Abstract
We present iCluster, a fast and efficient algorithm that clusters a set of images while co-registering them using a parameterized, nonlinear transformation model. The output of the algorithm is a small number of template images that represent different modes in a population. This is in contrast with traditional, hypothesis-driven computational anatomy approaches that assume a single template to construct an atlas. We derive the algorithm based on a generative model of an image population as a mixture of deformable template images. We validate and explore our method in four experiments. In the first experiment, we use synthetic data to explore the behavior of the algorithm and inform a design choice on parameter settings. In the second experiment, we demonstrate the utility of having multiple atlases for the application of localizing temporal lobe brain structures in a pool of subjects that contains healthy controls and schizophrenia patients. Next, we employ iCluster to partition a data set of 415 whole brain MR volumes of subjects aged 18 through 96 years into three anatomical subgroups. Our analysis suggests that these subgroups mainly correspond to age groups. The templates reveal significant structural differences across these age groups that confirm previous findings in aging research. In the final experiment, we run iCluster on a group of 15 patients with dementia and 15 age-matched healthy controls. The algorithm produces two modes, one of which contains dementia patients only. These results suggest that the algorithm can be used to discover sub-populations that correspond to interesting structural or functional “modes.”
Index Terms: Image Registration, Clustering, Population Analysis, Computational Anatomy, Segmentation
I. INTRODUCTION
Today, computational anatomy studies are mainly hypothesis-driven, aiming to identify and characterize structural or functional differences between, for instance a group of patients with a specific disorder and control subjects. This approach is based on two premises: accurate clinical classification of subjects and spatial correspondence across the images. In practice, achieving either can be challenging. First, the complex spectrum of symptoms of neuro-degenerative disorders like schizophrenia and overlapping symptoms across different types of dementia, such as Alzheimer’s disease, delirium and depression, make a diagnosis based on standardized clinical tests difficult [22]. Second, establishing across-subject correspondence in the images is a particularly hard problem constrained by the specifics of the application. A popular technique is to normalize all subjects into a standard space, such as the so-called Talairach space [47], by registering each image with a single, universal template image that represents an average brain [12]. However, the quality of such an alignment is limited by the accuracy with which the universal template represents the population in the study.
With the increasing availability of medical images, data-driven algorithms offer the ability to probe a population and potentially discover subgroups that may differ in unexpected ways. In this paper, we propose and demonstrate an efficient probabilistic clustering algorithm, called iCluster, that
computes a small number of templates that summarize a given population of images,
simultaneously co-registers all the images using a non-linear transformation model,
assigns each input image to a template that best describes the image.
The templates are guaranteed to live in an affine-normalized space, i.e., they are spatially aligned with respect to an affine transformation model. A preliminary version of iCluster was published at the International Conference on Medical Image Computing and Computer Assisted Intervention [42]. This article expands the conference paper with a more detailed the-oretical development and more extensive experimental work.
In our experiments, we demonstrate that the templates computed by the proposed algorithm can be used for various purposes, including constructing multiple atlases for improved segmentation and discovering structural modes of a population. On a data set of 50 brain MR images with manual labels for several temporal lobe structures, we illustrate that the subpopulations computed by iCluster manifest significantly improved average label alignment compared to the clinical sub-populations and the whole population. This result suggests that a multi-template strategy will yield improved segmentation accuracy in an atlas-based framework. In other experiments, we show that the modes of the population discovered by iCluster capture known structural differences and similarities. On a population of 415 brain MRI of subjects aged 18–96 years, the algorithm computed three unique templates that mainly comprised of young subjects (mean age 31), older middle aged subjects (mean age 69) and elderly subjects (mean age 79). In another setting, we demonstrate that the modes discovered by the algorithm reflect the two groups of subjects (with mild dementia and healthy) in the population. These results suggest that iCluster can be used to probe a population of images to discover important structural or functional “modes.”
The remainder of the paper is organized as follows. Section II includes an overview of the literature on atlas construction and inter-subject registration. In Section III, we introduce the generative model and develop our algorithm. Section IV reports experimental results. Section V discusses the advantages and drawbacks of the proposed algorithm, while pointing to future directions of research. Section VI concludes with a summary of contributions.
II. BACKGROUND AND PRIOR WORK
In medical imaging, the term atlas usually refers to a (probabilistic) model of a population of images, with the parameters learned from a training data set [14], [51]. In its simplest form, an atlas is a mean intensity image, which we call a template [6], [12], [53], [54]. Richer statistics, such as intensity variance or segmentation label counts, can also be included in the atlas model [19]. Atlases are used for various purposes including normalization of new subjects for structure and function localization, segmentation or parcellation of certain structures of interest, and group analysis that aims to identify pathology-related changes or developmental trends.
Atlas construction requires a dense correspondence across subjects. Earlier techniques used a single image – either a standard template [12], or an arbitrary subject from the training data set [25] – to initially align images using a pairwise registration algorithm. Other methods focused on determining the least biased template from the training set [31], [37]. A single template approach faces substantial methodological challenges when presented with a heterogeneous population, such as patients and matched normal control subjects in clinical studies. To circumvent this, more recent approaches co-register the group of images simultaneously without computing a group template [46], [58]. Even though these algorithms remove the requirement of a single template, they do not attempt to model the heterogeneity in the population. Recent work [9] presented a method that automatically identified the modes of a population using a mean-shift algorithm. This approach solved pairwise registrations to compute each inter-image distance, which slowed down the algorithm substantially. Furthermore, the multi-modality of the population was not modeled explicitly, making it difficult to extract a representation of the heterogeneous population. An alternative strategy to atlas-based segmentation is to use all training images as the atlas [27]. A new subject is registered with each training image and segmentation is based on a fusion of the manual labels in the training data. This approach is not suitable for anatomical variability studies, where a universal coordinate frame is necessary to identify and characterize group differences and study developmental and pathological trends.
There is a rich range of techniques used to characterize similarities and differences across sub-populations defined by attributes like gender, handedness and pathology. Volume-based [11], [39], [44], voxel-based [4], [15] and deformation-based [5] morphometry methods are commonly used to compare anatomical MRI scans of two or more groups of subjects. Other examples include statistical analysis of fMRI, PET and diffusion data to identify age and disease-related changes in the functional and structural organization of the brain [24], [33]. In these studies, inter-subject correspondence is typically achieved via one of the image registration algorithms discussed above. When faced with a heterogeneous group of healthy and pathological brains, however, establishing inter-subject correspondence is an ambiguous and more challenging problem due to dramatic structural changes associated with the pathology. For instance, defining a similarity measure when certain corresponding regions are missing or unclear, is not straightforward.
Probabilistic atlases are powerful tools used commonly for supervised segmentation [3], [13], [18], [55]. A probabilistic atlas can provide statistics about the frequency of a certain label at a particular location, and topological information like the frequency of two different labels neighboring each other at a particular location and with a certain orientation. Moreover, it can include information about the relationship between labels and image intensities. Given a new image, intensity models, such as a template image, are typically used for spatial normalization. Automatic segmentation is then formulated as an inference problem. Recent joint registration and segmentation frameworks [3], [38] integrate the two steps: spatial normalization is updated based on the current segmentation and vice versa. Most atlas-based segmentation approaches make a strong unimodal assumption on the intensity distribution either when building the atlas, or when segmenting the new image or at both stages. In other words, they assume a homogeneous population, where each subject can be modeled as a deformed and noisy version of a universal template. However, there is growing evidence that population-specific atlases can improve the quality of segmentation [48], [57]. This, we believe, highlights the limitations of a single-template atlas in segmentation applications and points toward a multi-template atlas strategy.
In this paper, we develop a probabilistic framework for joint registration of a set of images into a common coordinate frame, while clustering them into a small number of groups, each represented by a template image. We employ a mixture of Gaussians model and a maximum likelihood framework which we solve using the Generalized Expectation Maximization (GEM) algorithm. A similar approach was independently developed in [1], which provides a rigorous analysis of the maximum a posteriori estimate of the deformable templates using a Gaussian kernel based deformation parametrization. In [1] the application of the framework was limited to 2D images of handwritten digits. In contrast, we focus on high-resolution 3D medical data and employ a B-spline parametrization for the nonlinear transformation, as previously demonstrated in [41]. Furthermore, we present approximate solutions to the template estimation problem that yield fast algorithms applicable to large data sets. Our algorithm can also be viewed as an extension of the approach in [50], which solves the registration problem as an initial, separate step. Our framework leads to a fast, scalable and flexible algorithm that removes the sensitivity of the resulting atlas coordinate frame to the selected target. Moreover, it provides a novel, data-driven way to probe the population for different modes. Analyzing the discovered sub-populations and their representative templates promises to advance our understanding of dominant structural or functional changes due to pathology or development.
III. THE MODEL AND ALGORITHM
We assume that the input images are generated from a small number of templates , where K is known and fixed. Later, we will propose a strategy to automatically determine K from the data. Thus, for each n ∈ {1,…,N}, there exists k ∈ {1,…,K} such that
| (1) |
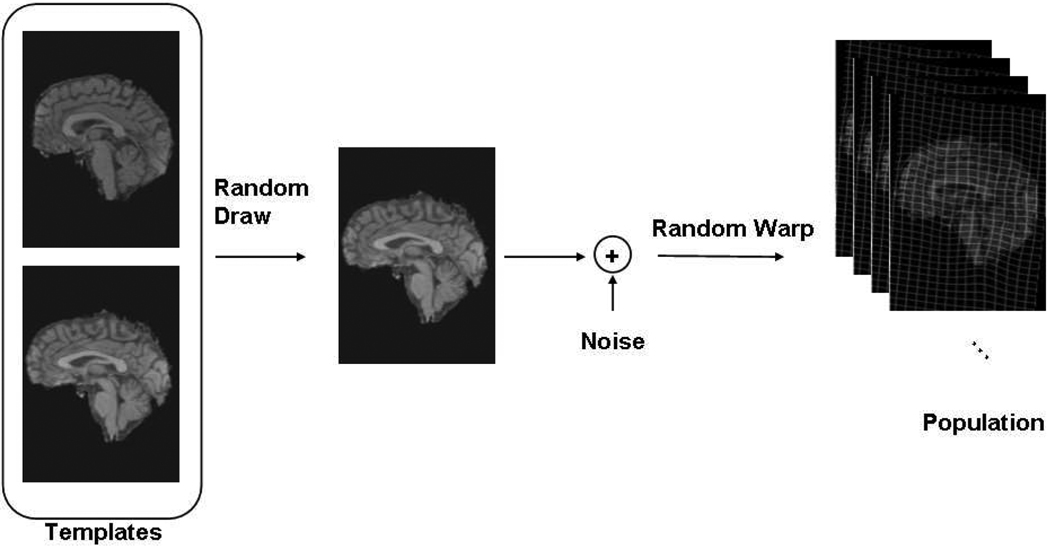
where Φn : ℝ3 ↦ ℝ3 is an admissible, invertible spatial warp, such as a parameterized nonlinear transformation, denotes its inverse, ∈n(·) is a spatially independent, non-stationary Gaussian noise field with zero mean and standard deviation σ(·). The last term models imaging noise, and the independent Gaussian assumption is a commonly used model in the literature [18]. We model the noise parameters in the coordinate frame of the template. Figure 1 illustrates this generative model for two templates.
Fig. 1.
Generative Model that assumes two templates.
Let pk(In; Tk, σ, Φn) denote the conditional probability of the image In given that it is generated by the k’th template, and with the fixed model parameters. This can be computed from Equation (1):
| (2) |
where 𝑁(·; μ, σ) is the Gaussian density with mean μ and standard deviation σ.
Let {πk} denote the prior probabilities of the templates. This distribution governs the initial random draw of templates shown in Figure 1 and models the possibly unbalanced sizes of the clusters. Thus the parameters for the whole model include the templates {Tk}, template priors {πk} and standard deviation image σ(·). The spatial transformations {Φn} can be viewed as hidden random variables, drawn independently for each image from a prior distribution that favors smoother transformations, for instance. In this paper, however, for simplicity we will treat {Φn} as model parameters. We use θ = {{Tk}, {πk}, σ, {Φn}} to denote the pooled set of model parameters and spatial transformations. Marginalizing over all possible template indices, we obtain the probability of observing a particular image In:
| (3) |
A. Generalized EM for Atlas Construction
We formulate the problem of atlas construction as a maximum likelihood estimation:
| (4) |
where L(θ) denotes the log-likelihood of the entire image set evaluated for the parameter θ. We use a Generalized Expectation Maximization (GEM) algorithm to solve Equation (4). For a fixed θ0 = {{Tk0}, {πk0}, σ0, {Φn0}}, using Jensen’s inequality we form a lower bound for L(θ):
| (5) |
where c is a constant that does not depend on θ and qk(In; θ0) is the posterior probability that the image In was generated from the template Tk:
| (6) |
Note that L(θ0) = Q(θ0; θ0). The GEM algorithm iteratively improves this lower bound. Let θ(i) be the guess of θ at iteration i. Computing Q(θ; θ(i)) – or, equivalently qk(In; θ(i)) – is the E-step of iteration i + 1. The M-step updates θ to increase Q(θ; θ(i)). In our formulation, we use a coordinate ascent strategy in the M-step and divide it into two sub-steps: the T-step (“T” stands for template) where we compute the closed form expressions of the template parameters {{Tk}, {πk}, σ} that maximize Q(·; θ(i)); and the R-step (“R” stands for registration) where we numerically solve for the transformation parameters {Φn}. We will use J(Φ, x⃗) to denote the Jacobian field of a transformation Φ(x⃗) with respect to the spatial coordinates and |J| will indicate the determinant of matrix J. Derivations for the T- and R-steps can be found in the Appendix. Here we summarize the algorithm.
- E-step: Given the model parameters from iteration i, the algorithm updates the posterior cluster probabilities:
- , where pk(·) is defined in Equation (2).
- Normalize qk to sum to 1:
(7)
- T-step: Given the posterior probability estimates {qk(In; θ(i))} and transformation parameters , the algorithm updates its estimates of the templates {Tk}, template priors {πk} and standard deviation image σ, for which we derive closed-form expressions:
(8) (9) (10) - R-step: Given the new template parameters , standard deviation image σ(i+1), and memberships {qk(·; θ(i))} the spatial transformations are updated:
(11)
where is the “effective template” (i.e., target image in registration) for image In at iteration (i + 1) and Rσ(·, ·) is the weighted sum of square differences (WSSD) objective function of the R-step. The effective template is a weighted average of the current templates and the weights are membership probabilities. A single, invertible transformation Φn is estimated for each image. Current membership probabilities determine which template the image should be aligning with.(12)
We employ a B-spline transformation model (on an 8 × 8 × 8 control point grid, unless specified otherwise) and a multi-resolution strategy. In general, this transformation model does not guarantee invertibility. In practice, the algorithm checks for invertibility by monitoring the Jacobian terms and terminates when there is a Jacobian determinant value below a certain small positive threshold. Rather than solving the non-convex optimization problem of Equation (11), we perform a single Brent’s method line search [10] based on gradient directions. The line search of each image is done in parallel, since the optimization for one image does not depend on other images. This strategy guarantees that the lower bound on the log-likelihood is improved, if not maximized, at each step; hence the name Generalized EM.
B. Initialization
The above GEM algorithm does not guarantee that the computed template images are in alignment. To introduce a notion of common coordinate frame, we use an initial affine normalization step that co-registers all images using a single dynamic mean image and an affine transformation model. This step is one of the popular co-registration algorithms used in practice. After affine normalization, the GEM algorithm starts with the E-step by computing membership probabilities according to Equation (7). We initialize the template images as a random selection of K different input images, where K is the pre-determined number of templates. In our experiments, we explore various values for K and only report results for the K values that produce robust results across multiple random initializations as discussed in Section IV-A. The template priors are initially assigned to be , and the variance image is initialized to be the sample variance at each voxel after affine normalization. Each R-step is initialized with the transformation parameters from the previous iteration.
C. Gradient Re-normalization
In group-wise registration, one needs to anchor the registration parameters to avoid global transformation drifts across subjects [8], [46], [58]. A natural common coordinate frame can be defined as the average of the population. This natural coordinate frame is computed implicitly by constraining the sum of all displacements across the subjects to be zero. We extend this strategy to the multi-template setting by constraining each point in the common coordinate frame to lie at the average location of corresponding points across the images in each cluster. To impose this constraint, we use the soft memberships qk(·):
| (13) |
Equivalently:
| (14) |
∀x⃗ ∈ Ω, and ∀k. Summing both sides of Equation (14) over k yields
| (15) |
which is the anchoring constraint used by other group-wise registration methods [8], [46], [58].
In a gradient descent optimization strategy, one way of imposing the constraint of Equation (15) is to re-normalize the gradients of the R-step objective function by subtracting the average gradient from all the individual image gradients. Let be a D dimensional row vector that denotes the gradient of the R-step objective function with respect to the transformation parameters of the image In at iteration i + 1. Then, before each update of the transformation parameters, one re-normalizes the gradients:
| (16) |
In the multi-template setting, we extend this re-normalization to satisfy the constraint of Equation (13). We stack all the gradient row vectors to create an N × D matrix G(i+1) and all the membership probabilities qk(In; θ(i)) to create an N × 1 column vector for each k = 1,…,K. First, using the Gram-Schmidt process, we obtain at most K orthonormal vectors from . Using this orthonormal basis, we re-normalize all the gradients as:
| (17) |
where IN × N denotes the N × N identity matrix, Gj denotes the jth column of G and u⃗T denotes the transpose of u⃗. After re-normalization each column of G is orthogonal to for all k. In other words .
D. Determining the optimal number of templates
Determining the optimal number of clusters is a classical problem in unsupervised machine learning, which unfortunately has no universal solution [35], [49]. The problem can be viewed as a specific case of model selection. In general, increasing the number of clusters provides a better fit to the observed data, yet this does not necessarily translate into improved generalization. A standard approach to controlling the generalization ability of the model is to penalize the model complexity. Bayesian Information Criterion (BIC) is a widely-used technique that attempts to achieve this balance [45]. In our setting, BIC (or equivalently Minimum Description Length) can be formulated as minimizing the penalized negative log-likelihood:
| (18) |
where p(In; θ(K)*) is the maximum value of the likelihood in Equation (3) for a fixed number of templates K and |θ(K)| is the total number of model parameters, which in our case is equal to K + KV + V + ND, where D is the number of transformation parameters and V is the number of voxels.
Alternatively, one can use the stability of the resulting model to quantitatively asses the structure in the clustered data, c.f. [7]. In practice, we found it useful to measure the stability of the output against different random initializations. For example, we observed that beyond a particular input K, the computed clustering is significantly less consistent across runs with different initializations. We quantify this consistency using a relative measure defined for each run as:
| (19) |
where qk(In; θ*(r) (K)) denotes the membership probabilities computed in run r and q̄k (In; θ(K)) is the average membership probability over all remaining runs for a fixed input K. To handle the ambiguity in cluster indexing, we maximized Equation (19) over all permutations of indexing of the templates in all runs. This procedure yields a relative consistency value for each run with a fixed input K. Based on the stability criterion, we propose to pick the highest value of K that yields a relatively high average consistency (e.g., the average over multiple runs exceeds 0.9).
We tested both BIC and the consistency criterion using synthetic data where ground truth was known. Our experiments, presented in Section IV-A, indicate that the consistency criterion yields an accurate prediction of the optimal number of templates.
E. Complexity
Each iteration of the algorithm has a computational complexity and memory requirement of 𝒪(NKV), where N is the number of input images, K is the number of templates and V is the number of voxels. We use multi-threading in ITK [30] to implement a parallelized version of iCluster. Similar to [2], [58], we employ a stochastic sub-sampling strategy to speed up the algorithm. At each iteration, a random sample of less than 1% of the voxels was used to compute the soft memberships, templates, template priors, standard deviation image and to update transformation parameters. In practice, we run the numerical optimization of the R-step as a single line search for each image, where the search directions are the normalized gradients. The effect of stochastic sub-sampling is investigated using synthetic data in Secion IV-A. Selecting a stopping criterion is not straightforward with the sub-sampling strategy, since a comparison of the objective function values across iterations is not possible. Instead, one can monitor the change in the parameters. In practice, the algorithm stops when the change in the class memberships and registration parameters falls below a pre-determined threshold. Figure 2 summarizes the iCluster algorithm.
Fig. 2.
iCluster: Pseudo-Code
IV. EXPERIMENTS
We validate the algorithm and investigate its behavior in four different experiments. In the first experiment, we use synthetic data to inform a choice of parameter settings, including the amount of sub-sampling. The availability of ground truth allows us to quantify the quality of results objectively and perform comparisons across different settings of parameters. The second experiment demonstrates the use of iCluster for building a multi-template atlas for a segmentation application. In the third experiment, we employ iCluster to compute multiple templates from a large data set that contains 415 brain MRI volumes. Our results demonstrate that these templates correspond to different age groups. In the last experiment, we use our algorithm on a smaller population that contains patients with dementia and healthy subjects. The results indicate that the templates computed by the algorithm correspond to the two clinical groups. We find the correlation between the image-based clustering and demographic and clinical characteristics particularly intriguing, given that iCluster does not have access to this information when constructing the model of heterogeneity in the population.
A. Synthetic Experiments
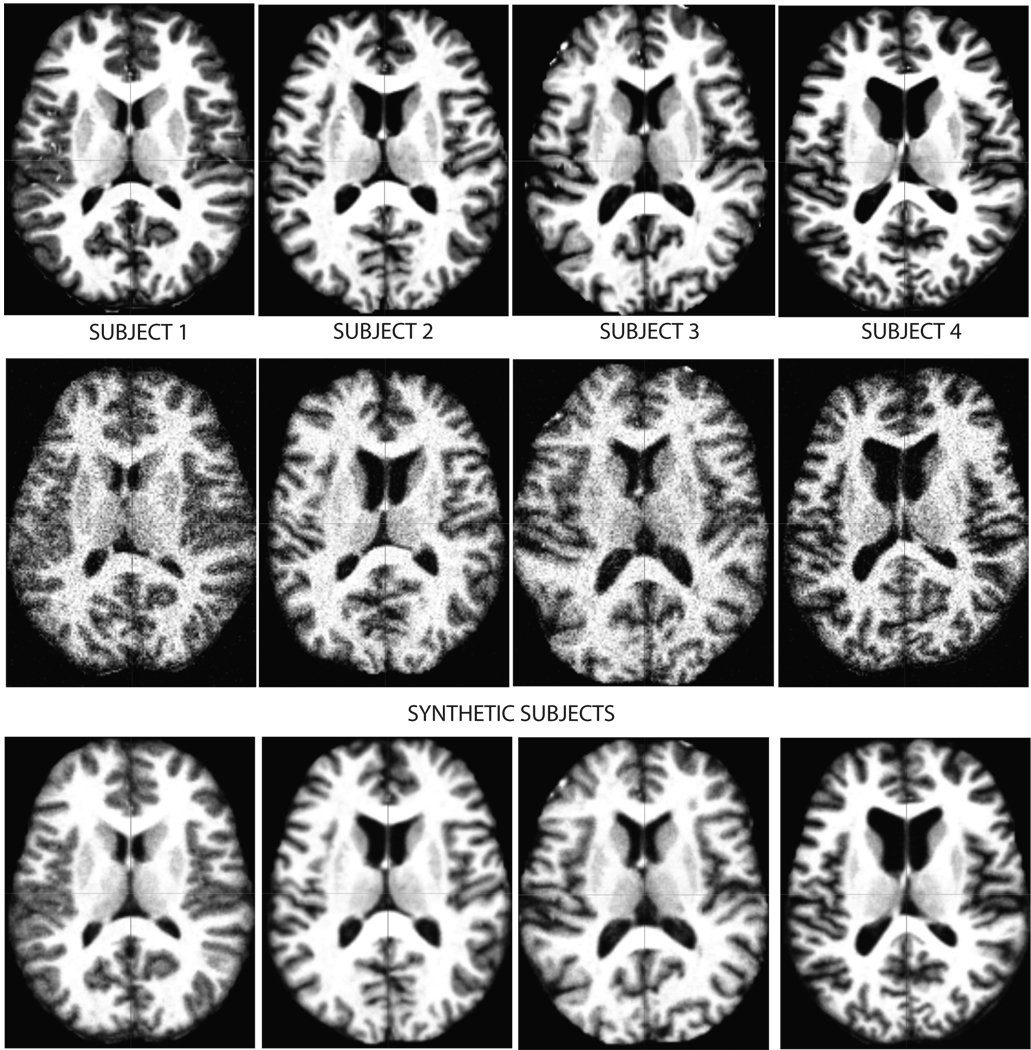
In this experiment, we synthesized three data sets from four whole brain MR images (obtained from the Oasis repository [34], with an image resolution of 176 × 208 × 176 voxels and voxel dimensions of 1 mm3). The subjects were warped by applying random transformations parameterized with a 8 × 8 × 8 B-spline model [41]. Each control point was displaced by an amount sampled uniformly from a 20 mm3 box around its original location. Furthermore, the warped images were corrupted with i.i.d. zero mean Gaussian noise with a variance equal to 10% of the maximum intensity value. Axial slices of the original images and representative synthetic images are shown in Figure 3. Table I summarizes the ground truth information for the synthetic data.
Fig. 3.
Top row: Axial slices of the original subject MRI’s used to synthesize data. Middle row: Axial slices of representative synthetic images. Bottom row: Axial slices of the four templates computed by iCluster with K = 4 and 0:5% sampling percentage.
TABLE I.
Summary of ground truth for the synthetic data
| Templates | True K | # Subjects | Relative Cluster Size | |
|---|---|---|---|---|
| Data Set 1 | Subj. 1,4 | 2 | 15 | 0.4, 0.6 |
| Data Set 2 | Subj. 1,2,4 | 3 | 20 | 0.35, 0.5, 0.65 |
| Data Set 3 | Subj. 1–4 | 4 | 30 | 0.2, 0.3, 0.3, 0.2 |
1) Effect of stochastic sub-sampling
First, we analyze the effect of stochastic sub-sampling on the quality of results. We ran iCluster on synthetic Data Set 3, with input K = 4. The four templates were initialized poorly as four different synthetic subjects that were all generated from the original subject 1. The quality of results was assessed using two measures: membership accuracy and error in the template images.
To define membership accuracy, we used the inner product between two membership probability matrices as a proxy for similarity. Formally, let denote a set of output membership probabilities and denote ground truth membership probabilities, with 1 corresponding to the template that generated the image and all remaining entries equal to zero. We define membership accuracy as:
| (20) |
where N is the number of input images. To resolve the ambiguity in the cluster indices, we maximize Equation (20) over all possible permutations of the ground truth template indices. We use this maximum value as a measurement of membership accuracy.
Let Tk(x⃗) denote the output template images and denote the ground truth templates, i.e., original subject MRIs. We define the average template error as:
| (21) |
where V is the number of voxels in Ω, K is the number of templates and the template indexing is determined by maximizing Equation (20) for output memberships.
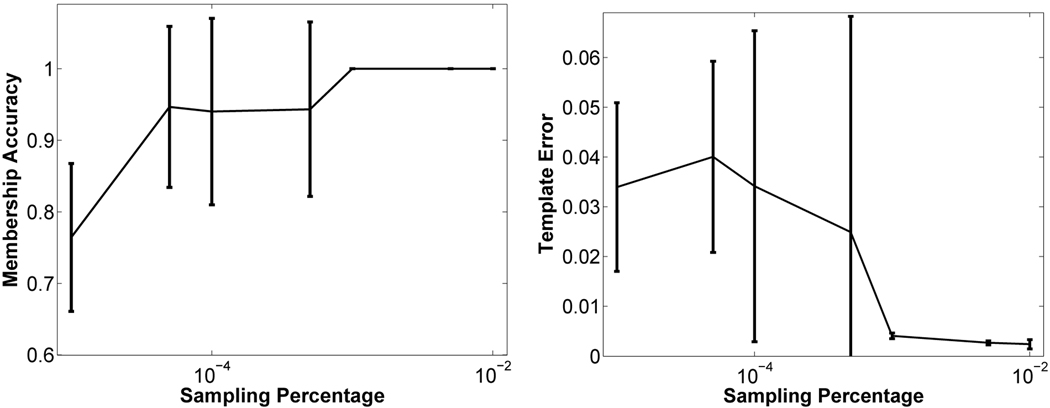
Figure 4 shows both the membership accuracy and template error values for a range of sampling percentages, where the sampling percentage is the ratio of the size of the stochastic set of voxels used at each iteration to the total number of voxels. For each parameter setting, we performed 10 runs of iCluster starting from the same poor initialization. Each run yielded a different output due to stochastic sub-sampling. For sampling percentage values larger than 0.1% membership accuracy was perfect and the template error reached its minimum for all ten runs. In practice, we chose 0.5% as the sampling percentage. This corresponds to using roughly 30,000 voxels at each iteration. The bottom row of Figure 3 shows the four templates computed by iCluster with input K = 4 and 0.5% sampling percentage. The output templates were computed using Equation (8) on the whole domain Ω with the estimated model parameters.
Fig. 4.
Output quality as a function of sampling percentage, i.e., the ratio of the size of stochastic set of voxels used at each iteration to the total number of voxels. Error bars indicate standard deviation.
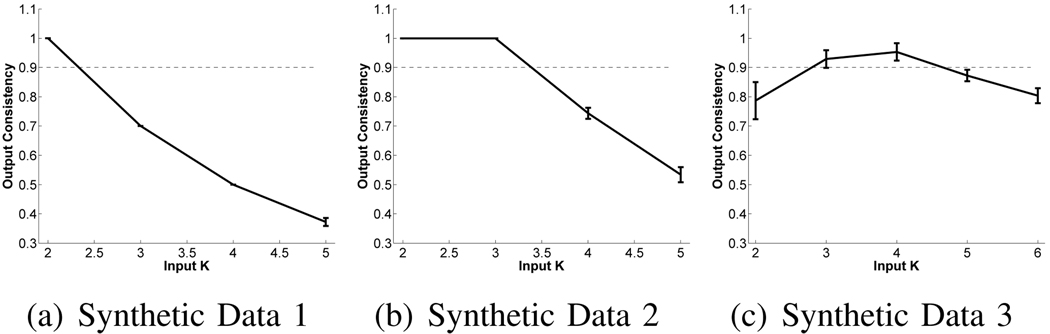
2) Determining the optimal number of templates
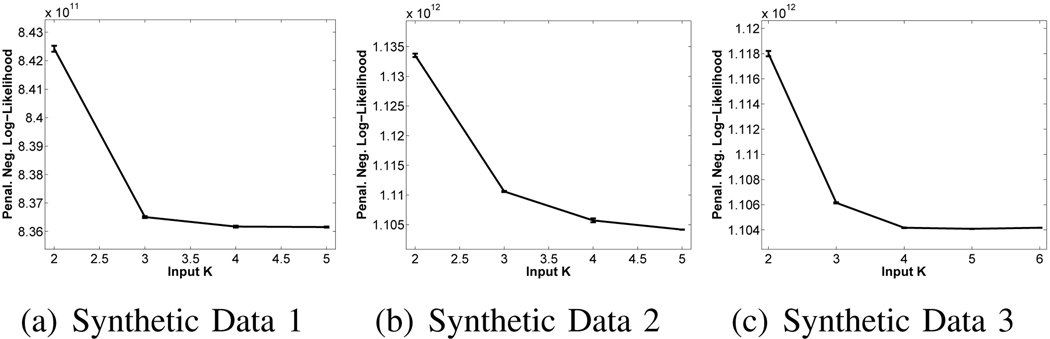
Here, we compare two methods for automatically determining the optimal number of templates. We ran iCluster on the three synthetic data sets with a range of input K values. For each setting, we ran the algorithm ten times with different random initializations to get a collection of outputs. Using Equation (18), we computed the negative penalized log-likelihood values for these outputs. Figure 5 plots these values as a function of input K for the three data sets. BIC determines the optimal number of templates as the value of K that minimizes the penalized log-likelihood of the data under the estimated model. According to this criterion, data sets 1,2 and 3 have at least 4, 5 and 4 underlying templates, respectively. The optimal K for data sets 1 and 2 should have been 2 and 3, respectively.
Fig. 5.
BIC: Penalized negative log-likelihood values for a range of input K values. Error bars indicate standard error.
Alternatively, we can look at the consistency of the resulting model to determine the optimal number of templates. We quantified the consistency of the model using the relative membership consistency measure defined in Equation (19). The average relative membership consistency values for each input K are shown in Figure 6. Based on the consistency criterion, we propose to select the highest value of K that yields a relatively high average consistency (e.g., the mean over multiple runs exceeds 0.9). According to this criterion, data sets 1, 2 and 3 have 2, 3 and 4 underlying templates, respectively, which agrees perfectly with the ground truth. In the remaining experiments, we used the consistency criterion to determine the optimal number of templates.
Fig. 6.
Consistency Criterion: The consistency of output membership probabilities for a range of input K values. Error bars indicate standard error.
B. Segmentation Label Alignment
In atlas-based segmentation, one typically normalizes the new subject by registering the image with a template. Segmentation is then achieved by inferring labels based on the intensities of the new image and the training images that contain manual labels. The training data is usually employed to establish a prior for segmentation. To assess the quality of this prior, one can measure its agreement with the ground truth label of a new subject. In the following experiment, we measure this agreement by quantifying the alignment between one (new) subject and the remaining (training) subjects. In the case of multiple atlases, this requires an assignment of the new subject to one of the atlases. If these atlases are constructed through an image-based clustering strategy, as the one proposed in this paper, one can use the same framework to determine this assignment. This means fixing the template images, noise variance image and template priors in the iCluster algorithm. The assignment of the new subject can then be computed using the same GEM algorithm, which iterates over the E and R-steps.
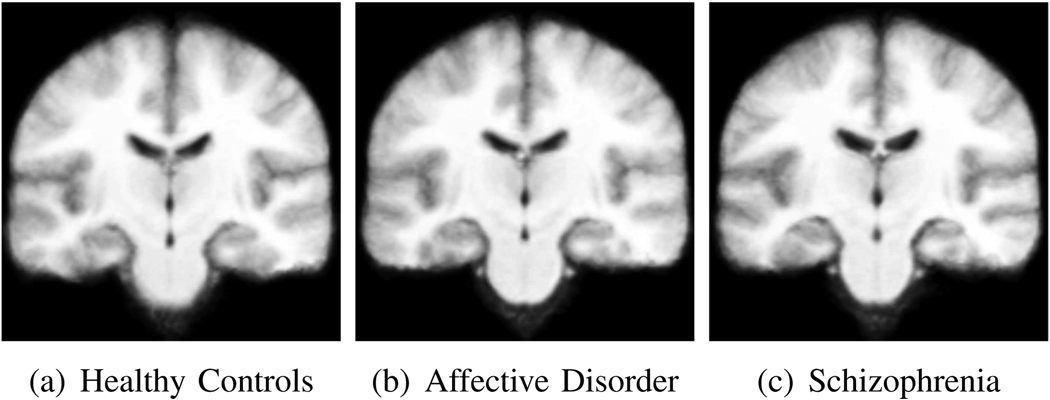
In this experiment, we used a data set of 50 whole brain MR brain images that contained 16 patients with first episode schizophrenia (SZ), 17 patients with first-episode affective disorder (AFF) and 17 age-matched healthy subjects (CON). The MRI volumes were obtained using a 1.5-T General Electric scanner (GE Medical Systems, Milwaukee). The acquisition protocol was a coronal series of contiguous images. The imaging variables were as follows: TR=35 msec, TE=5 msec, one repetition, 45 nutation angle, 24-cm field of view, NEX=1.0 (number of excitations), matrix = 256 × 256 (192 phase-encoding steps) × 124. The voxel dimensions were 0.9375 × 0.9375 × 1.5 mm. First episode patients are relatively free of chronicity-related confounds such as the long-term effects of medication, thus any structural differences between the three groups are subtle, local and difficult to identify in individual scans. Figure 7 shows coronal slices of the affine-normalized mean images for each clinical population. A detailed description of the data and related findings are reported in [28].
Fig. 7.
Mean images for each clinical population after affine normalization.
For these images, we also had manual delineations of eight temporal lobe structures: the (left and right) superior temporal gyrus (STG), hippocampus (HIP), amygdala (AMY) and parahippocampal gyrus (PHG). Prior MRI studies of schizophrenic patients revealed structural brain abnormalities, with low volumes of gray matter in the left posterior superior temporal gyrus and in medial temporal lobe structures. However, the specificity to schizophrenia and the roles of chronic morbidity and neuroleptic treatment in these abnormalities remain under investigation [28], [29]. Accurate segmentation tools for temporal lobe structures is thus important for schizophrenia research. We used manual labels to explore label alignment across subjects under different groupings: on the whole data set, on the clinical grouping, and on the image-based clustering as determined by iCluster.
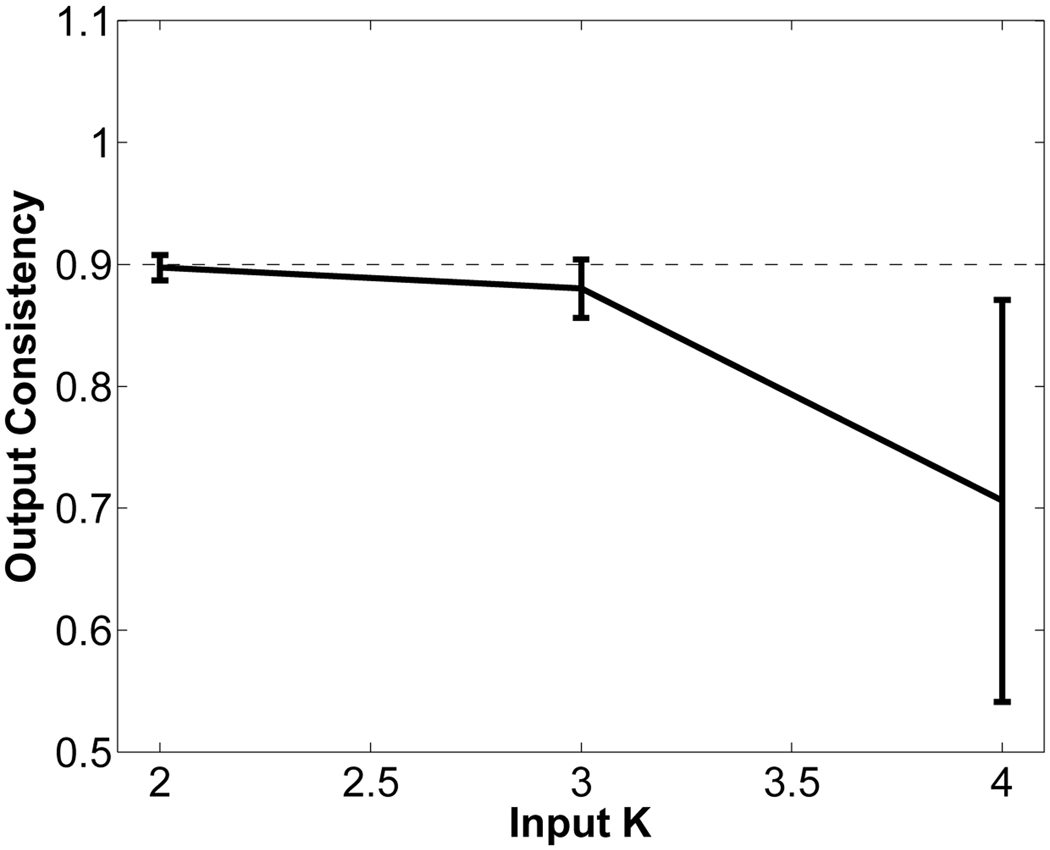
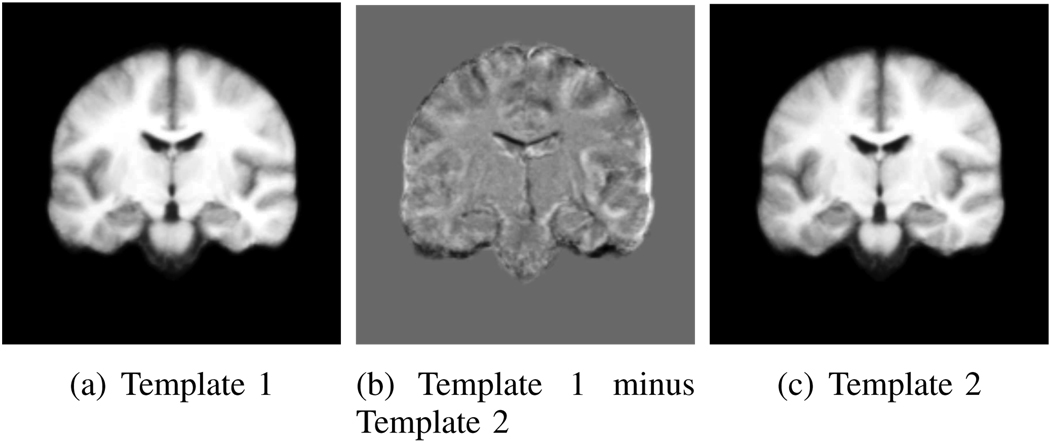
We ran iCluster on the 50 MR images for different values of input K. We emphasize that the algorithm did not have access to the clinical and manual label data. Figure 8 shows the iCluster output membership consistency, as defined in Section III-D. We ran the algorithm ten times for each value of input K. Based on our proposed consistency criterion, we determine K = 2 as the optimal number of templates. However, to provide a comparison with the clinical grouping (where there are three groups: SZ, AFF and CON), we present results for K = 3 as well. Table II and Table III show the relationship between the clustering of the algorithm and the clinical diagnosis. We observe that the clustering computed by the algorithm demonstrates no correlation with the clinical diagnosis. This result confirms the difficulty of identifying structural differences between these first-episode patients and control subjects on an individual basis. Figure 9 shows coronal views of the two templates discovered by iCluster and the difference image between these two. There are subtle structural differences between the two templates, especially around the cortical regions of the temporal lobes.
Fig. 8.
Consistency Criterion for the schizophrenia data set: The consistency of output membership probabilities for input K = 2, 3, 4. Error bars indicate standard error.
TABLE II.
Clinical composition of clusters for K=2
| Cluster | AFF | CON | SZ |
|---|---|---|---|
| 1 | 11 | 9 | 10 |
| 2 | 6 | 8 | 6 |
TABLE III.
Clinical composition of clusters for K=3
| Cluster | AFF | CON | SZ |
|---|---|---|---|
| 1 | 7 | 6 | 8 |
| 2 | 5 | 7 | 4 |
| 3 | 5 | 4 | 4 |
Fig. 9.
Two Templates computed by iCluster. In the difference image, gray is zero, darker (lighter) values correspond to negative (positive) values.
To measure the quality of alignment of a region of interest in two subjects, we employed two measures: (1) the Dice score which quantifies the overlap between the regions of interest in two subjects [55]; and (2) the modified Haussdorff distance [56], which is defined as the average Euclidean distance (in mm) between a boundary point and the closest corresponding boundary point in the other subject. The Dice score ranges between 0 and 1, where 1 indicates a perfect overlap. The Haussdorff distance achieves zero at perfect alignment; higher values indicate worse alignment.
We compared average label alignments for three strategies:
ALL: All subjects were co-registered with a single dynamic average template. This was achieved using the iCluster algorithm with K = 1 and a 32 × 32 × 32 B-spline grid. The average label alignment for each subject was then computed by averaging all pairwise measures of label alignment with the remaining subjects.
CLIN: Each clinical group was co-registered separately using iCluster with K = 1 and a 32 × 32 × 32 B-spline grid. The average label alignment for each subject was then computed by averaging all pairwise measures of label alignment with the remaining subjects with the same clinical diagnosis.
iC2 and iC3: We ran iCluster on all subjects with input K = 2 and 3, and a 32 × 32 × 32 B-spline grid. For each input K value, we report label alignment results for the run that yielded the highest relative consistency value as defined in Equation (19). The average label alignment for each subject was then computed by averaging all pairwise measures of label alignment with the remaining subjects in the same cluster.
Figure 10 shows the average Dice scores and Hausdorff distances for the individual ROIs. These values were computed in the atlas space, where the manual labels were interpolated using the transformations obtained from the registrations and the nearest neighbor interpolator. We performed a paired permutation test comparison between the average label alignments of the three scenarios. The p-values were computed by assessing the average difference between two sets of paired measurements based on a histogram of differences obtained by randomly shuffling the order of pairings. The comparisons for the Haussdorff distances are presented in Table IV. Dice score comparisons yield similar results. In summary, iCluster with input K = 2 yields the best label alignment results, where 6 out of 8 ROIs were significantly better aligned (with p < 0.05) compared to the first two strategies of co-registering all subjects (ALL) and clinical groups separately (CLIN). This result provides further evidence for the usefulness of the proposed consistency criterion that determines the optimal number of templates. ALL and CLIN yield statistically improved label alignment for only one ROI: the right Superior Temporal Gyrus.
Fig. 10.
Top row: Dice scores for each ROI. Bottom row: Haussdorff Distances in mm. Error bars indicate standard error.
TABLE IV.
Statistical comparison of average label alignment. improvement: +++ p < 0.01, ++ p < 0.05, + p < 0.1. Equivalent: =. Impairment: − − − p > 0.99. L and R denote left and right, respectively.
| l-AMY | r-AMY | l-HIP | r-HIP | l-STG | r-STG | l-PHG | r-PHG | |
|---|---|---|---|---|---|---|---|---|
| iC2 vs. ALL | +++ | +++ | +++ | ++ | + | − − − | ++ | +++ |
| iC2 vs. CLIN | +++ | +++ | +++ | + | +++ | − − − | ++ | +++ |
| iC3 vs. ALL | +++ | +++ | +++ | ++ | +++ | = | + | = |
| iC3 vs. CLIN | +++ | +++ | = | + | +++ | = | = | + |
These results suggest that, on average, for most ROIs we achieve a better agreement between the ground truth labels and a prior obtained via iCluster, than a prior computed by co-registering all subjects or subjects within a clinical population.
C. Age Groups in the OASIS Data Set
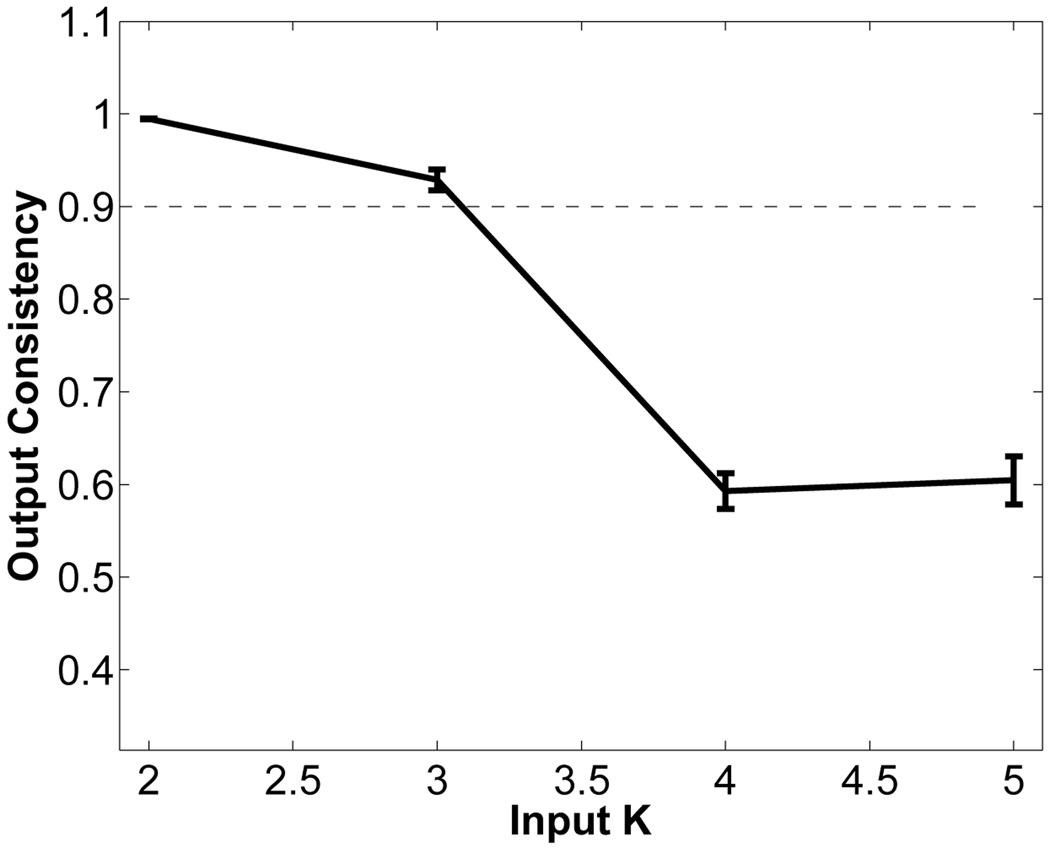
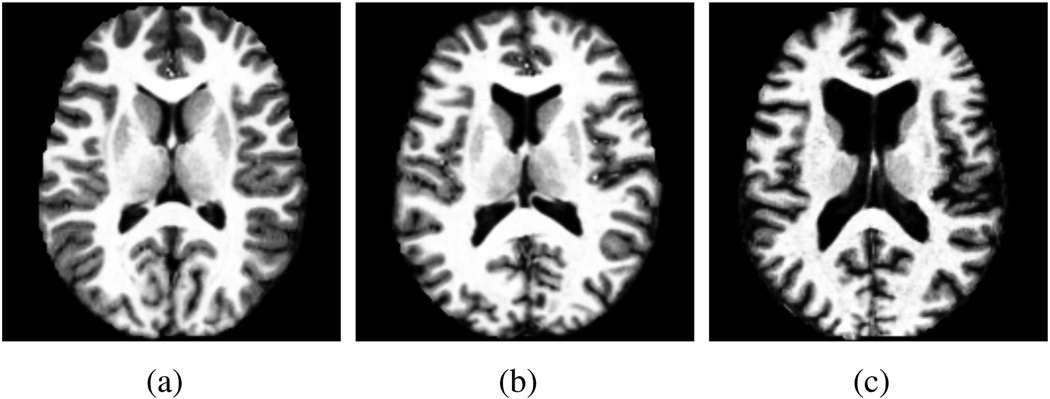
In this experiment, we used the OASIS data set [34] which consists of 415 pre-processed (skull stripped and gain-field corrected) brain MR images of subjects aged 18–96 years including individuals with early-stage Alzheimer’s disease (AD). We ran iCluster on the whole data set while varying the number of templates K from 2 through 5. Each run took 4–8 hours on a 16 processor PC with 128GB RAM. Figure 11 shows the output consistency against for different values of input K. For K = 4 and 5 the consistency values are significantly smaller than 0:9. We therefore report our results for K = 2 and K = 3. Figure 12 and Figure 13 show the two and three robust templates computed with K = 2 and K = 3, respectively. Figure 15 shows the age distributions determined via Parzen window estimator based on a Gaussian kernel with a standard deviation of 4 years.
Fig. 11.
Consistency Criterion for the Oasis data set: The consistency of output membership probabilities for a range of input K values. Error bars indicate standard error.
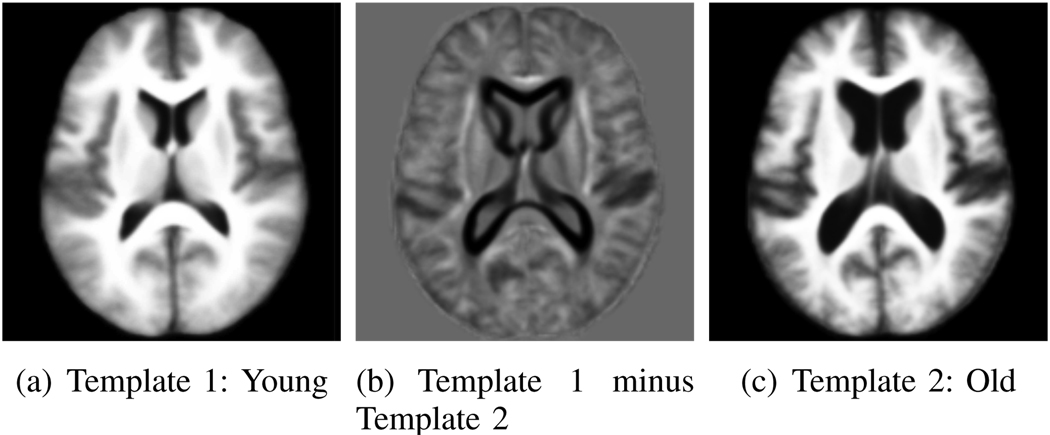
Fig. 12.
Two templates of the OASIS data. In the difference image, gray is zero, darker (lighter) values correspond to negative (positive) values.
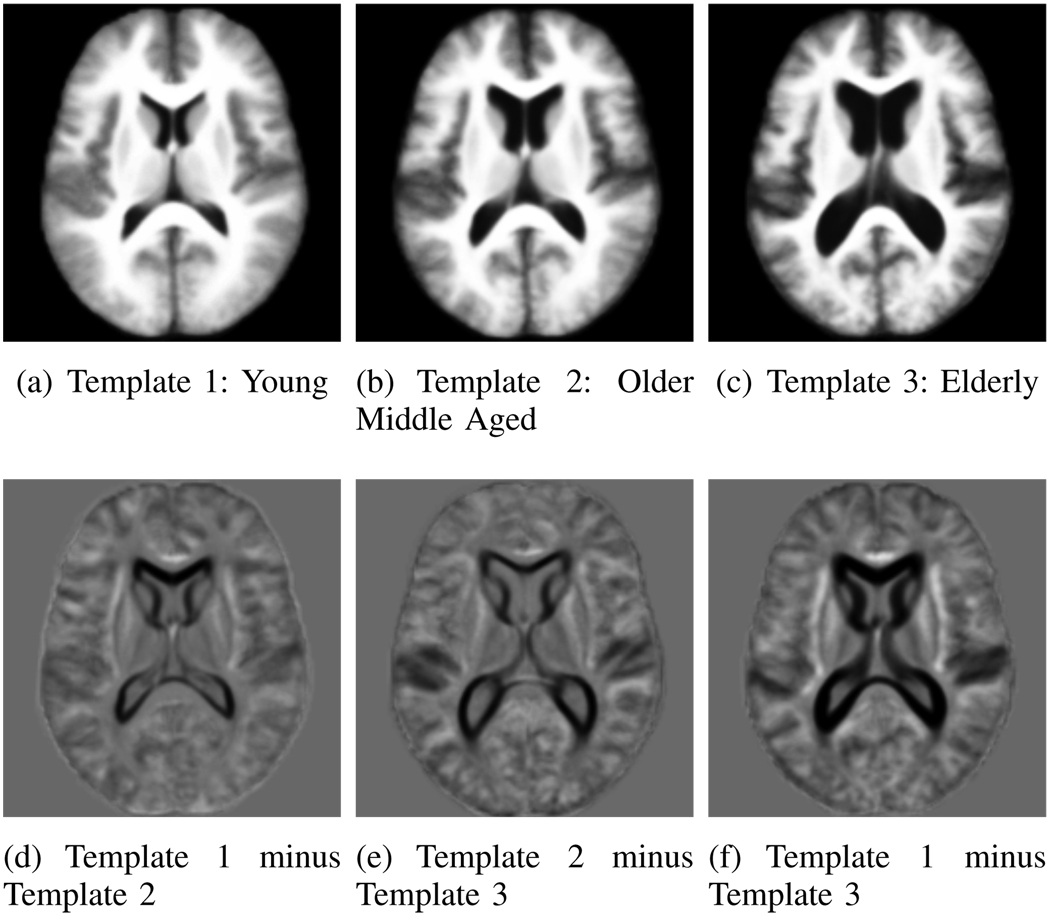
Fig. 13.
Top Row: Three templates of the OASIS data. Bottom Row: Difference images. Gray is zero, darker (lighter) values correspond to negative (positive) values.
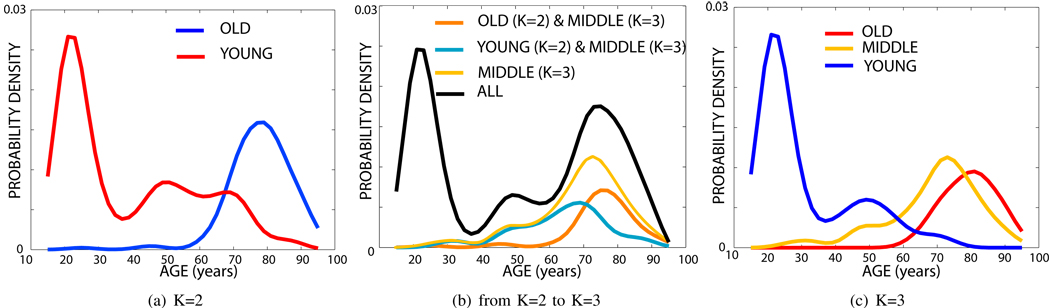
Fig. 15.
Age distributions of the OASIS data. (a) Age distributions for K=2, (b) the relationship between the ages of subjects in clusters identified for K=2 and for K=3, (c) Age distributions for K=3.
It is easy to see that each template corresponds to a unique age group: For K = 2, we identify a group of 268 young subjects (aged 39.1 ± 19.9 years) and a group of 147 elderly subjects (aged 77.8 ± 9.3 years). For K = 3 the algorithm detected 201 young subjects (Group 1, aged 31.2 ± 14:5 years), an older middle aged group of 127 subjects (Group 2, aged 68.9 ± 13.6 years) and elderly 87 subjects (Group 3, aged 79.6 ± 7.5 years). Figure 15-b illustrates the intersection between the middle aged distribution of K = 3 and the distributions of K = 2. This plot reveals that the middle aged group for K = 3 consists of two sub-populations: (1) a younger group of subjects that are in the young group for K = 2 and (2) an older age group in the elderly for K = 2. These results suggest that the dominant structural modes in this large population are mainly due to aging. Analyzing the decomposition of the whole age distribution (shown in black in Figure 15-b) indicates that iCluster does not simply find the three major age modes. Specifically, the small middle peak around 50 years is robustly included with the younger population in both K = 2 and K = 3. With three modes, the algorithm identifies an older middle aged group (Group 2) that has a significant overlap in age with the elderly group (Group 3).
We further analyzed the clinical dementia rating (CDR) [36] data to explore the differences across the image-based clusters. Table V summarizes the results. Group 1 (Figure 13-a) has almost no subjects with positive CDR (an indication of probable Alzheimer’s), whereas Group 2 (Figure 13-b) consists of 35% patients diagnosed with probable Alzheimer’s disease (AD) (i.e., has a CDR score of greater than zero), and 65% subjects with no dementia. Group 3 (Figure 13-c) includes 69% patients with probable AD and 31% healthy subjects with zero CDR. The difference between the patient percentage in each group is statistically significant at p < 10−4 as determined by a permutation test. This result indicates that the old-middle aged group computed by iCluster contains a majority of healthy individuals, whereas the elderly group is dominated by probable AD patients.
TABLE V.
Number (Percentage) of subjects with respect to their gender and clinical dementia score in each group computed by icluster with K = 3.
| Positive CDR | Zero CDR | |||
|---|---|---|---|---|
| Female | Male | Female | Male | |
| Group 1 | 1 (0.2) | 1 (0.2) | 119 (28.7) | 80 (19.3) |
| Group 2 | 28 (6.8) | 16 (3.9) | 58 (14.0) | 25 (6.0) |
| Group 3 | 30 (7.2) | 24 (5.8) | 19 (4.6) | 14 (3.4) |
An important question at this point is to what extent these dementia profiles are correlated with the age data of the individuals, since it is known that the rate of incidence of dementia increases with aging [21]. Moreover, we would like to explore the influence of gender on these structural modes. One important point to note is that approximately half of the subjects over 60 years old (100 subjects) were clinically diag-nosed with dementia, as summarized in Table VI. Examining this table reveals a difference between the two genders: healthy females without dementia are more likely to belong to Group 2 (Figure 13-b). On the other hand, males with positive CDR (i.e, with dementia) are more likely to belong to Group 3 (Figure 13-c). For the other two groups, i.e., males without dementia and females with dementia, there is no obvious relationship that these tables reveal.
TABLE VI.
Number (Percentage) of subjects aged 60 and older with respect to their gender and clinical dementia score data in each group computed by icluster with K = 3.
| Positive CDR | Zero CDR | |||
|---|---|---|---|---|
| Female | Male | Female | Male | |
| Group 1 | 1 (0.5) | 1 (0.5) | 7 (3.5) | 2 (1.0) |
| Group 2 | 28 (14.1) | 16 (8.1) | 46 (23.2) | 10 (5.1) |
| Group 3 | 30 (15.2) | 24 (12.1) | 19 (9.6) | 14 (7.1) |
To get a better insight into the characteristics of the discovered structural modes, we performed a multinomial logistic regression on the iCluster group memberships using age, gender and clinical data1 as regressors. Table VII reports the regression coefficients, assuming Group 2 to be the reference category. If we convert the estimated probabilities to group assignments, the total model achieves around 75% training accuracy and a likelihood ratio test estimates the significance of the full, fitted model at p < 0:01. The significance of each coefficient was determined with a Wald test [17]. These results suggest that the most significant factor that determines group assignment is age: with each year, the odds of a subject being assigned to the next, older group increases by approximately 0.1(≈ exp(0.1) − 1). Groups 2 and 3 are also differentiated by the clinical score and gender (with less significance). One point decrease in the MMSE score increases the odds of a subject belonging to Group 3, rather than Group 2, by 0.1(≈ exp(0.1) − 1). A female’s odds of belonging to Group 2 vs. Group 3 is roughly 2.5-fold (exp(0.94)) higher than a male’s.
TABLE VII.
Logistic regression coefficients: on the whole oasis data set.
| Age (years) | Clinical State (MMSE) | Gender (Male: 0, Female: 1) | |
|---|---|---|---|
| Group 1 vs. Group 2 | −0.12*** | 0.34 | 0.84 |
| Group 3 vs. Group 2 | 0.11*** | −0.10** | −0.94* |
p < 0.05,
p < 0.01,
p < 0.001
These results confirm that aging and dementia are both significant factors that influence major structural changes in the brain. Moreover, our results indicate that these factors may have different effects for the two genders. These findings demonstrate a qualitative similarity with the ones reported in [20], where aging and dementia are shown to correlate with brain atrophy in a similar manner. Furthermore, [20] reports that these effects have a tendency to be different in the two genders: males tend to demonstrate a higher rate of atrophy. The gender difference, however, does not reach statistical significance in the analysis of [20] and remains under debate in the literature [23], [32].
D. Patients with Dementia
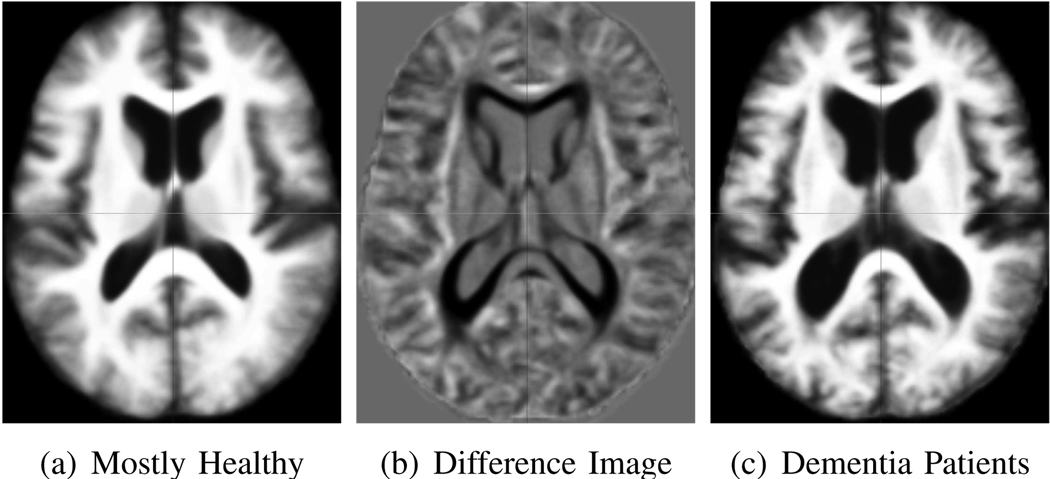
In the fourth experiment, we used a set of 30 subjects (aged between 65 and 84 years) from the OASIS data set. Fifteen of these had a positive CDR, i.e., were diagnosed with very mild to mild dementia and probable AD (aged 74.7 ± 5.5 years, with education level of 3 ± 1.2), while the other fifteen individuals were controls (aged 74.6 ± 5.4 years, with education level of 3.1 ± 1.5) with no sign of clinical dementia at the time of scanning. Figure 16 shows the consistency of iCluster outputs over a range of input K values. For K > 2, we observe that the membership consistency is less than 0.9, thus we report results for K = 2: Group 1 (Figure 17-a) consists of 25 subjects, 15 of which were CDR zero. Group 2 (its template shown in Figure 17-c) consists of 5 subjects, all of which have dementia.
Fig. 16.
Consistency Criterion for the 30 subject dementia data set: The consistency of output membership probabilities for a range of input K values. Error bars indicate standard error.
Fig. 17.
Two templates and their difference image for the 30 subject dementia data set.
We performed a multinomial logistic regression on the iCluster assignments using age, education data (1: less than high school, 2: high school, 3: some college, 4: college graduate, 5: beyond college), clinical score and gender data as regressors. Only the clinical score demonstrated significant relevance to differentiate the two groups: the first group’s average MMSE score was 25.2 ± 5.1, whereas group two’s score was 19.8 ± 2.9.
The fact that Group 2 comprised of dementia patients with significantly low MMSE scores is intriguing. Yet, the more interesting question is, what is special about the ten dementia patients assigned to Group 1? This clustering suggests that their anatomies are more similar to healthy subjects in the same age group. Clinical and demographic attributes of the patients in the two groups are virtually identical: (1) Age: 74.4 ± 4.9 versus 75.4 ± 7.2 ;(2) MMSE score: 19.8±3.9 versus 19.8 ± 2.9; and (3) Education level: 3 ± 1.2 versus 3 ± 1.4. Thus, based on the data we have, this question remains open and requires further investigation.
V. DISCUSSION
Our experiments demonstrate the use of iCluster in multiple settings. The synthetic experiments served to asses the effect of stochastic sub-sampling on the quality of results and informed the design of the method that automatically determines the optimal number of templates. In the second experiment presented in Section IV-B, we show that, using the proposed clustering strategy, one can compute a multi-template atlas for a segmentation application. Based on growing evidence that population-specific atlases yield more accurate segmentation, we can employ iCluster to discover coherent sub-populations in a large population of images and construct separate atlases for each sub-population. Our experiments suggest that a multi-template atlas can improve segmentation quality. The proposed approach promises significantly better segmentation than a disease-specific atlas, especially in the case of spectrum diseases such as schizophrenia.
In another setting, we demonstrate the utility of an image-driven approach for computational anatomy. This is in contrast with today’s popular techniques that rely on a clinical or demographic classification of the subjects. Our experiments show that iCluster can robustly identify structural modes in a population that are mainly determined by age and dementia. This type of analysis promises to provide insight into the major factors that drive structural change and, more importantly, characterize subtypes of a particular disorder.
In our experiments, enlarged ventricles are immediately obvious in the older and dementia templates when compared to the younger and healthy populations, respectively. Moreover, cortical thinning and anterior white matter changes are visible in the difference images shown in Figure 13. These types of structural changes due to aging and dementia have been well-documented in the literature [16], [26], [43]. Further analysis is required to understand the structural differences between the discovered modes. The intermediate group (the older middle aged in the first experiment) and the mixture group in the dementia experiment can provide interesting insights into structural changes due to aging and dementia.
With a single template, i.e., input K = 1, iCluster can be seen as an efficient unbiased template estimation algorithm, similar to the ones proposed in [14], [31], [58]. Yet, the main point of this paper is that a single template is not sufficient to summarize the variability in a large and heterogenous population of images. To that extent, iCluster is similar to the recent works on atlas stratification [9] and deformable templates [1]. In the atlas stratification framework of [9], the authors propose to use an off-the-shelf clustering algorithm on images to identify underlying homogeneous sub-populations. The framework does not explicitly model anatomical heterogeneity and yields a computationally inefficient algorithm, where one needs to perform 𝒪(N2) pairwise registration instances to analyze N input images. The generative model we developed in this paper is similar to the deformable templates model of [1]. Yet, in contrast with [1], our main focus is to propose a computationally efficient algorithm that can be employed on large collections of high resolution medical image data. Most importantly, however, we include a concrete demonstration of how an image-clustering approach can be used to construct multiple segmentation atlases and study the effects of clinical and demographic factors on neuroanatomy.
The image-based clustering approach can also be extended to descriptors of anatomical shape, such as volume [20] or surface-based representations [52]. Various shape descriptors have been used to study the effects of disease progression and aging on anatomy. Based on similarity measures defined for these different descriptors, one can potentially derive different shape clustering algorithms. One such algorithm was proposed in [50]. The main drawback of such a shape-based approach is the need for accurate segmentations, which limits the amount of data such a strategy can be applied to. An image-based clustering approach, on the other hand, has the advantage that it can be used with large collections of raw images. Furthermore, image-based clustering can potentially reveal modes in a population that differ in unexpected regions.
We view iCluster as a first step towards a more comprehensive image-driven population analysis framework. The current algorithm suffers from several limitations. Notably, the simple additive Gaussian noise model cannot handle significant intensity variations across images. Thus, the current algorithm can only be used with intensity corrected (e.g., histogram matched, bias field corrected) images of the same modality. Moreover, the algorithm constructs clusters based on a similarity measure computed over whole images. This makes the method less sensitive to subtle and local differences across groups of images. One solution is to use a similarity measure computed over a region of interest in the E-step of iCluster. In the following, we summarize the possible directions one can explore to extend iCluster to a broader set of problems:
Use an entropy-based similarity measure that is insensitive to intensity variations to compute memberships in the E-step and perform co-registration in the R-step.
Compute memberships within a region of interest or based on a different type of information, e.g., connectivity from diffusion data.
Use more sophisticated models of deformation, e.g., diffeomorphisms. Moreover, one can integrate a more sophisticated prior on the spatial transformations. Hence, the memberships will be a function of both a similarity measure based on image intensities and the deformation cost.
Rather than using an additive noise model on intensities, one could explicitly model the variance in warps which would lead to a clustering strategy based on deformations.
VI. CONCLUSION
We presented a fast and efficient image clustering algorithm for co-registering a group of images, computing multiple templates that represent different modes in the population, and determining template assignments. We demonstrated our algorithm in several experiments, which illustrated a multi-template atlas strategy for accurate image segmentation and revealed age and disease-related modes in a population. Our results confirm previous findings and lead to interesting insights that suggest future research directions in computational anatomy.
Fig. 14.
Typical Subjects: (a) Group 1: 24-year-old, healthy female, (b) Group 2: 52-year-old, healthy female, (c) Group 3: 76-year-old male with very mild dementia and probable AD.
TABLE VIII.
Logistic regression coefficients: on the icluster with input K = 2 memberships of the 30 subject dementia data set
| Age (years) | Clinical State (MMSE) | Gender (Male: 0, Female: 1) | Education | |
|---|---|---|---|---|
| Group 1 vs. Group 2 | −0.03 | −0.36* | −2.74 | 0.22 |
p < 0.05
ACKNOWLEDGMENTS
The authors would like to thank Bruce Fischl, Koen Van Leemput, B.T. Thomas Yeo and the anonymous reviewers for their helpful feedback. We would also like to extend our appreciation to Dr. Randy Buckner for making the OASIS dataset available. Support for this research is provided in part by the Department of Veterans Affairs Merit Awards, National Alliance for Medical Image Analysis (NIH NIBIB NAMIC U54-EB005149), the Neuroimaging Analysis Center (NIT CRR NAC P41-RR13218), the Morphometry Biomedical Informatics Research Network (NIH NCRR mBIRN U24-RR021382), the NIH NINDS R01-NS051826 grant, National Institute of Mental Health grant 5R01-MH050740-13 and the NSF CAREER 0642971 grant.
VII. APPENDIX
In this appendix, we provide derivations for the update equations of the T- and R-steps of the iCluster algorithm presented in Section III-A.
A. T-step
Given the posterior probability estimates {qk(In; θ(i)) and fixing the spatial transformations from the previous iteration, the template images {Tk}, template priors {πk} and standard deviation image σ are updated to maximize the lower bound Q(θ, θ(i)) of Equation (5):
| (22) |
| (23) |
such that ∑k πk = 1.
In Equation (23) all the template priors can be optimized independently. We introduce a Lagrange multiplier λ for the constraint;
| (24) |
differentiate Equation (24) with respect to πk and set the derivative to zero, obtaining
| (25) |
where .
We recall that
| (26) |
| (27) |
| (28) |
| (29) |
where |·| denotes matrix determinant, J(Φ, x⃗) is the Jacobian matrix of Φ that contains the partial derivatives of the warp field with respect to the coordinates and Ωc is a continuous and compact subset of ℝ3 that covers the discrete set Ω. Equation (27–Equation 29) assume a suitable interpolator for making I, {Tk} and σ spatially continuous. Equation (28) assumes the boundary condition Φn(∂Ωc) = ∂Ωc for all n, where ∂Ωc is the boundary of Ωc and uses a change of variables with .
Substituting Equation (29) into Equation (23), we obtain:
| (30) |
Differentiating the objective function in Equation (30) with respect to Tk(x⃗) and setting the derivative to zero yields
| (31) |
which is independent of σ(x⃗).
To determine σ(x⃗), we substitute Equation (29) into Equation (23) and obtain:
| (32) |
Differentiating the objective function of Equation (32) with respect to σ(x⃗) and setting the derivative to zero yields
| (33) |
B. R-step
Fixing the model parameters computed in the previous T-step, the R-step updates the transformations {Φn} to improve the lower bound Q(θ, θ(i)) of Equation (5). Substituting Equation (29) into Equation (23) and focusing on the terms that depend on Φn yields
| (34) |
| (35) |
| (36) |
where in Equation (35) and Equation (36) we dropped and added terms that do not depend on Φ.
Footnotes
Mini-Mental State Exam scores [40] that ranged from 14 (poor mental health) to 30 (good mental health).
Contributor Information
Mert R. Sabuncu, Email: msabuncu@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Serdar K. Balci, Email: serdar@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Martha E. Shenton, Surgical Planning Laboratory, Harvard Medical School and Brigham and Womens Hospital, Boston, MA 02115 USA, with the Psychiatry Neuroimaging Laboratory, Department of Psychiatry, Brigham and Womens Hospital, Harvard Medical School, Boston, MA 02115, USA, and also with the Clinical Neuroscience Division, Laboratory of Neuroscience, VA Boston Healthcare System and Harvard Medical School, Brockton, MA 02301, USA.
Polina Golland, Email: polina@csail.mit.edu, Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA..
REFERENCES
- 1.Allassonniere A, Amit Y, Trouve A. Towards a coherent statistical framework for dense deformable template estimation. Journal of the Royal Statistical Society: Series B. 2007;69:3–29. [Google Scholar]
- 2.Viola P, Wells WM. Alignment by maximization of mutual information. International Journal of Computer Vision. 1997;24(2):137–154. [Google Scholar]
- 3.Ashburner J, Friston K. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner J, Friston KJ. Voxel-based morphometry - the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: Deformation-based morphometry. Human Brain Mapping. 1998;6:348–357. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburner J, Neelin P, Collins DL, Evans A, Friston K. Incorporating prior knowledge into image registration. NeuroImage. 1997;6(4):344–352. doi: 10.1006/nimg.1997.0299. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Hur A, Elisseeff A, Guyon I. A stability based method for discovering structure in clustered data. Pacific Symposium on Biocomputing. 2002;7(6):6–17. [PubMed] [Google Scholar]
- 8.Bhatia KK, Hajnal JV, Puri BK, Edwards AD, Rueckert D. Consistent groupwise non-rigid registration for atlas construction. Biomedical Imaging: Nano to Macro, 2004. IEEE International Symposium on. 2004;1:908–911. [Google Scholar]
- 9.Blezek D, Miller J. Atlas stratification. Medical Image Analysis. 2007;11(5):443–457. doi: 10.1016/j.media.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brent RP. Algorithms for Minimization without Derivatives. Englewood Cliffs, NJ: Prentice-Hall; 1973. [Google Scholar]
- 11.Clifford JR, Petersen RC, O’Brien PC, Tangalos EG. MR-based hippocampal volumetry in the diagnosis of Alzheimer’s disease. Neurology. 1992;42 doi: 10.1212/wnl.42.1.183. [DOI] [PubMed] [Google Scholar]
- 12.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. Journal of Computer Assisted Tomography. 1994;18(2):192–205. [PubMed] [Google Scholar]
- 13.Collins DL, Zijdenbos AP, Baaré WFC, Evans AC. ANIMAL+INSECT: Improved Cortical Structure Segmentation. In Proceedings Information Processing in Medical Imaging 1999. 1999;1613:210–223. [Google Scholar]
- 14.De Craene M, d Aische AB, Macq B, Warfield SK. Multi-subject registration for unbiased statistical atlas construction. In Proceedings MICCAI 2004: Medical Image Computing and Computer-Assisted Intervention. 2004;3216:655–662. LNCS. [Google Scholar]
- 15.Davatzikos C, Genc A, Xua D, Resnick SM. Voxel-based morphometry using the ravens maps: Methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 16.DeCarli C, Haxby JV, Gillette JA, Teichberg D, Rapoport SI, Schapiro MB. Longitudinal changes in lateral ventricular volume in datients with dementia of the alzheimer type. Neurology. 1992;42(10):2029–2036. doi: 10.1212/wnl.42.10.2029. [DOI] [PubMed] [Google Scholar]
- 17.Harrell FE., Jr. Regression modelling strategies. 2001 doi: 10.1002/sim.4780030207. [DOI] [PubMed] [Google Scholar]
- 18.Fischl B, Salat D, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale A. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 19.Fischl B, van der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat D, Busa E, Seidman LJ, Goldstein J, Kennedy D, Cavinnes V, Makris N, Rosen B, Dale A. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- 20.Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and ad. Neurology. 2005;64:1032–1039. doi: 10.1212/01.WNL.0000154530.72969.11. [DOI] [PubMed] [Google Scholar]
- 21.Gao S, Hendrie HC, Hall KS, Hui S. The relationships between age, sex, and the incidence of dementia and alzheimer disease: A meta-analysis. Archives of General Psychiatry. 1998;55:809–815. doi: 10.1001/archpsyc.55.9.809. [DOI] [PubMed] [Google Scholar]
- 22.Geldmacher DS, Whitehouse PJ. Differential diagnosis of alzheimer’s disease. Neurology. 1997;48(5) doi: 10.1212/wnl.48.5_suppl_6.2s. [DOI] [PubMed] [Google Scholar]
- 23.Gooda CD, Johnsrude IS, Ashburner J, Henson RNA, Friston KJ, Frackowiak RSJ. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001;14(1):21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 24.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proceedings of the National Academy of Sciences. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guimond A, Meunier FJ, Thirion JP. Average brain models: A convergence study. Computer Vision and Image Understanding. 2000;77(2):192–210. [Google Scholar]
- 26.Head D, Buckner RL, Shimony JS, Williams LE, Akbudak E, Conturo TE, McAvoy M, Morris JC, Snyder AZ. Differential vulnerability of anterior white matter in nondemented aging with minimal acceleration in dementia of the alzheimer type: Evidence from diffusion tensor imaging. Cerebral Cortex. 2004;14:410–423. doi: 10.1093/cercor/bhh003. [DOI] [PubMed] [Google Scholar]
- 27.Heckemann RA, Hajnal J, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain mri segmentation combining label propagation and decision fusion. NeuroImage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 28.Hirayasu Y, Shenton ME, Salisbury DF, Dickey CC, Fischer IA, Mazzoni P, Kisler T, Arakaki H, Kwon JS, Anderson JE, Yurgelun-Todd D, Tohen M, McCarley RW. Lower left temporal lobe mri volumes in patients with first-episode schizophrenia compared with psychotic patients with first-episode affective disorder and normal subjects. American Journal of Psychiatry. 1998;155(10):1384–1391. doi: 10.1176/ajp.155.10.1384. [DOI] [PubMed] [Google Scholar]
- 29.Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia: A meta-analysis of voxel-based morphometry studies. The American Journal of Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- 30.Ibanez L, Schroeder W, Ng L, J Cates, and the Insight Software Consortium. The ITK Software Guide. 2005 [Google Scholar]
- 31.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphism atlas construction for computational anatomy. NeuroImage. 2004;23:151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Lemieux L, Bell GS, Sisodiya SM, Shorvon SD, Sander JWAS, Duncan JS. A longitudinal study of brain morphometrics using quantitative magnetic resonance imaging and difference image analysis. Neuroimage. 2003;20:22–33. doi: 10.1016/s1053-8119(03)00219-2. [DOI] [PubMed] [Google Scholar]
- 33.Makris N, Worth AJ, Sorensen AG, Papadimitriou GM, Wu O, Reese TG, Wedeen VJ, Davis TL, Stakes JW, Caviness VS, Kaplan E, Rosen BR, Pandya DN, Kennedy DN. Morphometry of in vivo human white matter association pathways with diffusion-weighted magnetic resonance imaging. Annals of Neurology. 1997;42(6):951–962. doi: 10.1002/ana.410420617. [DOI] [PubMed] [Google Scholar]
- 34.Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. Journal of Cognitive Neuroscience. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 35.Milligan GW, Cooper MC. An examination of procedures for determining the number of clusters in a data set. Psychometrika. 1985;50(2):159–179. [Google Scholar]
- 36.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 37.Park H, Bland PH, Hero AO, Meyer CR. Least biased target selection in probabilistic atlas construction. In Proceedings MICCAI2005: Medical Image Computing and Computer-Assisted Intervention. 2005;3750:419–426. doi: 10.1007/11566489_52. LNCS. [DOI] [PubMed] [Google Scholar]
- 38.Pohl KM, Fisher J, Grimson W, Kikinis R, Wells WM. A bayesian model for joint segmentation and registration. NeuroImage. 31;2006:228–239. doi: 10.1016/j.neuroimage.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 39.Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC. Volumetry of hippocampus and amygdala with high-resolution mri and three-dimensional analysis software: Minimizing the discrepancies between laboratories. Cerebral Cortex. 2000;10(4):433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- 40.Rubin EH, Storandt M, Miller JP, Kinscherf DA, Grant EA, Morris JC, Berg L. A prospective study of cognitive function and onset of dementia in cognitively healthy elders. Archives of Neurology. 1998;55:395–401. doi: 10.1001/archneur.55.3.395. [DOI] [PubMed] [Google Scholar]
- 41.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Transaction of Medical Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 42.Sabuncu MR, Balci SK, Golland P. Discovering modes of an image population through mixture modeling. In Proceedings MICCAI 2008: Medical Image Computing and Computer-Assisted Intervention. 2008;5242:381–389. doi: 10.1007/978-3-540-85990-1_46. LNCS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scahill RI, Frost C, Jenkins R, Whitwell JL, Rossor MN, Fox NC. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Archives of Neurology. 2003;60:989–994. doi: 10.1001/archneur.60.7.989. [DOI] [PubMed] [Google Scholar]
- 44.Schulz JB, Skalej M, Wedekind D, Luf AR, Abele M, Voigt K, Dichgans J, Klockgether T. Magnetic resonance imaging-based volumetry differentiates idiopathic parkinson’s syndrome from multiple system atrophy and progressive supranuclear palsy. Annals of Neurology. 2002;45(1):65–74. [PubMed] [Google Scholar]
- 45.Schwarz G. Estimating the dimension of a model. Annals of Statistics. 1978;6(2):461–464. [Google Scholar]
- 46.Studholme C, Cardenas V. A template free approach to volumetric spatial normalization of brain anatomy. Pattern Recognition Letters. 2004;25:1191–1202. [Google Scholar]
- 47.Talairach J, Tornoux P. Co-planar stereotaxic atlas of the human brain. Thieme Medical Publishers. 1998 [Google Scholar]
- 48.Thompson PM, Woods RP, Mega MS, Toga AW. Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain. Human Brain Mapping. 2000;9(2):81–92. doi: 10.1002/(SICI)1097-0193(200002)9:2<81::AID-HBM3>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;63(2):411–423. [Google Scholar]
- 50.Tsai A, Wells WM, Warfield SK, Willsky AS. An EM algorithm for shape classification based on level sets. Medical Image Analysis. 2005;9:491–502. doi: 10.1016/j.media.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Twining CJ, Cootes T, Marsland S, Petrovic V, Schestowitz R, Taylor C. A unified information-theoretic approach to groupwise non-rigid registration and model building. In Proceedings Information Processing in Medical Imaging 2005. 2005;3565:1–14. doi: 10.1007/11505730_1. LNCS. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Swank S, Glick IE, Gado MH, Miller MI, Morris JC, Csernansky JG. Changes in hippocampal volume and shape across time distinguish dementia of the alzheimer type from healthy aging. NeuroImage. 2003;20(2):667–682. doi: 10.1016/S1053-8119(03)00361-6. [DOI] [PubMed] [Google Scholar]
- 53.Woods RP, Dapretto M, Sicotte NL, Toga AW, Mazziotta JC. Creation and use of a Talairach-compatible atlas for accurate, automated, nonlinear intersubject registration, and analysis of functional imaging data. Human Brain Mapping. 1999;8(2–3):73–79. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<73::AID-HBM1>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woods RP, Grafton ST, Watson JD, Sicotte NL, Mazziotta JC. Automated Image Registration: II. Intersubject Validation of Linear and Nonlinear Models. Comp. Assisted Tomography. 1998;22(1):153–165. doi: 10.1097/00004728-199801000-00028. [DOI] [PubMed] [Google Scholar]
- 55.Yeo BTT, Sabuncu MR, Desikan R, Fischl B, Golland P. Effects of registration regularization and atlas sharpness on segmentation accuracy. Medical Image Analysis. 2008;12(5):603–615. doi: 10.1016/j.media.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeo BTT, Sabuncu MR, Mohlberg H, Amunts K, Zilles K, Golland P, Fischl B. What data to co-register for computing atlases; Proceedings of the International Conference on Computer Vision, IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis; 2007. pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zöllei L, Jenkinson M, Timonerm S, Wells WM. A marginalized MAP approach and EM optimization for pair-wise registration. In Proceedings Information Processing in Medical Imaging. 2007;4584:662–674. doi: 10.1007/978-3-540-73273-0_55. LNCS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zöllei L, Learned-Miller E, Grimson E, Wells W. Efficient population registration of 3d data. Computer Vision for Biomedical Image Applications. 2005;3765:291–301. LNCS. [Google Scholar]