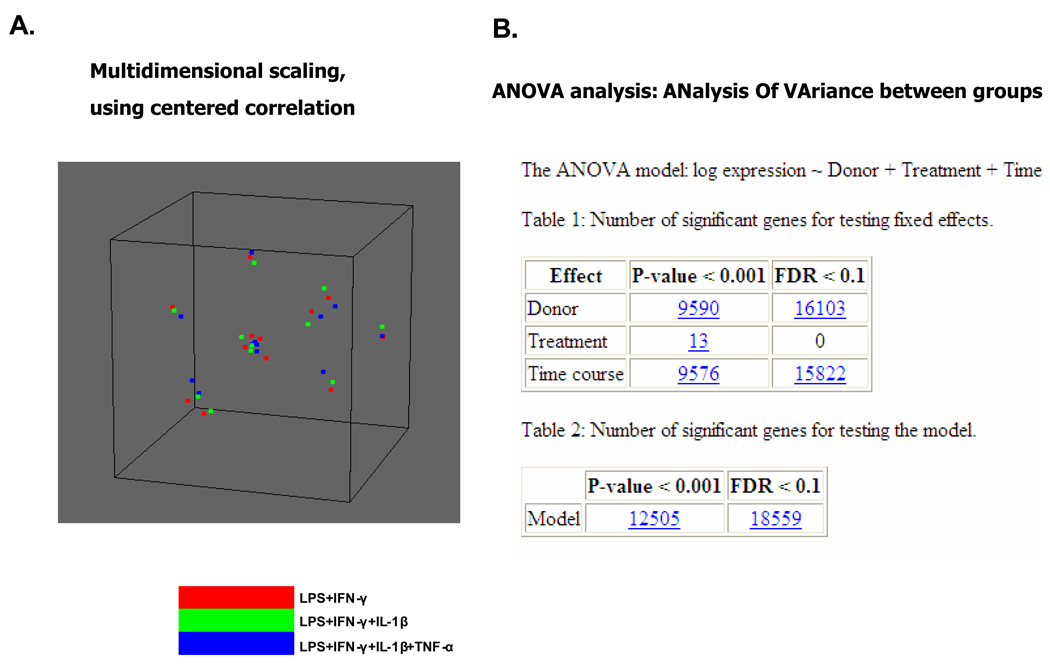
We analyzed the DCs prepared from the 6 donors by global gene expression profiling. The DCs were analyzed before and after the addition of the 3 maturation cocktails. Among the more than 36,000 probes in the array, only those expressed by >80% of DCs were selected for analysis. The resulting 14,609 genes were analyzed by unsupervised hierarchical clustering. The 36 DC samples cells were grouped into 2 clusters: one with all 18 iDC samples (0 h of maturation) and the other with all 18 mDC samples (24 h of maturation) (Figure 2). Among the mDCs, there was no segregation of samples according to maturation cocktail (Figure 2). Similarly, a multidimensional scaling analysis (BRB-ArrayTools) did not group DCs according to maturation agent (Figure 3A). ANOVA analysis revealed that differences in the expression of 9,576 genes could be attributed to DC maturation (time course), 9,590 due to differences among donors, and only 13 to the type of maturation (treatment).This shows that donor effects and time course effects are much more significant than differences in maturation cocktails (p≤0.001) (Figure 3B). Among the 13 genes whose expression was related to type of maturation cocktail, none were related to DC function.
Figure 2.
Global gene expression analysis of immature and mature DCs. Immature DCs (iDCs) and mature DCs (mDCs) produced from 6 healthy subjects were analyzed with by an oligonucleotide expression microarray with greater than 36,000 probes. The 14,609 genes that remained after selecting only those expressed by >80% of samples were analyzed by hierarchical clustering of Eisen. One cluster (shown by the purple bar) contained all of the iDCs (0h) and the other all of the mDCs (24h). The maturation cocktails are C1 = LPS plus IFN-γ; C2 = LPS, IFN-γ, plus IL-1β; C3 = LPS, IFN-γ, IL-1β, plus TNF-α.
Figure 3.
Panel A. A multidimensional scaling plot of gene expression profiles of mature DCs. Mature DCs produced from 6 healthy subjects were analyzed with multidimensional scaling by using BRB-Arrays Tools. Each spot represents mDCs produced by each of the 3 maturation cocktails from the 6 donors. The DCs did not group according to type of maturation cocktail.
Panel B. ANalysis Of VAriace(ANOVA).Number of genes whose expression differed among maturation cocktails, donors and duration of treatment with the maturation cocktails. Immature DCs and mature DCs produced from 6 healthy subjects were analyzed with by an oligonucleotide expression microarray with greater than 36,000 probes.
We further compared the gene expression between iDC and mDC. There were 2,905 differently expressed genes; 1,218 genes we up-regulated in mDCs and 1,687 were up-regulated in iDCs. Thirty representative mononuclear phagoctye genes whose expression differed significantly between iDCs and mDCs are shown in table 3. The genes encoding co-stimulatory molecules CD80, CD83 and CD86 were all up-regulated in mDCs and the expression of CD83 was increased the most. The expression of HLA Class I genes were also increased in mDCs, but those of HLA Class II genes were decreased (Table 3). The expression of two other markers of mDCs, CCR7 and CD40, were also increased in mDCs.
Table 3.
Mononuclear phagocyte genes that were significantly up- or down-regulated in mature DCs (mDCs) produced with 3 different maturation cocktails: LPS and IFN-γ (C1); LPS, IFN-γ and IL-1β (C2); and LPS, IFN-γ, IL-1β and TNF-α (C3)
| Genes whose expression was increased in mDCs | Genes whose expression was decreased in mDCs | ||||||
|---|---|---|---|---|---|---|---|
| Gene expression fold change* | Gene expression fold change* | ||||||
| Gene | C1 | C2 | C3 | Gene | C1 | C2 | C3 |
| CCL5 | 104.00 | 86.82 | 75.33 | CCR1 | −2.31 | −2.43 | −2.90 |
| CXCL10 | 20.85 | 19.38 | 18.31 | CCR2 | −3.98 | −3.92 | −3.92 |
| CXCL11 | 3.84 | 2.09 | 3.59 | AP2A2 | −2.36 | −2.66 | −2.61 |
| CCR7 | 19.98 | 15.83 | 16.53 | CD206/ MRC1 | −26.25 | −25.88 | −28.97 |
| CXCR4 | 4.10 | 2.80 | 2.73 | HLA-DRA | −3.16 | −2.96 | −3.36 |
| IL1B | 4.52 | 4.45 | 4.15 | HLA-DRB1 | −2.21 | −2.49 | −2.45 |
| IL6 | 3.73 | 5.86 | 5.19 | HLA-DMA | −3.08 | −3.19 | −3.47 |
| CD40 | 2.42 | 2.58 | 2.65 | HLA-DPA1 | −4.18 | −4.06 | −4.83 |
| CD80 | 2.84 | 3.40 | 4.26 | HLA-DPB1 | −3.86 | −4.22 | −5.01 |
| CD83 | 22.01 | 17.37 | 23.58 | HLA-DQB1 | −2.06 | −1.94 | −2.02 |
| CD86 | 1.45 | 1.80 | 1.84 | CD209/CDSIGN | −2.15 | −2.15 | −2.35 |
| ICAM1 | 5.19 | 4.64 | 4.25 | CLEC10A | −38.08 | −37.76 | −36.62 |
| LAMP3 | 6.44 | 4.97 | 5.78 | CD1B | −5.34 | −5.70 | −6.22 |
| HLA-A | 1.51 | 1.63 | 1.53 | CD1C | −45.51 | −67.60 | −64.27 |
| HLA-B | 2.82 | 2.23 | 2.56 | CD14 | −1.66 | −1.71 | −2.10 |
C1 = LPS and IFN-γ
C2 = LPS, IFN-γ, and IL-1β
C3 = LPS, IFN-γ, IL-1β and TNF-α
compared to iDCs
The expression of the antigen processing and presentation molecules ICAM1/CD54 and LAMP3/CD208 were increased in all three types of mDCs. In addition, the expression of several inflammatory cytokines genes were up-regulated in all three types of mDCs including: CCL5, CXCL10, CXCL11, IL1B and IL6. Two chemokine receptors that are important for migration of DC to T-cell area of lymphoid tissue, CCR7 and CXCR4, were also up-regulated in mDCs.
In contrast the expression for inflammatory cytokine receptor genes CCR1 and CCR2 were down-regulated in mDCs. Several molecules involved with antigen uptake were also down-regulated including: AP2A2, MRC1/CD206, CLEC10A/CD301, and CDSIGN/CD209. The expression of CD1 the non-polymorphic MHC-class I-like amphipathic lipid antigen presenting molecular was decreased on mDCs as was the endotoxin receptor CD14.
Gene expression of quantitative RT-PCR
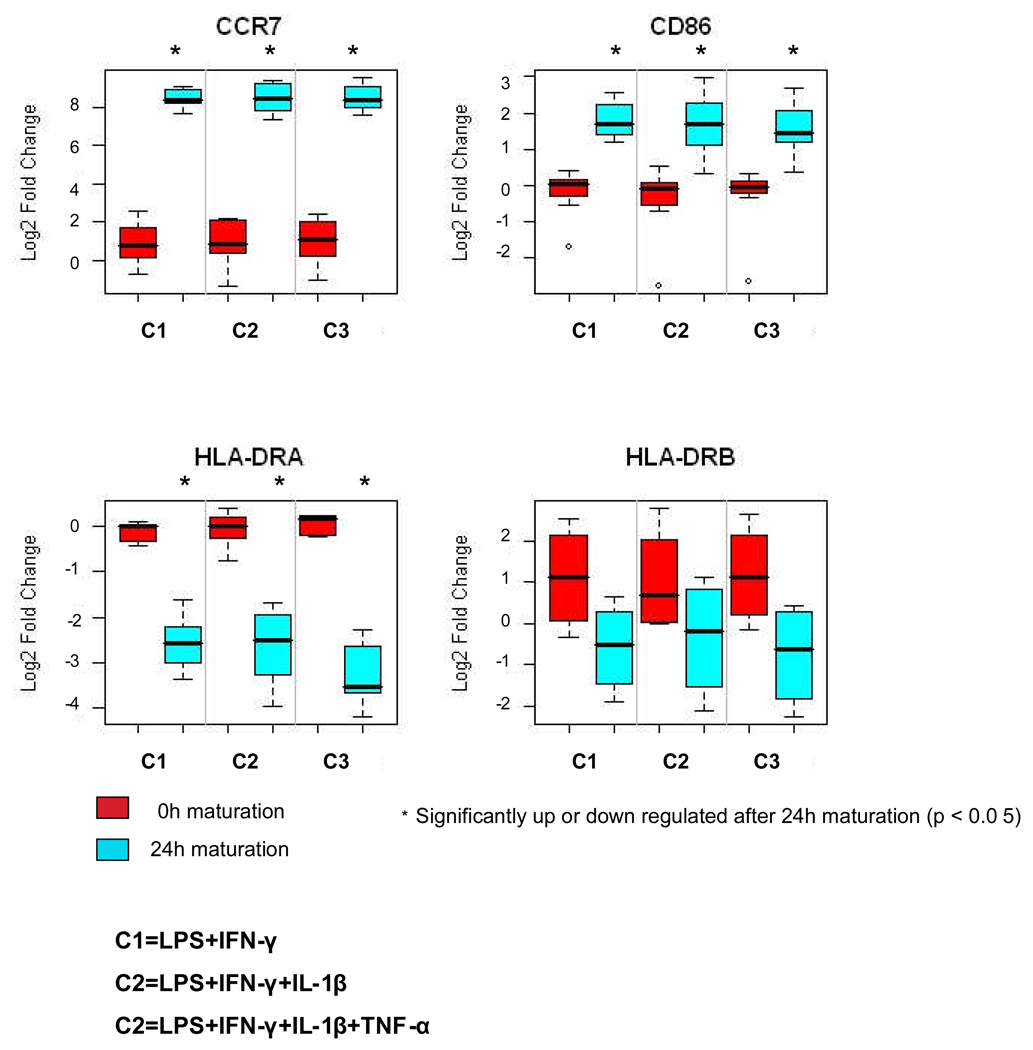
To validate our microarray data, we measured the expression of representative genes using quantitative RT-PCR. The relative quantities of CCR7 and CD86 transcripts produced by mDCs were significantly greater than those produced by iDCs as measured by qRT-PCR (Figure 4). In contrast, the levels of HLA-DRA transcripts in mDCs were significantly less than in iDCs. The levels of HLA-DRB did not differ between iDCs and mDCs (Figure 4). The expression of CCR7, CD86, HLA-DRA and HLA-DRB1 showed no difference among the three types of mDCs (ANOVA p=0.314 for CCR7; p=0.511 for CD86; p=0.256 for HLA-DRA; p=0.758 for HLA-DRB). These qRT-PCR results were consistent with those obtained with the oligonucleotide gene expression microarray.
Figure 4.
Analysis of the expression of CCR7, CD86, HLA-DRA, and HLA-DRB genes by immature and mature DCs by quantitative real-time PCR (RT-PCR). Quantitative real-time PCR confirmed the expression of 2 genes found by microarray analysis to be up-regulated after 24h maturation: CCR7 and CD86, and 2 genes down-regulated after 24h maturation: HLA-DRA and HLA-DRB. The results were normalized with endogenous control GAPDH and the fold change relative to the calibrator was calculated by the equation2 −ΔΔCt. The y-axis indicates the Log2-transformed fold change. The levels of expression of these genes by immature DCs (iDCs) is shown by the red bars and by mature DCs (mDCs) produced by each of the 3 maturation cocktails by the blue bars. The expression of CCR7, and CD86 was significantly greater in mDCs, but there was no difference in the expression of CCR7 and CD86 among the three types of mDCs. The expression of HLA-DRA and HLA-DRB were significantly less in mDCs compared to iDCs, but there was no difference in expression of HLA-DRA and HLA-DRB among the three types of mDCs. The maturation cocktails are C1 = LPS plus IFN-γ; C2 = LPS, IFN-γ, plus IL-1β; C3 = LPS, IFN-γ, IL-1β, plus TNF-α.
Discussion
We compared mDCs produced with the maturation cocktail LPS and IFN-γ with those produced with these agents plus IL-1β or both IL-1β and TNF-α to determine if more potent mDCs could be produced. All assays we used found no difference in mDCs produced with the three DC maturation cocktails. All three maturation cocktails produced mDCs that expressed greater levels of commonly used markers of DC maturation: CD80, CD83, CD86 and CCR7. The expression of co-stimulator molecules CD80, CD83 and CD86 as measured by both gene expression profiling and flow cytometry were all increased in mDCs compared to iDCs and there were no differences in the expression of these proteins and genes among the three types of iDCs. Global gene expression analysis also found that the expression of CCR7 was increased in mDCs. We also confirmed the increased levels mDC of gene expression of CD86 and CCR7 by qRT-PCR. In addition, there were no differences in the production of IL-12p70 and IL-10 by the mDCs produced with the 3 different cocktails.
The expression of HLA antigens are also of used as markers of DC maturation. The expression of HLA Class I genes were increased in all three types of mDCs. The expression of HLA-DR antigen as assessed by flow cytometry was increased in all 3 mDCs. In contrast the expression of several HLA Class II genes was decreased in the mDCs. However, it has had been reported that MHC II is stored in late endocytic compartments of iDCs and during maturation, MHC II synthesis is down-regulated but MHC II is recruited to the plasma membrane and its expression is increased. 27 23
Although the traditional assays used to assess DCs, flow cytometry and ELISA, did not reveal any differences between the three groups of mDCs, the function of DCs is complex and not completely understood. It is likely that DC function is influenced by many other factors in addition to the expression of CD80, CD83, CD86, HLA Class I, HLA Class II and CCR7 and the production of IL-12p70 and IL-10. To more thoroughly compare the mDCs produced with the three maturation cocktails, we analyzed the mDCs using gene expression microarrays with more than 36,000 probes. Global gene expression analysis allows for the comparison of the expression of genes known to affect the function of DCs and those which may affect the function of DCs but have not been yet identified as important DC genes. While we found that 9,576 genes were differentially expressed between iDCs and mDCs, the expression of only 13 genes differed between the three groups of mDCs. Multivariate analysis also suggested that there were more difference between DCs from different donors than between maturation cocktails.
The levels of IL-12 produced by the mDCs were lower than in some reports but similar to those produced by DCs matured with conventional maturation cocktails.28 These differences may be due to differences in the methods that were used to isolate peripheral blood monocytes and to produce the DCs since IL-12p70 production by mDCs is dependent on method of monocyte isolation, culture media, the presence of serum and donor/patient factors.28 We isolated monocytes from peripheral blood leukocytes by elutriation, while many other studies isolated monocytes from peripheral blood leukocytes by adhesion to plastic surfaces. We also produced iDC by culture with GM-CSF and IL-4 for 3 days compared to 5 to 7 days for some studies. The duration of exposure of DCs to LPS and IFN-γ also effects production of IL-12p70: after 8 hours of exposure IL-12p70 production begins to fall.29;30
Although some studies have used functional assays such as mixed leukocyte reactions in addition to the measurement costimulatory molecule expression and IL-12p70 production to assess DCs, we elected to use gene expression analysis in place of functional analysis. Our microarrays provide a broad analysis of the transcriptome. The fact that results of our comparison of the three DC maturation cocktails by analysis of costimulatory molecule expression and IL-12p70 production and gene expression profiling yielded similar results suggests that gene expression profiling is an effective method for comparing the potency of DCs. However, further comparisons of DC analysis by gene expression profiling with expression of costimulatory molecules, IL-12p70 production and other function assays are needed.
These results show that there is no benefit of adding IL-1β and TNF-α to the LPS plus IFN-γ DC maturation cocktail. There was no difference among three mDC as assessed by any of the measures we used. The lack of an effect of the addition of TNF-α and IL-1β may be due to a much greater DC maturation effect of LPS and IFN-γ compared to TNF-α and IL-1β. Alternatively, added IL-1β and TNF-α may provide no additional benefit since the production of both of these factors was induced by LPS and IFN-γ stimulation. The expression of IL-1β and TNF-α was up-regulated (4.3-fold and 1.5-fold respectively) during DC maturation for all three cocktails. In addition, the expression of several TNF-α and IL-1β induced genes were increased by LPS and IFN-γ stimulation including TNFAIP3/PTX (8.8-fold) and TNFAIP6 (10.7-fold). These findings support the concept that autogenous TNF-α and IL-1β are produced by DCs stimulated with cocktails containing LPS and INF-γ and exogenous TNF-α and IL-1β may not be necessary.
Acknowledgements
The authors thank the staff of the Dowling Apheresis Clinic, DTM, Clinical Center, NIH, for collecting the cells and the Cell Processing Laboratory, DTM, Clinical Center, NIH for preparing the elutriated cells.
Footnotes
Financial Disclosure: The authors have declared there are no financial conflicts of interests in regards to this article.
Reference List
- 1.Banchereau J, Palucka AK. Dendritic cells as therapeutic vaccines against cancer. Nat.Rev.Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 2.Young JW, Merad M, Hart DN. Dendritic cells in transplantation and immune-based therapies. Biol.Blood Marrow Transplant. 2007;13:23–32. doi: 10.1016/j.bbmt.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Randolph GJ, Jakubzick C, Qu C. Antigen presentation by monocytes and monocyte-derived cells. Curr.Opin.Immunol. 2008;20:52–60. doi: 10.1016/j.coi.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicolette CA, Healey D, Tcherepanova I, et al. Dendritic cells for active immunotherapy: optimizing design and manufacture in order to develop commercially and clinically viable products. Vaccine. 2007;25 Suppl 2:B47–B60. doi: 10.1016/j.vaccine.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Dauer M, Obermaier B, Herten J, et al. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J.Immunol. 2003;170:4069–4076. doi: 10.4049/jimmunol.170.8.4069. [DOI] [PubMed] [Google Scholar]
- 6.Li YP, Paczesny S, Lauret E, et al. Human mesenchymal stem cells license adult CD34+ hemopoietic progenitor cells to differentiate into regulatory dendritic cells through activation of the Notch pathway. J.Immunol. 2008;180:1598–1608. doi: 10.4049/jimmunol.180.3.1598. [DOI] [PubMed] [Google Scholar]
- 7.Felzmann T, Witt V, Wimmer D, et al. Monocyte enrichment from leukapharesis products for the generation of DCs by plastic adherence, or by positive or negative selection. Cytotherapy. 2003;5:391–398. doi: 10.1080/14653240310003053. [DOI] [PubMed] [Google Scholar]
- 8.Berger TG, Strasser E, Smith R, et al. Efficient elutriation of monocytes within a closed system (Elutra) for clinical-scale generation of dendritic cells. J.Immunol.Methods. 2005;298:61–72. doi: 10.1016/j.jim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Gilboa E. DC-based cancer vaccines. J.Clin.Invest. 2007;117:1195–1203. doi: 10.1172/JCI31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albert ML, Jegathesan M, Darnell RB. Dendritic cell maturation is required for the cross-tolerization of CD8+ T cells. Nat.Immunol. 2001;2:1010–1017. doi: 10.1038/ni722. [DOI] [PubMed] [Google Scholar]
- 11.Dieu MC, Vanbervliet B, Vicari A, et al. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J.Exp.Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mailliard RB, Wankowicz-Kalinska A, Cai Q, et al. alpha-type-1 polarized dendritic cells: a novel immunization tool with optimized CTL-inducing activity. Cancer Res. 2004;64:5934–5937. doi: 10.1158/0008-5472.CAN-04-1261. [DOI] [PubMed] [Google Scholar]
- 13.Ten BA, Karsten ML, Dieker MC, Zwaginga JJ, van Ham SM. The clinical grade maturation cocktail monophosphoryl lipid A plus IFNgamma generates monocyte-derived dendritic cells with the capacity to migrate and induce Th1 polarization. Vaccine. 2007;25:7145–7152. doi: 10.1016/j.vaccine.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Snijders A, Kalinski P, Hilkens CM, Kapsenberg ML. High-level IL-12 production by human dendritic cells requires two signals. Int.Immunol. 1998;10:1593–1598. doi: 10.1093/intimm/10.11.1593. [DOI] [PubMed] [Google Scholar]
- 15.Zobywalski A, Javorovic M, Frankenberger B, et al. Generation of clinical grade dendritic cells with capacity to produce biologically active IL-12p70. J.Transl.Med. 2007;5:18. doi: 10.1186/1479-5876-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figdor CG, de V, I, Lesterhuis WJ, Melief CJ. Dendritic cell immunotherapy: mapping the way. Nat.Med. 2004;10:475–480. doi: 10.1038/nm1039. [DOI] [PubMed] [Google Scholar]
- 17.Jonuleit H, Kuhn U, Muller G, et al. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur.J.Immunol. 1997;27:3135–3142. doi: 10.1002/eji.1830271209. [DOI] [PubMed] [Google Scholar]
- 18.Nakahara T, Moroi Y, Uchi H, Furue M. Differential role of MAPK signaling in human dendritic cell maturation and Th1/Th2 engagement. J.Dermatol.Sci. 2006;42:1–11. doi: 10.1016/j.jdermsci.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Akira S. TLR signaling. Semin.Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Vieira PL, de Jong EC, Wierenga EA, Kapsenberg ML, Kalinski P. Development of Th1-inducing capacity in myeloid dendritic cells requires environmental instruction. J.Immunol. 2000;164:4507–4512. doi: 10.4049/jimmunol.164.9.4507. [DOI] [PubMed] [Google Scholar]
- 21.Dohnal AM, Witt V, Hugel H, et al. Phase I study of tumor Ag-loaded IL-12 secreting semi-mature DC for the treatment of pediatric cancer. Cytotherapy. 2007;9:755–770. doi: 10.1080/14653240701589221. [DOI] [PubMed] [Google Scholar]
- 22.Czerniecki BJ, Koski GK, Koldovsky U, et al. Targeting HER-2/neu in early breast cancer development using dendritic cells with staged interleukin-12 burst secretion. Cancer Res. 2007;67:1842–1852. doi: 10.1158/0008-5472.CAN-06-4038. [DOI] [PubMed] [Google Scholar]
- 23.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat.Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 24.Wang E, Miller LD, Ohnmacht GA, et al. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2002;62:3581–3586. [PMC free article] [PubMed] [Google Scholar]
- 25.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc.Natl.Acad.Sci.U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 27.Wilson NS, El-Sukkari D, Villadangos JA. Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood. 2004;103:2187–2195. doi: 10.1182/blood-2003-08-2729. [DOI] [PubMed] [Google Scholar]
- 28.Butterfield LH, Gooding W, Whiteside TL. Development of a potency assay for human dendritic cells: IL-12p70 production. J.Immunother. 2008;31:89–100. doi: 10.1097/CJI.0b013e318158fce0. [DOI] [PubMed] [Google Scholar]
- 29.Langenkamp A, Messi M, Lanzavecchia A, Sallusto F. Kinetics of dendritic cell activation: impact on priming of TH1, TH2 and nonpolarized T cells. Nat.Immunol. 2000;1:311–316. doi: 10.1038/79758. [DOI] [PubMed] [Google Scholar]
- 30.Huttner KG, Breuer SK, Paul P, et al. Generation of potent anti-tumor immunity in mice by interleukin-12-secreting dendritic cells. Cancer Immunol.Immunother. 2005;54:67–77. doi: 10.1007/s00262-004-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]