Abstract
Xeroderma pigmentosum (XP) is caused by defects in the nucleotide excision repair (NER) pathway. NER removes helix-distorting DNA lesions, such as UV–induced photodimers, from the genome. Patients suffering from XP exhibit exquisite sun sensitivity, high incidence of skin cancer, and in some cases neurodegeneration. The severity of XP varies tremendously depending upon which NER gene is mutated and how severely the mutation affects DNA repair capacity. XPF-ERCC1 is a structure-specific endonuclease essential for incising the damaged strand of DNA in NER. Missense mutations in XPF can result not only in XP, but also XPF-ERCC1 (XFE) progeroid syndrome, a disease of accelerated aging. In an attempt to determine how mutations in XPF can lead to such diverse symptoms, the effects of a progeria-causing mutation (XPFR153P) were compared to an XP–causing mutation (XPFR799W) in vitro and in vivo. Recombinant XPF harboring either mutation was purified in a complex with ERCC1 and tested for its ability to incise a stem-loop structure in vitro. Both mutant complexes nicked the substrate indicating that neither mutation obviates catalytic activity of the nuclease. Surprisingly, differential immunostaining and fractionation of cells from an XFE progeroid patient revealed that XPF-ERCC1 is abundant in the cytoplasm. This was confirmed by fluorescent detection of XPFR153P-YFP expressed in Xpf mutant cells. In addition, microinjection of XPFR153P-ERCC1 into the nucleus of XPF–deficient human cells restored nucleotide excision repair of UV–induced DNA damage. Intriguingly, in all XPF mutant cell lines examined, XPF-ERCC1 was detected in the cytoplasm of a fraction of cells. This demonstrates that at least part of the DNA repair defect and symptoms associated with mutations in XPF are due to mislocalization of XPF-ERCC1 into the cytoplasm of cells, likely due to protein misfolding. Analysis of these patient cells therefore reveals a novel mechanism to potentially regulate a cell's capacity for DNA repair: by manipulating nuclear localization of XPF-ERCC1.
Author Summary
XPF-ERCC1 is a nuclease that plays a critical role in DNA repair. Mutations in XPF are linked to xeroderma pigmentosum, characterized by sun sensitivity, high incidence of skin cancer, and neurodegeneration, or XFE progeroid syndrome, a disease of accelerated aging. Herein we report the unexpected finding that mutations in XPF cause mislocalization of XPF-ERCC1 to the cytoplasm. Recombinant mutant XPF-ERCC1 derived from XP– and XFE–causing alleles are catalytically active and if delivered to the nucleus of cells restore DNA repair. This demonstrates that protein mislocalization contributes to defective DNA repair and disease arising as a consequence of mutations in XPF. It also illustrates a novel mechanism of regulating a cell's capacity for DNA repair: by manipulating nuclear localization of XPF-ERCC1 to enhance or inhibit repair and to prevent cancer or tumor resistance to chemotherapy, respectively.
Introduction
Xeroderma pigmentosum (XP) is a rare autosomal recessive disease characterized by photosensitivity and a greater than a 1000-fold increased risk of skin cancer in sun-exposed areas of the skin [1]. In approximately 20% of patients, there is also progressive neurodegeneration leading to loss of coordinated motion, vision and hearing [2],[3]. XP is caused by mutations in genes that encode proteins required for nucleotide excision repair (NER) of DNA. Eight complementation groups of XP have been identified based on fusion studies with XP patient cells. These complementation groups include XP-A through XP-G and a variant, XP-V. The severity of XP varies tremendously, with diagnosis occurring anywhere from infancy to adulthood [1]. The severity of the disease is determined largely by which gene is mutated and to what extent the mutation affects NER.
NER removes helix-distorting lesions in DNA, for example cyclobutane pyrimidine dimers (CPDs) and pyrimidine pyrimidone photoproducts (6–4PPs) caused by the ultraviolet (UV) component of sunlight [4]. There are two ways by which DNA damage is recognized in NER. Lesions anywhere in the genome can be recognized by the complex XPC-RAD23B [5],[6]. For some lesions, this is facilitated by a second complex XPE/DDB2-DDB1 [7]. Alternatively, lesions that occur in the coding strand of DNA, within transcribed regions, can trigger NER if they stall progression of RNA polymerase II [8],[9]. This requires CSA, CSB and XAB2 [10]–[12]. Once the damage is recognized, the subsequent steps of damage excision are believed to be uniform. The basal transcription factor TFIIH is recruited to the site of helix-distortion to unwind the DNA around the lesion, using two of its ten subunits, XPB and XPD [8],[13]. RPA and XPA bind the unwound repair intermediate to stabilize it and recruit subsequent factors. The damaged strand of DNA is then incised by two structure-specific endonucleases, the heterodimer of XPF-ERCC1 and XPG, which cut 5′ and 3′ of the lesion, respectively [14]–[17]. This leads to removal of the lesion as part of a 24–32 base oligonucleotide. The resultant gap is filled by the replication machinery including RPA, PCNA, RF-C, DNA polymerase δ/ε, and the backbone is sealed by DNA LIGI or LIGIIIα-XRCC3 [18]–[20].
XPF-ERCC1 is a highly conserved endonuclease that nicks double-stranded DNA 5′ to a junction with single-stranded DNA [21]. In addition to NER, XPF-ERCC1 is involved in the repair of DNA interstrand crosslinks (ICL) [22] and double-strand breaks [23]. XPF and ERCC1 are paralogs thought to have arisen by gene duplication of the conserved nuclease and helix-hairpin helix protein-interaction domains [24]. The proteins interact via their C-terminal helix-hairpin-helix domains and this interaction is required to stabilize both proteins in vivo [25],[26]. XPF contains the nuclease catalytic domain [27], whereas ERCC1 mediates the interaction with XPA and recruitment to NER complexes [28],[29]. Unlike NER-specific proteins XPA and XPC, XPF-ERCC1 appears to be essential for human development or viability since no patients have yet been identified who are homozygous for early nonsense or frameshift mutations in either gene.
XP-A and XP-C are among the most common complementation groups in XP [30]. XP-C patients have severe skin abnormalities but generally lack neurological symptoms [31]. In contrast, XP-A patients show profound neurodegeneration, in addition to cutaneous features [1]. XP-F patients typically have very mild cutaneous features of XP, including late onset of skin cancer, but often have complications due to neurodegeneration as adults. It was recently discovered that mutations in XPF can lead to a second disease, XFE progeroid syndrome (short for XPF-ERCC1), characterized by spontaneous, accelerated aging of multiple tissues, including the nervous system [32]. Herein, we attempted to understand the molecular basis for how mutations in XPF could lead to such diverse outcomes. This led to the surprising discovery that mutation of XPF promotes mislocalization of XPF-ERCC1 to the cytoplasm of cells.
Results
Characterization of R153P-XPF-ERCC1 activity in vitro
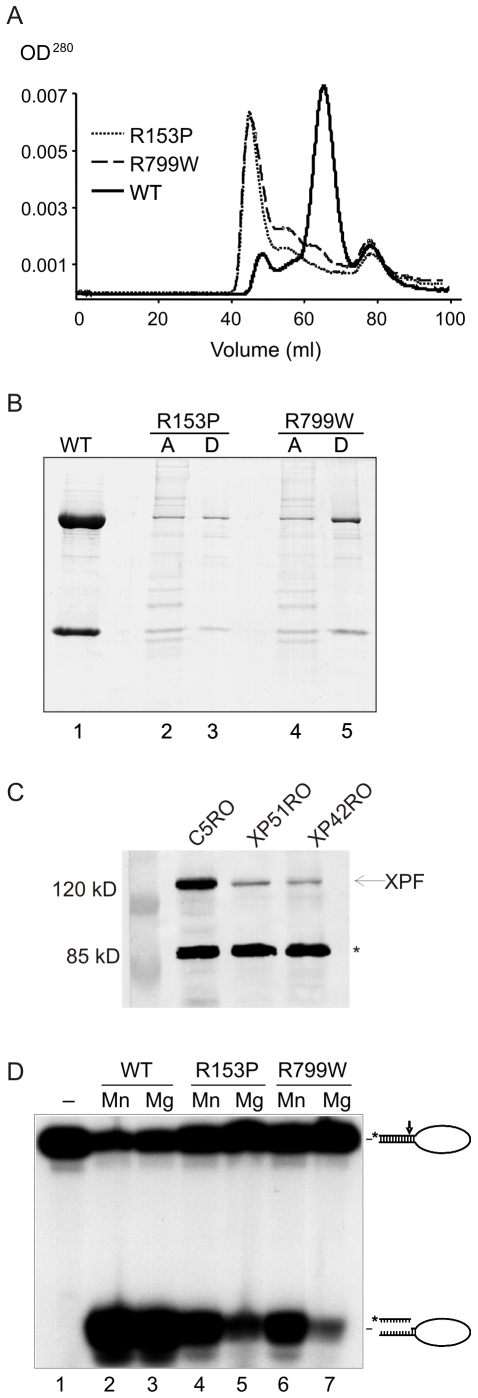
We first asked if mutations in XPF that cause mild or severe disease differentially affect the biochemical properties of XPF-ERCC1. To answer this, we compared the biochemical properties of XPF-ERCC1 from two patients, XP42RO (a patient with mild XP, homozygous for a mutation causing an R799W substitution in XPF [33]) and XP51RO (a patient with XFE progeroid syndrome, homozygous for a mutation causing an R153P substitution in XPF [32]) to that of wild type XPF-ERCC1. Recombinant XPFWT-ERCC1, XPFR153P-ERCC1 and XPFR799W-ERCC1 were purified from baculovirus-infected Sf9 insect cells using a His6 tag on ERCC1. We previously reported [27] that our purified preparations of XPFWT-ERCC1 elute from a gel-filtration column in three fractions: (1) a minor fraction in the void volume (∼45 ml) containing aggregated, inactive protein; (2) active heterodimeric XPF-ERCC1 at ∼65 ml, which corresponds to a molecular weight of ∼200 kD, as expected, and (3) monomeric ERCC1, which peaks at ∼78 mL, which corresponds to ∼50 kD. Recombinant XPFWT-ERCC1 eluted as expected (Figure 1A). Both mutant protein complexes eluted with similar profiles that differed substantially from that of XPFWT-ERCC1. The majority of the mutant complexes eluted at ∼45 mL rather than at 65 ml, indicating that they were aggregated. The peak at 78 ml, corresponding to free ERCC1 was identical for both mutant and WT XPF-ERCC1 preps. These results suggest that the mutations in XPF that cause both mild and severe disease lead to protein misfolding that does not interfere with ERCC1 binding, but does lead to protein aggregation.
Figure 1. Biochemical characterization of XPFR153P-ERCC1 and XPFR799W-ERCC1 mutants.
(A) Gel filtration profiles from the purification of recombinant XPF-ERCC1, XPF R153P-ERCC1 and XPF R799W-ERCC1 from baculovirus-infected Sf9 insect cells using a His6 tag on ERCC1. Aggregated proteins elute at ∼45 ml in the void volume of the column; heterodimeric XPF-ERCC1 elutes at ∼65 ml, corresponding to ∼200 kD, and monomeric ERCC1 elutes at ∼78 ml (∼50kD). (B) SDS PAGE analysis of purified protein complexes. Lane 1, 3 and 5 (D): XPF-ERCC1, XPF R153P-ERCC1 and XPF R799W-ERCC1, respectively, after purification over NTA-agarose, gel filtration and heparin columns. Lanes 2 and 4 (A) show the proteins present in the fractions eluting at 45 ml in the gel filtration column step of XPF R153P-ERCC1 and XPF R799W-ERCC1, respectively. (C) Immunodetection of XPF in normal (C5RO) and XPF mutant cells. The star indicates the migration of a cross-reactive band demonstrating equal loading [32]. (D) Incision activities of XPF-ERCC1, XPF R153P-ERCC1 and XPF R799W-ERCC1 (200 fmol) on a 5′-32P-labeled stem-loop DNA substrate (100 fmol) in the presence of either 0.4 mM MnCl2 (lanes 2, 4 and 6) or 2 mM MgCl2 (lanes 3, 5 and 7). Reactions were analyzed on a 15% denaturing polyacrylamide gel. The 46-mer substrate and 9–10-mer products are indicated.
We were able to purify a small amount of XPFR153P-ERCC1 and XPFR799W-ERCC1 from the fractions eluting at 65 ml, indicating that at least some of the mutant proteins are likely to be properly folded. SDS-PAGE analysis of the complexes after an additional purification step over a heparin column revealed dramatically reduced yields of the complexes of XPFR153P-ERCC1 and XPFR799W-ERCC1 compared to XPFWT-ERCC1 (Figure 1B). Similarly, the amount of XPF protein detectable by immunoblot in whole cell extracts of human fibroblasts harboring the XPFR153P and XPFR799W mutations (XP51RO and XP42RO, respectively) was reduced compared to normal cells (C5RO) (Figure 1C).
The catalytic activity of the purified heterodimers was investigated by measuring their ability to incise a 32P-end-labeled stem–loop DNA substrate at the single-strand:double-strand DNA junction in the presence of 0.4 mM MnCl2 or 2 mM MgCl2 at a 2-fold molar excess of protein over substrate (Figure 1D). With XPFWT-ERCC1, >80% of the stem-loop substrate was cleaved. Both XPFR153P-ERCC1 and XPFR799W-ERCC1 also incised the DNA substrate, demonstrating that both mutant complexes retain catalytic activity (Figure 1D, lanes 5 & 7). Incision by both mutant complexes was reduced compared to the WT complex. This may simply reflect the fact that preparations of mutant heteroduplexes were less concentrated than XPFWT-ERCC1 (Figure 1B), inevitably leading to differences in the buffering conditions between incision reactions.
We previously observed that mutant XPF-ERCC1 complexes tend to be more active in the presence of Mn2+ than Mg2+ since this metal has less stringent requirements for the proper alignment of the active site residues [27]. Consistent with this, incision by XPFR153P-ERCC1 and XPFR799W-ERCC1 was increased ∼2-fold in the presence of Mn2+ compared to Mg2+, whereas the cation had no effect on incision by XPFWT-ERCC1 (Figure 1D). These data support the conclusion that even monomeric XPFR153P and XPFR799W are to some extent misfolded. Notably, there was not a dramatic difference in the enzymatic activity of XPFR153P-ERCC1 and XPFR799W-ERCC1 on stem-loop DNA substrates, indicating that the biochemical basis for the more severe phenotype associated with the R153P mutation is unlikely to be simply a loss of catalytic activity.
Immunolocalization of XPF protein
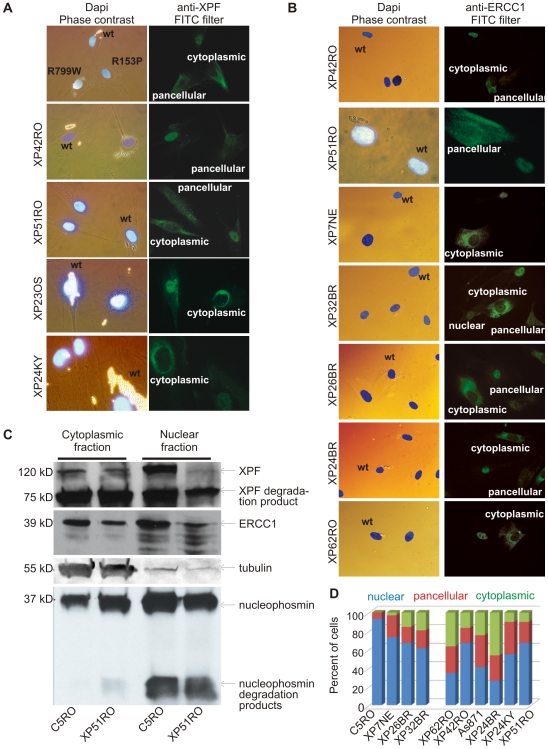
R153P is situated in a lysine-rich domain of XPF that might be part of a complex nuclear localization sequence (NLS). Thus we next asked if XPFR153P-ERCC1 is mislocalized in cells. Differential immunofluorescence was used to identify the subcellular localization of XPF-ERCC1 in XPF mutant cell lines that were co-cultured with normal cells as an internal control [34]. XPF was detected exclusively in the nucleus of normal fibroblasts (Figure 2A, upper panel). In the same sample, XPF was detected in the cytoplasm of XP51RO cells (harboring XPFR153P). In XP42RO cells (harboring XPFR799W), the XPF signal was pancellular so that the nucleus and cytoplasm could not be distinguished from one another. To explore this further, we used immunofluorescence to detect XPF in additional XP-F patient cell lines and to our surprise discovered that in all mutant cell lines XPF was frequently detected in the cytoplasm. In all cases, XPF mutant cell lines could be discriminated from normal fibroblasts by the staining pattern of the cell population: reduced nuclear XPF-ERCC1 and the presence of cells in which the heterodimer was exclusively cytoplasmic. This was true irrespective of the antibody used for analysis [monoclonal 3F2 (Cancer Research UK), monoclonal Ab-1 (Neomarkers), or polyclonal anti-XPF (Erasmus Medical Centre)] (data not shown). This indicates that mislocalization of XPF in the cytoplasm is a common consequence of XPF mutation.
Figure 2. Differential immunofluorescence of cells from patients with XPF mutations.
Fibroblasts from patients with mutations in XPF and a normal control were grown in the presence of different size beads. After 24 hr the cultures were washed to remove extracellular beads, mixed and co-plated on glass coverslips. The next day, the cells were fixed and immunostained as indicated. Cells were stained with Dapi to identify nuclei and examined by phase contrast microscopy to identify the cell type by their bead content and by fluorescence microscopy for immunodetection of XPF or ERCC1. (A) Analysis of XPF protein sub-cellular localization. Cells from an unaffected individual were labeled with 2 µM beads; XPF mutant cells were labeled with 0.8 µM beads. (B) Analysis of ERCC1 subcellular localization in patients with mutations in XPF. (C) Immunodetection of XPF and ERCC1 in nuclear and cytoplasmic fractions of normal fibroblasts (C5RO) and XPF mutant cells (XP51RO). Tubulin is used as a loading control of the cytoplasmic fraction. Nucleophosmin is used as a loading control for the nuclear fraction. (D) Quantitation of the fraction of cells containing exclusively nuclear XPF-ERCC1, XPF-ERCC1 in the nucleus and cytoplasm (pancellular) or exclusively cytoplasmic complex, as determined from immunofluorescence images (n≥100 cells per cell line).
This raised the possibility that misfolding of mutant XPF caused abnormal subcellular localization of the protein. Therefore, next we asked if these mutant XPF proteins still interacted with their obligate binding partner ERCC1. ERCC1 was detected in the nucleus of normal (wt) fibroblasts, as expected. However, like XPF, ERCC1 was frequently detected in the cytoplasm of XPF mutant cells (Figure 2B). All eight XPF mutant cells lines tested were readily distinguished from wt cells by their ERCC1 staining pattern. Furthermore, there was a strong correlation between the staining pattern for ERCC1 and XPF in all cell lines. This observation indicates that ERCC1 can interact with each of these mutant XPF proteins. Furthermore, the results suggest that normally ERCC1 enters the nucleus as a heterodimer with XPF, and is retained in the cytoplasm with XPF when XPF is misfolded.
To rule-out the possibility that the abnormal subcellular localization of XPF-ERCC1 was an artifact of immunofluorescence, wt and XPFR153P fibroblasts were fractionated and XPF-ERCC1 was detected by immunoblot in the nuclear and cytoplasmic fractions (Figure 2C). In normal cells (C5RO), XPF-ERCC1 is predominantly nuclear. XPF-ERCC1 was also detected in the cytoplasmic fraction, but to an extent that could be attributed to nuclear contamination, as determined by immunodetection of the nuclear protein nucleophosmin. In contrast, substantially more XPF and ERCC1 were detected in the cytoplasm than in the nucleus of XP51RO cells. Similar results were obtained for other XPF mutant cell lines (XP24BR, XP26BR and XP32BR) and using other antibodies against XPF and ERCC1 (data not shown). Degradation products of both ERCC1 and XPF are commonly detected on immunoblot, but were not overrepresented in the mutant cells. These data confirm the immunofluorescence data and support the conclusion that mislocalization of XPF-ERCC1 in the cytoplasm occurs in XPF mutant cells.
Remarkably, cytoplasmic XPF-ERCC1 is not detected by immunofluorescence in all XPF mutant cells within a population. To quantify the phenomena, the fractions of cells with exclusively nuclear, exclusively cytoplasmic, or pancellular XPF-ERCC1 were determined from immunofluorescence images (Figure 2D). In wt fibroblasts, 93% of cells have XPF-ERCC1 only in the nucleus. Seven percent of cells show pancellular XPF-ERCC1. But never is the complex seen exclusively in the cytoplasm. In all of the XPF mutant cell lines, XPF-ERCC1 was detected exclusively in the cytoplasm of a fraction of cells ranging from 3–46% of the total population. Thus all known XPF mutations lead to a reduction in nuclear XPF-ERCC1 and an increase in the amount of the complex detected in the cytoplasm.
Direct detection of XPFR153P
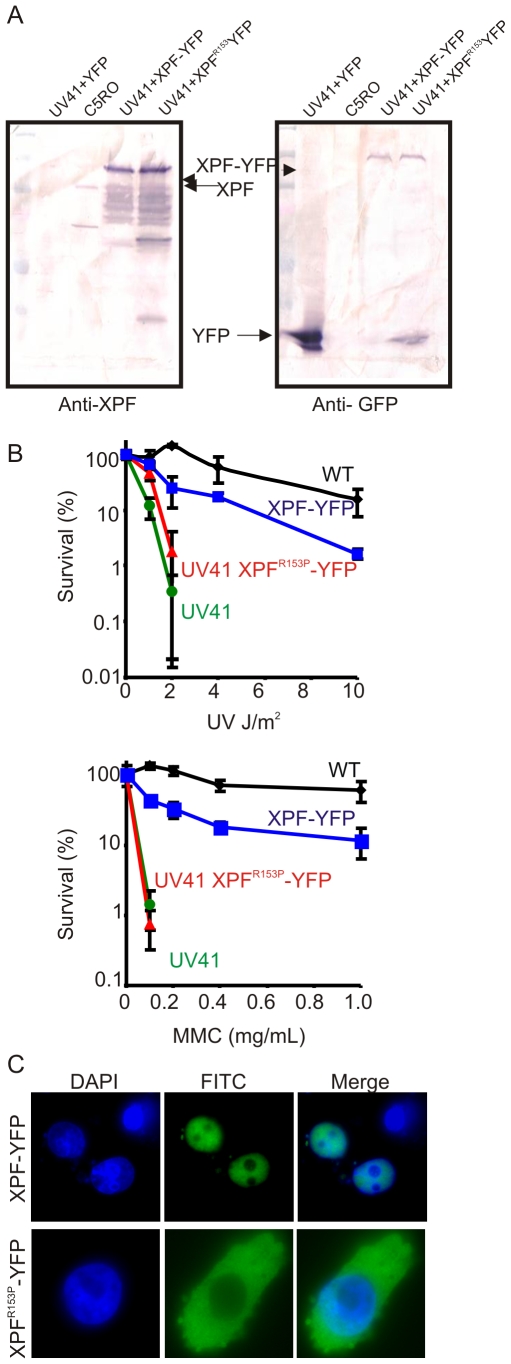
To further rule out the possibility that the cytoplasmic XPF-ERCC1 detected was an artifact generated by non-specific antibodies, human XPFR153P and XPFWT were tagged with YFP and expressed in Xpf mutant hamster cells (UV41) for direct detection of XPF protein. The expression of fusion proteins was confirmed by immunoblot using antibodies against human XPF and GFP (Figure 3A). Immunodetection of XPF revealed overexpression of both fusion proteins relative to endogenous XPF protein levels in normal fibroblasts (C5RO). Numerous breakdown products of XPF were also observed, likely due to its overexpression. But only a single fusion protein migrating at the expected molecular mass of full length XPF-YFP was detected using an antibody that detects GFP. To determine if the fusion proteins were functional, transiently transfected cells were tested for their sensitivity to UV to measure NER and mitomycin C (MMC) to measure interstrand crosslink repair (Figure 3B). Wild-type XPF-YFP yielded near complete correction of the hypersensitivity of UV41 mutant cells to UV and MMC. By contrast, despite the fact that XPFR153P_YFP was overexpressed to the same extent as XPFWT-GFP, this protein was unable to correct either DNA repair defect (Figure 3B), as expected based on the hypersensitivity of the XP51RO patient cell lines [32]. To determine the sub-cellular localization of XPFR153P, cells expressing the YFP-tagged protein were plated on glass coverslips and the protein detected by fluorescence microscopy (Figure 3C). XPFWT-YFP was exclusively in the nucleus. However, XPFR153P_YFP was detected in the cytoplasm of 95% of the transfected cells. This confirms the immunodetection data indicating that mutant XPF is cytoplasmic.
Figure 3. Characterization of XPF-YFP and XPF153-YFP in CHO cells.
(A) Western blot analysis of XPF-YFP expressed in Xpf mutant cells. XPF-deficient hamster cell line, UV41, was transiently transfected with wild type XPF-YFP or XPF153-YFP and the fusion proteins were detected using an antibody against XPF or GFP. C5RO was used as positive control for the XPF blot and as a negative control for the GFP blot. UV41 cells transfected with YFP alone was used as a negative control for XPF blot and as a positive control for GFP blot. (B) Clonogenic survival of wild-type (wt), XPF-deficient CHO cell line UV41, and UV41 transfected with wild type XPF-YFP and XPF153-YFP after UV and MMC treatment. Colonies were counted 7–10 days after treatment and results are plotted as mean 3 independent experiments. (C) Subcellular localization of wild type XPF-YFP and XPF153-YFP after transient transfection in XPF-deficient the CHO cell line UV41 detected by fluorescence microscopy.
Characterization of XPFR153P-ERCC1 activity in living cells
Unscheduled DNA synthesis (UDS) measures the incorporation of radiolabeled nucleotides into the genome of non-S phase cells after exposure to UV radiation and is a direct measure of NER [35]. Previously, UDS in cells from patient XP42RO (XPFR799W) and XP51RO (XPFR153P) was reported to be 20% and <5% of that in normal fibroblasts, respectively (Table 1). UV-induced UDS was measurable in all of the mutant XPF cell lines except XP51RO. This demonstrates that all of the mutant XPF proteins, with the exception of XPFR153P, retain catalytic activity in vivo.
Table 1. Characteristics of XPF mutant cell lines.
| Patient | Mutation Allele 1 | Mutation Allele 2 | Age (yr) | Skin Cancer | Clinical features | UDS | UV sensitivity | % of cells with non-nuclear XPF-ERCC1 | Ref |
| C5RO | none | none | – | normal | 100% | 1X | 7% | [33] | |
| Father of XP42RO | R799W | none | – | photosensitivity without skin lesions | 100% | 1X | rare | [33] | |
| XP23OS | 455fs | ?* | 45 | – | mild XP | 45% | 4X | rare | [60] |
| XP7NE | P379S | silent | 28 | – | mild XP | 30% | 2X | 27% | [61] |
| XP62RO° | R799W | R799W | mild XP with late onset neurodegeneration | 20% | not reported | 65% | |||
| XP42RO° | R799W | R799W | 62 | + | mild XP with late onset neurodegeneration | 20% | 2X | 33% | [33] |
| XP2YO | T567A ?♦ | 657fs | 65 | + | mild XP | 17% | 3X | n.d. | [46] |
| AS871 | R589W | del exon3 | severe XP with neurodegeneration | 15% | 2X | 59% | |||
| XP26BR | R799W | R799W | mild XP | 15% | not reported | 33% | |||
| XP32BR | R589W | P379S | 12 | – | mild XP | 10% | 2X | 39% | |
| XP24BR | R799W | R589W | 29 | – | severe XP with neurodegeneration | 5% | 3X | 74% | [61] |
| XP24KY | R799W | 537fs + 7bp | 50 | – | XP with late onset neurodegeneration | 7% | 3X | 45% | [46] |
| XP51RO | R153P | R153P | 16 | – | neurodegeneration severe progeria | <5% | 10X | >33% | [30] |
UDS unscheduled DNA synthesis
*The patient had normal levels of XPF transcript, suggesting one allele encodes a full-length mRNA.
♦: Mutation could not be confirmed on genomic DNA.
° Siblings.
n.d. = not determined
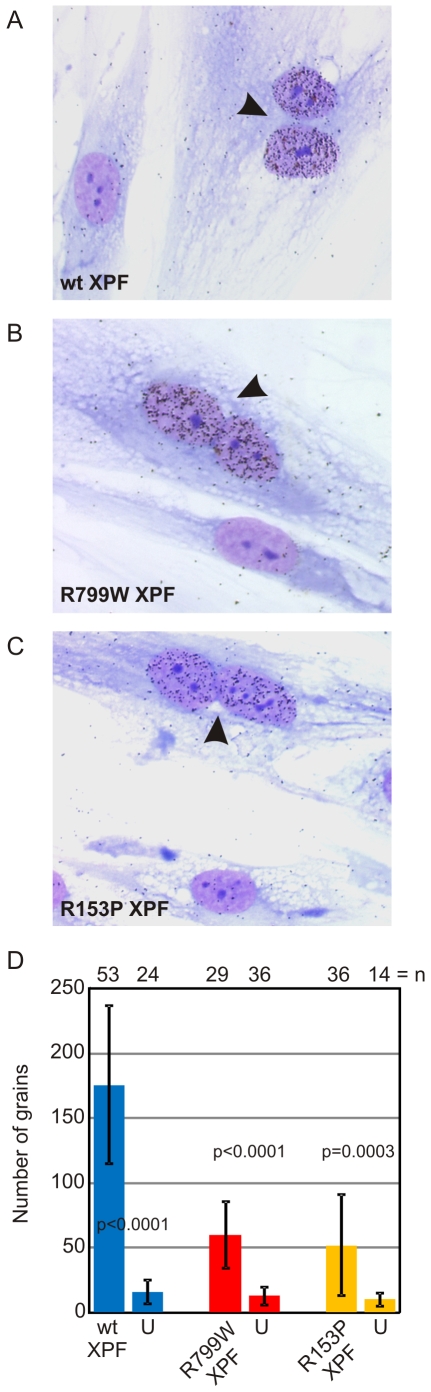
To ask if XPFR153P is also catalytically active in vivo, recombinant, purified XPFR153P-ERCC1 was microinjected into the nuclei of NER-deficient XP51RO primary fibroblasts to determine if UV-induced UDS could be restored. XP51RO cells were first fused on slides by treatment with inactivated Sendai virus to produce homopolykaryons (multinucleate cells). Only homopolykaryons were injected with protein, to permit identification of those cells that were injected with protein. The slides were irradiated with 10 J/m2 UV-C, cultured in the presence of 3H-thymidine and nuclear grains indicating sites of thymidine incorporation in non-S phase cells measured (Figure 4). As expected, delivery of XPFWT-ERCC1 to the nuclei of cells led to a significant increase in the number of grains detected in homopolykaryons relative to individual cells in the same culture (Figure 4A). A significant increase in UV-induced UDS was also detected in homopolykaryons injected with XPFR153P-ERCC1 (Figure 4C). This confirms the in vitro activity data and establishes that XPFR153P is catalytically active in vivo if it is delivered to the nucleus. UDS levels were not recovered to the same extent as when WT protein was injected (Figure 4D). But injection of XPFR153P-ERCC1 or XPFR799W-ERCC1 led to a similar increase in UDS. This suggests that the incomplete recovery of DNA repair is due to decreased concentration of the recombinant mutant proteins relative to XPFWT-ERCC1 (Figure 1). These data support the conclusion that mislocalization of XPF-ERCC1 contributes to the DNA repair defect and symptoms caused by XPF mutations.
Figure 4. Correction of XPF mutant cell NER defect by microinjection of XPF-ERCC1.
Primary fibroblasts from XFE progeroid patient XP51RO were fused to create homopolykaryons by treatment with inactivated Sendai virus then plated on glass coverslips. Only homopolykaryons were injected with recombinant XPF-ERCC1 protein complex (A) wild-type (B) XPFR799W-ERCC1 (C) XPFR153P-ERCC1. The cultures were irradiated with 10 J/m2 UV-C and 3H-thymidine was added to the culture. UV-induced unscheduled DNA synthesis was detected by autoradiography. Homopolykaryons are indicated with arrows. (D) Histogram indicating the average number of radiographic grains in nuclei injected with each of the recombinant protein complexes and uninjected cells in the same sample. Error bars indicate the standard deviation. N indicates the number of nuclei analyzed in each population. P values for the comparison between injected and uninjected cells were calculated using an unpaired two-tailed Student's t-test.
Clinical correlation
To determine if the severity of disease associated with a particular mutation in XPF could be predicted by the amount of XPF-ERCC1 detected in cell nuclei, the results in Figure 2D were compared to the clinical information available about the patients from which the cells were derived. In Table 1, the cell lines are listed in order of decreasing UDS. Patients with mild disease tend to have greater UDS or DNA repair. Patients XP32BR and XP26BR could be exceptions, but they are too young to know the full extent of their disease. In Figure 2D, the cell lines are clustered into those from patients in which severe disease/neurodegeneration was documented (right) or not yet observed (left). There is a trend towards those with severe disease to have more cells with non-nuclear XPF-ERCC1, but this trend did not reach significance (p = 0.06, unpaired Student's t-test), likely due to the small sample size. Therefore, the detection of cells with cytoplasmic XPF-ERCC1, while maybe useful to screen for patients with XPF mutations is not sufficient to predict patient prognosis.
Discussion
Classically, inherited mutations in a gene are associated with a single disease. However, mutations in several genes involved in the NER pathway can result in more than one disease. The most prominent example is XPD, which if mutated can cause the cancer-prone disease XP but also Cockayne Syndrome (CS) characterized by photosensitivity, growth retardation, developmental abnormalities and profound neurodegeneration, as well as trichothiodystrophy (TTD), which is similar to CS, but also involves the skin and nails [36]. Similarly, mutations in XPB can also cause XP, TTD and a combined XP-CS [37] and mutations in XPG can lead to XP or XP-CS [38]. Of all the genes whose products are required for NER, only XPB, XPD and XPG are required for the proper function and stability of the basal transcription factor TFIIH [39]. Thus the more severe symptoms of CS and TTD are attributed to a combined defect in NER as well as transcription [36], [40]–[42]. Mutations in XPF were recently linked to a second disease in addition to XP, a disease of systemic accelerated aging termed XFE progeroid syndrome [32]. In this study, we sought to determine how mutations in XPF can lead to such a wide variety of symptoms.
Mutations in XPF do not ablate catalytic activity
Since CS and TTD are attributed to defects in transcription [34],[43], the prediction is that mutations in XPB, XPD or XPG that cause CS or TTD should affect basal transcription in addition to NER, whereas mutations that affect only NER cause XP. Indeed, mutations in the catalytic domain of XPG, for example A792V, disrupt the endonuclease activity of XPG, but not its interaction with TFIIH and therefore causes classical XP [44]. Similarly, a mutation in the helicase domain of XPD, D234N, affects NER, but not basal transcription and therefore leads to XP [45]. By analogy, we examined the enzymatic activity of XPFR153P and XPFR799W, which cause XFE progeroid syndrome and XP respectively, and discovered that neither mutation ablates the catalytic activity of the protein. Recombinant protein complexes harboring either mutation are able to incise a stem-loop substrate in vitro (Figure 1) and to restore NER in vivo (Figure 4). This is in keeping with the fact that patients with XP-F have residual UDS or NER (Table 1). Intriguingly, XP-F patients tend to have much milder photosensitivity and later onset skin cancer than XP patients from other complementation groups with the same level of UDS [46]. One explanation for this is that NER occurs in XP-F cells but at a much slower rate [47], making UDS a relatively poor reflection of the true DNA repair capacity of a cell. In total, these data provide clear evidence that viable mutations in XPF do not ablate catalytic activity of the XPF-ERCC1 nuclease. Of note, all XP patients for which the mutation in XPF was confirmed by sequencing genomic DNA harbor one of three recurrent point mutations (R799W, R589W or P379S). The rarity and limited repertoire of only hypomorphic point mutations in patients strongly suggests that XPF-ERCC1 nuclease activity is essential for normal embryonic development.
Stability of XPFR799W-ERCC1 and XPFR153P-ERCC1 is reduced
Mutations in a single gene could lead to diverse clinical outcomes if mutations differentially affect the stability of the gene product. For example, mutations affecting the stability of the TFIIH complex are linked with TTD but not XP [34]. Total cellular XPF and ERCC1 are dramatically reduced in cells from a patient with XFE progeroid syndrome (Figure 2C). However, XPF levels are reduced to the same extent in whole cell extracts from a patient with mild disease and 20% of the normal level of NER (Figure 1C). Therefore, mutations in XPF clearly affect protein level, which undoubtedly contributes to reduced DNA repair and disease. However, the level of XPF-ERCC1 in patient cells, as detected by immunoblotting, is inadequate to explain the differences in the severity in the DNA repair defect and disease between patients with different mutations in XPF. Interestingly, in at least a subset of XP-F patients, XPF mRNA levels are normal, but XPF protein level is low [46], indicating that mutant XPF is unstable.
Mutations in XPF affect protein subcellular localization
The novel and unexpected finding is that mutation in XPF leads to increased cytoplasmic localization of the XPF-ERCC1 nuclease complex (Figure 2 and Figure 3) and that this aberrant subcellular localization is what prevents XPF-ERCC1 from participating in DNA repair (Figure 4). This was demonstrated by immunofluorescence detection of the complex using multiple antibodies. The results were confirmed by examining the subcellular localization of fluorescently tagged recombinant XPF (Figure 2). In further support of this, ERCC1 is also mislocalized to the cytoplasm of cells from the one patient reported with a mutation in ERCC1 [48].
Cytoplasmic localization of XPF-ERCC1 is not observed in all cells harboring XPF or ERCC1 mutations, suggesting the possibility that mutations affect proper folding of XPF-ERCC1 and that misfolded proteins are preferentially sequestered in the cytoplasm through interactions with other proteins or preferentially degraded. Alternatively, there may be tremendous selection for cells with nuclear XPF-ERCC1. This is consistent with the notion that the repair complex is essential for viability. Indeed, continuous passaging of XP51RO cells over years leads to a striking increase in the fraction of cells with nuclear XPF-ERCC1 and reduced sensitivity to the crosslinking agent mitomycin C (Ahmad, Bhagwat and Niedernhofer, unpublished data). Thus the fraction of cells with cytoplasmic XPF-ERCC1 may be underrepresented in Figure 2D, although only early passage cells were used in this study.
Disease caused by protein mislocalization
Many human diseases are caused by misrouting or mislocalization of proteins, ranging from metabolic disorders to cancer. Mislocalization of the tumor suppressors p53 [49], FOXO [50], p27Kip1 [51] and β-catenin [52] into the cytoplasm rather than the nucleus, leads to a loss of protein function and is associated with cancer. In contrast, mislocalization of NF-κB [53], BRCA1 and BARD1 [53],[54] from the cytoplasm into the nucleus is also associated with a variety of tumors. A classic example of a disease caused by protein mislocalization is cystic fibrosis which is caused by retention of the cystic fibrosis transmembrane conductance regulator (CFTR) protein in the endoplasmic reticulum, instead of its localizing to the cell surface [55],[56]. In addition, nephrogenic diabetes inspidus, retinitis pigmentosa, emphysema and α1-antitrypsin deficiency liver disease are also caused by mislocalized proteins [55].
Mislocalization of proteins may result from a mutated nuclear localization sequence (NLS) or nuclear export sequence (NES). Remarkably, the majority of the missense mutations in XPF is at arginine residues and leads to conversion of the arginine to a noncharged residue. So these mutations could affect a complex NLS. All of the point mutations (R→P, R→W and P→S) are also predicted to alter protein structure, supporting the notion that XPF mutations affect protein folding and/or protein:protein interactions that are critical for nuclear localization.
Our data add XPF to the list of proteins that if mislocalized contribute to disease. While this leads to novel insight into the regulation of XPF-ERCC1 and DNA repair in cells, the extent of XPF-ERCC1 mislocalization, as measured by immunodetection, does not predict the level of NER (UDS) or disease severity (Table 1). This could be because each mutation differentially affects folding of the protein and thereby differentially affects protein expression, protein degradation and/or cellular localization. Another possibility is that there are modifier proteins that influence disease severity, in particular in patients with homozygous mutations. However, we believe the former is of primary importance based on the observation that titration the level of expression of ERCC1-XPF in mice directly impacts lifespan and the severity of symptoms [57],[58].
In the case of XPF, it is the absence of XPF and its binding partner ERCC1 in the nucleus leading to reduced repair of genomic DNA that is disease-causing, rather than toxicity of mislocalized protein. Our data illustrate a novel mechanism by which the DNA repair capacity of a cell is determined: by nuclear localization of XPF-ERCC1. The identification of proteins that regulate this could lead to novel targets for improving DNA repair to treat patients with mutations in XPF or reduce cancer risk after exposure to genotoxic agents. Alternatively, these proteins would be excellent targets for small molecule inhibitors that would reduce repair and thereby prevent tumor resistance to genotoxic cancer therapies.
Materials and Methods
Biochemical characterization of XPFR153P-ERCC1 and XPFR799W-ERCC1
Purification of recombinant XPF–ERCC1 was performed essentially as previously reported [27] from baculovirus-infected Sf9 insect cells using a His6 tag on ERCC1. In brief, plasmids pFastBac1-XPF and pFastBac1-ERCC1-His were used to transfect Sf9 insect cells, and to amplify the virus according to the manufacturer's instructions (BAC TO BAC system; Life Technologies). Cell extracts were prepared 65 hr after infection with an MOI of 5 and highly purified protein was obtained using chromatography on Ni−agarose, gel-filtration and heparin columns. Only XPF-ERCC1 eluting as proper heterodimer on the gel filtration column at ∼65 ml of eluant was collected. The aggregated protein, eluting in the void volume (∼40–50 ml), was not used in experiments.
The endonuclease activity of wild-type and mutant XPF-ERCC1 was performed using a stem−loop substrate also as previously described [27]. A stem12−loop22 oligonucleotide (GCCAGCGCTCGGT22CCGAGCGCTGGC) was 5′-32P end-labeled. Nuclease reactions were performed on 100 fmol of DNA substrate and 20–200 fmol of XPF-ERCC1 protein in a total volume of 15 µl in optimized nuclease buffer (25 mM HEPES pH 8.0, 40 mM NaCl, 10% glycerol, 0.5 mM β-mercaptoethanol, 0.1 mg/ml bovine serum albumin and 0.4 mM MnCl2 or 2 mM MgCl2). The reactions were incubated at 30°C for 2 h and stopped by adding 15 µl of 90% formamide/10 mM EDTA and heating at 95°C for 5 min. Samples were loaded onto 15% denaturing polyacrylamide gels and reaction products were visualized by autoradiography and quantified on a PhosphorImager (STORM860; Molecular Dynamics).
Cell lines and culturing
Human fibroblasts immortalized with hTert were cultured in Ham's F10 with 10% fetal calf serum and antibiotics and incubated at 3% oxygen as described previously [32]. Cell lines included those derived from a normal individual (C5RO) [59], the parent of a patient, heterozygous for a mutation in XPF [33], XP-F patients (XP42RO) [33], XP23OS [60], XP24KY [46], XP7NE [61], XP32BR, XP26BR, XP24BR [61], and XP62RO, and a patient with XFE progeroid syndrome caused by a mutation in XPF (XP51RO) [32]. Unscheduled DNA synthesis (UDS) in these cells lines was previously reported as referenced above and confirmed in mixed cultures (XP-F cells co-cultured with normal cells using a more accurate click-staining method, as recently described [62].
Immunodetection of XPF in patient cells
Cells were trypsinized, washed twice with PBS and lysed with 1 ml NETT buffer (100 mM NaCl, 50 mM Tris base pH 7.5, 5 mM EDTA pH 8.0, 0.5% Triton X-100) containing Complete™ mini protease inhibitor cocktail and phosphatase inhibitor cocktail (Roche Molecular Biochemicals). Then the lysates were freeze-thawed twice in liquid nitrogen to disrupt nuclear membranes. From each sample, 50 µg of protein was resolved on 10% SDS-PAGE gels after boiling for 10 min in the presence of loading buffer. XPF was detected using a human XPF monoclonal antibody (clone 219; Neomarkers, Fremont, CA) at a dilution of 1∶1000.
Differential immunofluorescence of fibroblasts isolated from patients with mutations in XPF
Cultures of primary human fibroblasts from patients with mutations in XPF or a normal individual were grown in the presence of different size beads (2 µm or 0.8 µm; Sigma). After 24 hr the cultures were trypsinized and washed extensively with phosphate-buffered saline to remove any extracellular beads. The cells were then mixed in various combinations and co-plated on glass coverslips to provide internal controls of normal XPF-ERCC1 protein levels [34]. After 16 hr, the cells were fixed with 2% paraformaldehyde in sodium phosphate buffer, pH 7.4, for 15 min then permeabilized with 0.1% Triton X-100 in PBS. The samples were immunostained with polyclonal anti-ERCC1 (1∶2000; [63]) or polyclonal anti-XPF (1∶1000; [16]) followed by goat anti-rabbit ALEXA 488 (1∶500; Molecular Probes). Samples were stained with Dapi to identify nuclei and examined by phase contrast microscopy to identify the genotype of the cells according to their bead content and by fluorescence microscopy for immunodetection of repair proteins.
Cell fractionation
Cells were fractionated into nuclear and cytosolic fractions as described [64], with minor modification. In brief, cells were trypsinized, pelleted and washed twice with PBS. The pellet was vortexed at maximum speed for 15 sec with 200 µl of CERI reagent from the Pierce NE-PER fractionation kit (Pierce Biotech) with Complete™ mini protease inhibitor cocktail, then incubated on ice for 10 min. This was followed by addition of 11 µl of CERII, vortexing for 5 sec and incubation on ice for 1 min. The mixture was spun at 13,000 rpm for 5 min and the supernatant collected as the cytosolic fraction. The nuclei in the pellet were suspended in extraction buffer (15 mM Tris-HCl pH 7.3, 1 mM EDTA, 0.4 M NaCl, 1 mM MgCl2, 10% glycerol, 10 mM β-mercaptoethanol and Complete™ mini protease inhibitor cocktail), mixed for 1 hr at 4°C and spun down at 13,000 rpm for 10 min. The supernatant was collected as the soluble nuclear fraction.
Subcellular localization of XPF–YFP in Chinese hamster Ovary (CHO) cells
XPF cDNA was cloned into pYFP-N1 (BD Biosciences Clontech, Palo Alto, CA) such that YFP was expressed as fusion protein at the C-terminus of XPF. This construct, pXPF-YFP-N1, was then used to create XPFR153P-YFP by QuickChangeR Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX) according to the manufacturer's instructions. The wild type and mutant constructs were transfected in XPF-deficient CHO cell lines UV41 or UV47 using lipofectamine 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Cells expressing YFP were flow sorted using Dako Cytomation MoFLo high-speed cell sorter (Dako North America, Carpinteria, CA) 24–48 hrs after transfection.
To study the subcellular localization of XPF, YFP-positive CHO cells were plated on glass coverslips and grown to 95% confluency. The next day, the samples were fixed with 2% paraformaldehyde in sodium phosphate buffer, pH 7.4, for 15 min. The cells were permeabilized with 0.1% Triton X-100 in phosphate-buffered and nuclei were stained with Dapi-vector shield (Vector Laboratories, Inc. Burlingame, CA). XPF-YFP was visualized using an Olympus BX51 fluorescent 4 microscope at 60–100X magnification.
Clonogenic survival assays of wild-type and mutant CHO cells
Wild type (AA8), XPF-deficient (UV41), XPF-YFP and XPFR153P-YFP cells were seeded in 6 cm dishes in triplicate at 103–104 cells per plate, depending on the dose of genotoxin. After 16 hr, the cells were irradiated with UV-C or exposed to mitomycin C (MMC). After approximately one week, the cultures were fixed and stained with 50% methanol, 7% acetic acid and 0.1% Coomassie blue. Colonies, consisting of at least 10 cells, were counted using a Nikon SMZ 2B 15 stereomicroscope microscope with 10X eyepiece. The data were plotted as the number of colonies that grew on the treated plates relative to untreated plates ± the standard error of the mean for 2–3 independent experiments.
Immunoblotting of XPF in Wt and mutant CHO cells
Whole cell extracts were prepared from C5RO and UV41 cells transfected with vectors expressing YFP, XPF-YFP or XPFR153P-YFP. Proteins were separated by SDS PAGE using a 10% gel. XPF was detected using a human XPF monoclonal antibody (clone 219; Neomarkers, Fremont, CA) at a dilution of 1∶1000. YFP was detected using a GFP monoclonal antibody (Clones 7.1 and 13.1; Roche, Indianapolis, IN) at a dilution of 1∶1000.
Correction of XPF mutant cell UV sensitivity by micro-injection of recombinant XPF-ERCC1
Microinjection of purified proteins was performed as previously described [65],[66]. Briefly, primary human fibroblasts from XP51RO were fused by treating cultures with inactivated Sendai virus and then plated on glass coverslips. Subsequently, purified, recombinant XPF-ERCC1 protein complex (wild type or containing the R799W or R153P substitution in XPF) was injected into the nuclei of homopolykaryons. The cultures were irradiated with 10 J/m2 UV-C and pulse labeled for 3 hrs with 3H-thymidine. Unscheduled DNA synthesis (UDS) was detected by autoradiography.
One to ten femtoliters of a 10–100 nM solution was injected into the nuclei of 10–20 homopolykaryons for each of the three recombinant proteins and the number of radiographic grains counted in at least 20 nuclei of the homopolykaryons and a similar number of nuclei of single cells in the same sample. The mean and standard deviation of the number of grains was calculated for each of the three proteins. An unpaired, two-tailed Student's t-test was used to determine if there was a significant difference in unscheduled DNA synthesis between cells that were injected with recombinant XPF-ERCC1 and cells that were not injected.
Footnotes
The authors have declared that no competing interests exist.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was supported by grants ES016114 and CA121411 to LJN and by CA092584 and GM080454 to ODS.
References
- 1.Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- 2.Nouspikel T. Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 2008;7:1155–1167. doi: 10.1016/j.dnarep.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Robbins JH, Brumback RA, Mendiones M, Barrett SF, Carl JR, et al. Neurological disease in xeroderma pigmentosum. Documentation of a late onset type of the juvenile onset form. Brain. 1991;114 ( Pt 3):1335–1361. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- 4.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 5.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 6.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 7.Sugasawa K. UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. J Mol Histol. 2006;37:189–202. doi: 10.1007/s10735-006-9044-7. [DOI] [PubMed] [Google Scholar]
- 8.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 10.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, et al. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 11.Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell. 1995;82:555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 12.Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem. 2000;275:34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- 13.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 15.Bardwell AJ, Bardwell L, Tomkinson AE, Friedberg EC. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 16.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 17.Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shivji MK, Podust VN, Hubscher U, Wood RD. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry. 1995;34:5011–5017. doi: 10.1021/bi00015a012. [DOI] [PubMed] [Google Scholar]
- 19.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 21.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J Biol Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 22.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28:5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Laat WL, Sijbers AM, Odijk H, Jaspers NG, Hoeijmakers JH. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 1998;26:4146–4152. doi: 10.1093/nar/26.18.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 1993;12:3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. Embo J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, et al. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J. 2007;26:4768–4776. doi: 10.1038/sj.emboj.7601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci U S A. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng L, Quilliet X, Chevallier-Lagente O, Eveno E, Sarasin A, et al. Vol. 4. Gene Ther; 1997. Retrovirus-mediated gene transfer corrects DNA repair defect of xeroderma pigmentosum cells of complementation groups A, B and C. pp. 1077–1084. [DOI] [PubMed] [Google Scholar]
- 31.Moriwaki S, Kraemer KH. Xeroderma pigmentosum–bridging a gap between clinic and laboratory. Photodermatol Photoimmunol Photomed. 2001;17:47–54. doi: 10.1034/j.1600-0781.2001.017002047.x. [DOI] [PubMed] [Google Scholar]
- 32.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 33.Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, et al. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110:832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- 34.Vermeulen W, Bergmann E, Auriol J, Rademakers S, Frit P, et al. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat Genet. 2000;26:307–313. doi: 10.1038/81603. [DOI] [PubMed] [Google Scholar]
- 35.Hoeijmakers JH, Eker AP, Wood RD, Robins P. Use of in vivo and in vitro assays for the characterization of mammalian excision repair and isolation of repair proteins. Mutat Res. 1990;236:223–238. doi: 10.1016/0921-8777(90)90007-r. [DOI] [PubMed] [Google Scholar]
- 36.Taylor EM, Broughton BC, Botta E, Stefanini M, Sarasin A, et al. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc Natl Acad Sci U S A. 1997;94:8658–8663. doi: 10.1073/pnas.94.16.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermeulen W, Scott RJ, Rodgers S, Muller HJ, Cole J, et al. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am J Hum Genet. 1994;54:191–200. [PMC free article] [PubMed] [Google Scholar]
- 38.Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG. A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci U S A. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 42.Takayama K, Salazar EP, Broughton BC, Lehmann AR, Sarasin A, et al. Defects in the DNA repair and transcription gene ERCC2(XPD) in trichothiodystrophy. Am J Hum Genet. 1996;58:263–270. [PMC free article] [PubMed] [Google Scholar]
- 43.van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. Embo J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarkson SG. The XPG story. Biochimie. 2003;85:1113–1121. doi: 10.1016/j.biochi.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Dubaele S, Proietti De Santis L, Bienstock RJ, Keriel A, Stefanini M, et al. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 46.Matsumura Y, Nishigori C, Yagi T, Imamura S, Takebe H. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum Mol Genet. 1998;7:969–974. doi: 10.1093/hmg/7.6.969. [DOI] [PubMed] [Google Scholar]
- 47.Zelle B, Berends F, Lohman PH. Repair of damage by ultraviolet radiation in xeroderma pigmentosum cell strains of complementation groups E and F. Mutat Res. 1980;73:157–169. doi: 10.1016/0027-5107(80)90144-x. [DOI] [PubMed] [Google Scholar]
- 48.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moll UM, Riou G, Levine AJ. Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc Natl Acad Sci U S A. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 51.Min YH, Cheong JW, Kim JY, Eom JI, Lee ST, et al. Cytoplasmic mislocalization of p27Kip1 protein is associated with constitutive phosphorylation of Akt or protein kinase B and poor prognosis in acute myelogenous leukemia. Cancer Res. 2004;64:5225–5231. doi: 10.1158/0008-5472.CAN-04-0174. [DOI] [PubMed] [Google Scholar]
- 52.Rosin-Arbesfeld R, Cliffe A, Brabletz T, Bienz M. Nuclear export of the APC tumour suppressor controls beta-catenin function in transcription. Embo J. 2003;22:1101–1113. doi: 10.1093/emboj/cdg105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 54.Fabbro M, Rodriguez JA, Baer R, Henderson BR. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- 55.Edwards SW, Tan CM, Limbird LE. Localization of G-protein-coupled receptors in health and disease. Trends Pharmacol Sci. 2000;21:304–308. doi: 10.1016/s0165-6147(00)01513-3. [DOI] [PubMed] [Google Scholar]
- 56.Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 57.Weeda G, Donker I, de Wit J, Morreau H, Janssens R, et al. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–439. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 58.Dolle ME, Busuttil RA, Garcia AM, Wijnhoven S, van Drunen E, et al. Increased genomic instability is not a prerequisite for shortened lifespan in DNA repair deficient mice. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 59.Poot M, Verkerk A, Koster JF, Esterbauer H, Jongkind JF. Influence of cumene hydroperoxide and 4-hydroxynonenal on the glutathione metabolism during in vitro ageing of human skin fibroblasts. Eur J Biochem. 1987;162:287–291. doi: 10.1111/j.1432-1033.1987.tb10598.x. [DOI] [PubMed] [Google Scholar]
- 60.Arase S, Kozuka T, Tanaka K, Ikenaga M, Takebe H. A sixth complementation group in xeroderma pigmentosum. Mutat Res. 1979;59:143–146. doi: 10.1016/0027-5107(79)90202-1. [DOI] [PubMed] [Google Scholar]
- 61.Berneburg M, Clingen PH, Harcourt SA, Lowe JE, Taylor EM, et al. The cancer-free phenotype in trichothiodystrophy is unrelated to its repair defect. Cancer Res. 2000;60:431–438. [PubMed] [Google Scholar]
- 62.Limsirichaikul S, Niimi A, Fawcett H, Lehmann A, Yamashita S, et al. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 2009;37:e31. doi: 10.1093/nar/gkp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Vuuren AJ, Appeldoorn E, Odijk H, Humbert S, Moncollin V, et al. Partial characterization of the DNA repair protein complex, containing the ERCC1, ERCC4, ERCC11 and XPF correcting activities. Mutat Res. 1995;337:25–39. doi: 10.1016/0921-8777(95)00009-9. [DOI] [PubMed] [Google Scholar]
- 64.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Jonge AJ, Vermeulen W, Klein B, Hoeijmakers JH. Microinjection of human cell extracts corrects xeroderma pigmentosum defect. Embo J. 1983;2:637–641. doi: 10.1002/j.1460-2075.1983.tb01477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weeda G, van Ham RC, Vermeulen W, Bootsma D, van der Eb AJ, et al. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990;62:777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]