Abstract
One of the pivotal functions of endogenous tumor suppression is to oppose aberrant cell survival, but the molecular requirements of this process are not completely understood. Here, we show that caspase 2, a death effector with largely unknown functions, represses transcription of the survivin gene, a general regulator of cell division and cytoprotection in tumors. This pathway involves caspase 2 proteolytic cleavage of the NFκB activator, RIP1. In turn, loss of RIP1 abolishes transcription of NFκB target genes, including survivin, resulting in deregulated mitotic transitions, enhanced apoptosis, and suppression of tumorigenicity, in vivo. Therefore, caspase 2 functions as an endogenous inhibitor of NFκB-dependent cell survival, and this mechanism may contribute to tumor suppression in humans.
Keywords: Caspase-2, survivin, NFκB, RIP1, tumor growth, gene expression
INTRODUCTION
The process of malignant transformation is almost invariably associated with a heightened cell survival threshold (Luo et al., 2009), which in turn contributes to disease progression, metastatic dissemination, and resistance to conventional or targeted therapy. Defying cell death under these conditions typically involves aberrant expression and function of anti-apoptotic mechanisms coordinated by Bcl-2 or Inhibitor of Apoptosis (IAP) gene families, which oppose mitochondrial dysfunction (Cory and Adams, 2002), or antagonize caspase activity (Srinivasula and Ashwell, 2008), respectively. Conversely, intrinsic pro-cell death signals can be activated to oppose these cytoprotective processes, by silencing the expression of survival factors (Accili and Arden, 2004), antagonizing their functions (Du et al., 2000), or upregulating the cellular levels of death effectors (Vogelstein et al., 2000).
Such endogenous cell death pathways are likely to play an important role against malignant transformation, as multiple tumor suppression mechanisms overlap in their ability to remove a cell’s survival advantage and execute apoptosis (Lowe et al., 2004). One of the targets of disparate tumor suppression networks is survivin (Altieri, 2008), a unique IAP protein with essential roles in the control of mitosis and protection from apoptosis, whose expression in cancer is required to maintain the malignant phenotype. In this context, strategies to mimic or (re)activate an endogenous cell death machinery are being intensely pursued for novel cancer therapeutics (Fesik, 2005), and targeting survivin may provide an attractive approach to lower a global anti-apoptotic and proliferative threshold in tumor cells (Mita et al., 2008).
Caspases are pivotal effectors of the intrinsic cell death machinery. This has been historically linked to their ability to dismantle the cellular architecture (Shi, 2002), but many of these molecules have been recently found intercalated in multiple signaling mechanisms of cell proliferation, migration and differentiation (Li and Yuan, 2008) that may also contribute to cell death regulation. There is correlative evidence that, at least in some cases, caspase signaling in cell proliferation may antagonize tumor growth, in vivo. Accordingly, loss or inactivation of caspase 3 (Soung et al., 2004), caspase 10 (Shin et al., 2002), or caspase 8 (Stupack et al., 2006) has been documented in several types of human tumors, and potentially associated with disease dissemination and unfavorable outcome. In this context, caspase 2 is an evolutionary conserved apical caspase (Krumschnabel et al., 2009), whose pleiotropic signaling properties have been associated with endoplasmic reticulum stress (Upton et al., 2008), cytoskeletal disruption (Ho et al., 2008), mitotic catastrophe (Castedo et al., 2004), p53-dependent DNA damage (Baptiste-Okoh et al., 2008), and, more recently, tumor suppression, in vivo (Ho et al., 2009). Although caspase 2 knockout mice show no overt phenotype of (Bergeron et al., 1998), caspase 2−/− fibroblasts are more prone to oncogenic transformation, resist apoptosis, and exhibit accelerated tumor growth in mice (Ho et al., 2009). However, the mechanistic underpinning of how caspase 2 may function in endogenous tumor suppression have not been elucidated (Ho et al., 2009).
In this study, we investigated mechanisms of cell death regulation in tumor cells, and we unraveled a novel pathway of caspase 2-mediated tumor suppression centered on acute silencing of the survivin gene.
MATERIALS AND METHODS
Cells and culture conditions
Human colorectal p53+/+ and p53−/− HCT116 cancer cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD). For generation of stable clones, HCT116 p53+/+ cells were transfected with wild type (WT) HA-tagged caspase 2 or caspase 9 cDNA, and selected in 1 mg/ml G418 (GIBCO). Colonies were picked after 2 weeks, and confirmed for expression of HA or caspase 9 by Western blotting. Breast adenocarcinoma MCF-7 cells stably transfected with survivin were described previously (Ghosh et al., 2008).
Plasmids and antibodies
A full length wild type caspase 2 cDNA (Invitrogen) was amplified by PCR with primers 5′-ATATACTCGAGTAAGCGGGAAATGGCGGCGCCG-3′ (forward) and 5′-ATAGAGTCTAGATCATGTGGGAGGGTGTCCTGG-3′ (reverse), digested with XbaI and XhoI and inserted in HA-tagged pcDNA3.1. For recombinant protein expression, a full length caspase 2 cDNA was cloned into pGEX-4T vector (Amersham Biosciences) using SmaI and XhoI restriction sites. Primers used for amplification of recombinant caspase 2 (C2) were 5′-ATATACCCGGGTAAGCGGGAAATGGCGGCGCCG-3′ (forward) and 5′-ATAGAGCTCGAGTCATGTGGGAGGGTGTCCTGG-3′ (reverse). A constitutively active caspase 2 cDNA (Casp. 2 152 Ac) was generated by PCR by removal of the prodomain to mimic the processed caspase using primers 5′-GCCTGTCGACAGATACTGTGGAACACTCC-3 (forward) and 5′-ATAGAGCTCGAGTCATGTGGGAGGGTGTCCTGG-3′ (reverse). Full length or truncated caspase 2 mutants were generated by replacing the active site Cys320 to Ala (C320A) using site-directed mutagenesis (Stratagene). The catalytic activity of the various caspase 2 constructs was determined using a colorimetric assay kit (Calbiochem) in the presence of VDVAD-pNA as a substrate.
An 830 nt mouse survivin promoter construct fused upstream of GFP (ms-830-GFP) was characterized previously (Xia and Altieri, 2006). A putative NFκB consensus site at position - 150 nt in ms-830-GFP was mutated using forward primer 5′-GGCGTGGGGCctGACTaTCCCGGCTCG-3′ (NFκBΔ). A RIP1 cDNA was the gift of Dr. Michelle Kelliher (University of Massachusetts Medical School). Wild type or mutant (S529A) p65 NFκB cDNA was the gift of Dr. Neil Silverman (University of Massachusetts Medical School). A truncated RIP1 NH2 fragment (residues 1-350) was generated using primers 5′-GGATCCCCGGAATTCAGAATGCAACCAGACATG-3′ (forward) and 5′-TTACTCCTCGAGAGGACCCTACCCAAGTCCCTG-3′ (reverse). A RIP1 COOH terminus fragment (residues 351-672) was generated using primers 5′-TCCCAGGAATTCGGGATGGGTCCTGTGGAGGAG-3′ (forward) and 5′-CGGCCGCTCGAGTTAGTTCTGGCTGACGTAAAT-3′ (reverse). Both RIP1 fragments were amplified and digested with EcoR1 and XhoI and inserted into pcDNA3.1 vector. The antibodies against caspase 2, 3, 8 and 9 were from Cell Signaling. Antibodies to the p65 subunit of NFκB, Bcl-xL (Santa Cruz), survivin (NOVUS Biologicals) or RIP1 (BD Biosciences) were used.
Protein and RNA analysis
Recombinant caspase 2 fused to GST was expressed in BL-21 E.coli strain, as described (Kang and Altieri, 2006). All RIP1 cDNA constructs (full length, NH2 terminus and COOH terminus fragments) were transcribed and translated in vitro using T7, TNT coupled rabbit reticulocyte lysate system (Promega). Aliquots of 35S methionine-labeled RIP1 were incubated with recombinant caspase 2 at 37oC for 1 h, separated by SDS gel electrophoresis, and analyzed by autoradiography. Changes in survivin mRNA expression were analyzed by semi-quantitative RT-PCR using primers for survivin, 5′-GCATGGGTGCCCCGACGTTG-3′ (forward) and 5′-GCTCCGGCCAGAGGCCTCAA-3′ (reverse) and GAPDH as described (Xia and Altieri, 2006), or real time PCR (QR-PCR), using fluorescent TaqMan and Applied Biosystem’s gene expression assays Hs00153353_m1BIRC5 (survivin) and Hs9999905_m1GAPDH. Total RNA was extracted using RNeasy (Qiagen), and reverse transcribed using first strand cDNA synthesis kit (Invitrogen). Analysis of gene expression was done using a relative quantification ddCt method.
Transfections and reporter assays
β-galactosidase-normalized survivin promoter (pLuc-1430c, pLuc-649c, pLuc-441c, and pLuc-230c) luciferase activity was quantified as described (Li and Altieri, 1999). Differential ms-830-GFP expression in transfected cells was analyzed by fluorescence microscopy and Western blotting, as described (Xia and Altieri, 2006). Gene silencing experiments by small interfering RNA (siRNA) directed to caspase 2, 3, 8 or RIP1 (Dharmacon) were carried out as described (Lee et al., 2008). Two independent siRNA oligos targeting caspase 2 were tested to eliminate possibility of off-target effects (data not shown). A non-targeted siRNA characterized previously (Lee et al., 2008) was used as control.
Flow cytometry
Transfected HCT116 cells were treated with the apoptotic stimulus, staurosporine (STS, 0.8–1 μM), and analyzed by multiparametric flow cytometry using CaspaTag caspase 3 activity kit (Intergen). In some experiments, transfected cells were treated with STS and analyzed for nuclear morphology of apoptosis after 14–16 h, by fluorescence microscopy. Cell cycle analysis was carried out in thymidine-synchronized HCT116 stable transfectants. Cells were treated with 1μM thymidine for 16 hours to induce a G1 cell cycle arrest followed by release and collection of cells at various timepoints. Analysis of DNA content was done by propidium iodide staining and flow cytometry.
Electrophoretic mobility shift assay (EMSA)
Nuclear fractions were purified from HCT116 cells using NucBuster kit (Novagen). DNA probes were synthesized using the survivin promoter sequence containing the NFκB site (5′-GTGGGGCGGGACTTTCCCGGCTC-3′) and end-labeled with [γ-32P] deoxyadenosine triphosphate, 1 μl T4 kinase, and 2.5 μl PNK for 15 min at 37°C and then 15 min at 65°C. The labeled probes were purified using nucleic acid purification columns (Bio-Rad), and incubated with 15 μg of nuclear extract as described (Lee et al., 2008). To determine binding specificity, 100-fold excess of unlabeled competitor, mutant competitor or antibody to p65 subunit of NFκB (Santa Cruz) was used as indicated. The reactions were resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography.
Analysis of tumorigenicity
Stably transfected HCT116 cells were cultured in soft agar for 14 days at 37°C, and colonies (>50 cells) were counted by light microscopy. All experiments involving animals wereapproved by an Institutional Animal Care and Use Committee at the University of Massachusetts Medical School. HCT116 transfectants were injected (2×106 in 100 μl of PBS) subcutaneously into the flanks of 6 to 8 week-old female CB17 severe combined immunodeficient (SCID)/beige mice (3 mice per group, 2 tumors per mouse, 2 independent experiments). Tumor growth was monitored every other day, and tumor size was calculated with a caliper according to the formula L × W2/2 (mm3).
Statistical analysis
Data were analyzed using the unpaired t test on a GraphPad software package for Windows (Prism 4.0). A p-value of 0.05 was considered as statistically significant.
RESULTS
Caspase 2 activity represses survivin gene expression in tumor cells
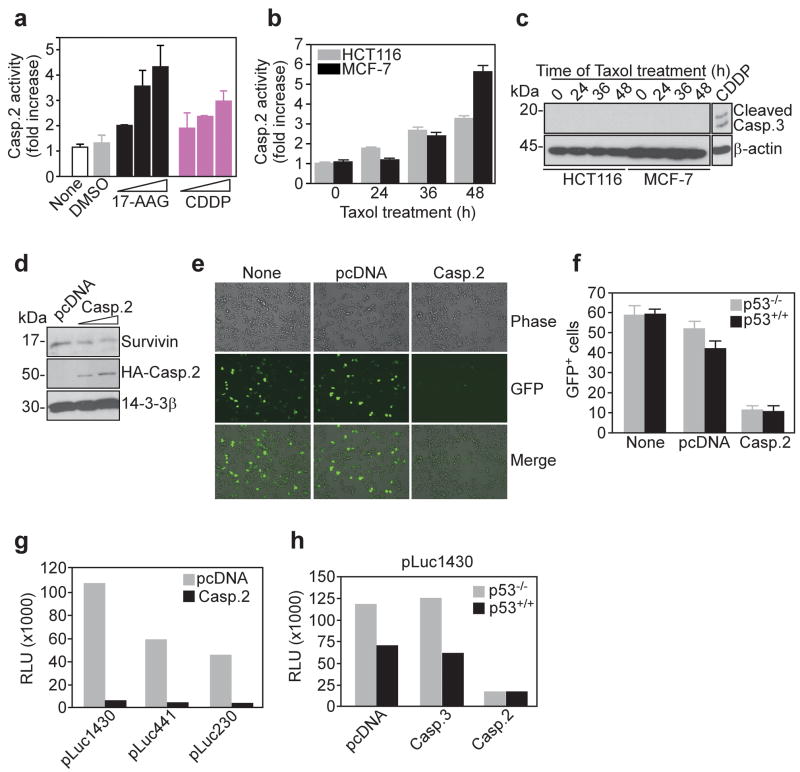
We began this study by testing the effect of anticancer agents on caspase activity, and we focused on caspase 2 for its role at the interface between cell proliferation signaling and apoptosis (Troy and Shelanski, 2003). Treatment of HCT116 colorectal cancer cells with the small molecule Heat Shock Protein-90 (Hsp90) inhibitor, 17-allylaminogeldanamycin (17-AAG), or the DNA-damaging agent, cisplatin (CDDP), resulted in concentration-dependent increase in caspase 2 activity (Figure 1a). To test the specificity of this response, we next treated breast adenocarcinoma MCF-7 cells or HCT116 cells with the chemotherapeutic agent, taxol, and monitored caspase 2 activity side-by-side with cleavage of caspase 3. Taxol treatment induced robust and time-dependent increase in caspase 2 activity (Figure 1b). However, this was not associated with detectable caspase 3 proteolytic activation (Figure 1c), indicating that caspase 2 activity was specific for this caspase. Conversely, proteolytic cleavage of caspase 3 was detectable at high concentrations of CDDP (Figure 1c) and 17-AAG (data not shown). To test whether a potential caspase 2 response affected cell survival pathways, we next transfected HCT116 cells with a caspase 2 cDNA, and looked at a potential modulation of IAP proteins (Srinivasula and Ashwell, 2008). In these experiments, transfection of caspase 2 in HCT116 cells did not significantly reduce cell viability (see below). Conversely, caspase 2-expressing cells exhibited concentration-dependent loss of endogenous survivin levels (Figure 1d), whereas a related IAP, XIAP (Srinivasula and Ashwell, 2008), was not affected (see below).
Figure 1.
Caspase 2 represses survivin gene expression. (a) Caspase 2 enzymatic activity in p53+/+ HCT116 cells treated with 17-AAG (0.1, 1 or 10 μM) or cisplatin (CDDP, 10, 30 μM or 50 μM) for 48 h. (b) Caspase 2 enzymatic activity in p53+/+ HCT116 or MCF-7 cells treated with 0.2 μM taxol. (c) Western blot analysis of cleaved caspase 3 in 0.2 μM taxol treated HCT116 or MCF-7 cells. CDDP was used at 50 μM. (d) Western blotting of HCT116 cells transfected with pcDNA or HA-tagged caspase 2. (e) Fluorescence microscopy of GFP expression in HCT116 cells transfected with ms-830-GFP plus pcDNA or caspase 2. Representative fields are shown. (f) The conditions are as in (e), and the number of GFP+ cells was counted for p53+/+ or p53−/− HCT116 cells. (g) β-galactosidase-normalized survivin promoter (pLuc1430, pLuc441, pLuc230) luciferase activity in transfected HCT116 cells. (h) Analysis of survivin promoter luciferase activity in p53+/+ or p53−/− HCT116 cells. RLU, relative luciferase units. For panels g and h, data are representative of at least two independent experiments.
To determine whether caspase 2 modulation of survivin occurred at the level of gene transcription, we next transfected HCT116 cells with the proximal 830 nt of the mouse survivin promoter fused to a GFP reporter gene (ms-830-GFP) (Xia and Altieri, 2006). In the presence of a control plasmid, or no plasmid, transfected cells expressed GFP under the control of the survivin promoter, by fluorescence microscopy (Figure 1e). In contrast, transfection of caspase 2 nearly completely abolished the GFP+ population of HCT116 cells (Figure 1e–f), suggesting that transcription of the survivin gene was repressed under these conditions. Similar results were obtained in p53+/+ or p53−/− HCT116 cells (Figure 1f), demonstrating that p53 was not involved in survivin gene modulation by caspase 2 (Hoffman et al., 2002; Mirza et al., 2002). Consistent with this model, transfection of caspase 2 in HCT116 cells suppressed transcription of several human survivin promoter constructs extending up to -1430 nt from the transcription start site(s), by luciferase reporter assay (Figure 1g). This response was specific for caspase 2, as expression of caspase 3 had no effect on survivin gene expression in p53+/+ or p53−/− HCT116 cells (Figure 1h).
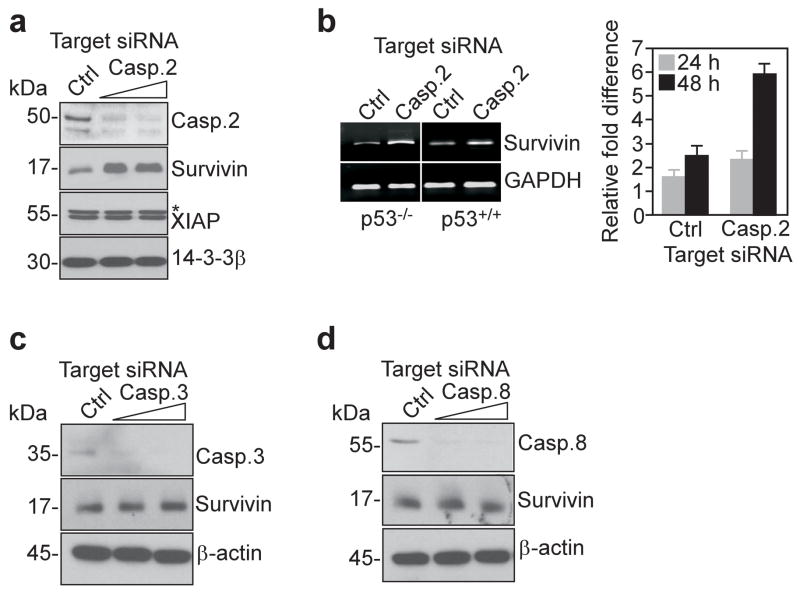
Caspase 2 targeting modulates endogenous survivin expression in tumor cells
In complementary experiments, we next acutely silenced caspase 2 expression in HCT116 cells by small interfering RNA (siRNA), and looked at potential changes in endogenous survivin levels. Caspase 2-directed siRNA efficiently suppressed caspase 2 protein levels in HCT116 cells, as compared with control siRNA (Figure 2a), and this resulted in upregulation of endogenous survivin expression, by Western blotting (Figure 2a), and semi-quantitative and quantitative PCR (Figure 2b). Conversely, caspase 2 silencing had no effect on XIAP expression in HCT116 cells (Figure 2a). As control, acute knockdown of endogenous caspase 3 (Figure 2c), or caspase 8 (Figure 2d) did not modulate survivin levels in HCT116 cells, thus confirming the specificity of the caspase 2 response, and a non-targeted siRNA was ineffective (Figure 2c, d).
Figure 2.
Transcriptional regulation of survivin by caspase 2. (a) Western blotting of HCT116 cells transfected with control (Ctrl) or caspase 2-directed siRNA. *, nonspecific. (b) Semi-quantitative (left), or quantitative (right) real time PCR amplification of survivin or GAPDH mRNA in transfected HCT116 cells. Right, GAPDH-normalized quantification of survivin mRNA expression. (c, d) Western blotting of HCT116 transfected with control (Ctrl), caspase-3 (c)- or caspase-8 (d)-directed siRNA.
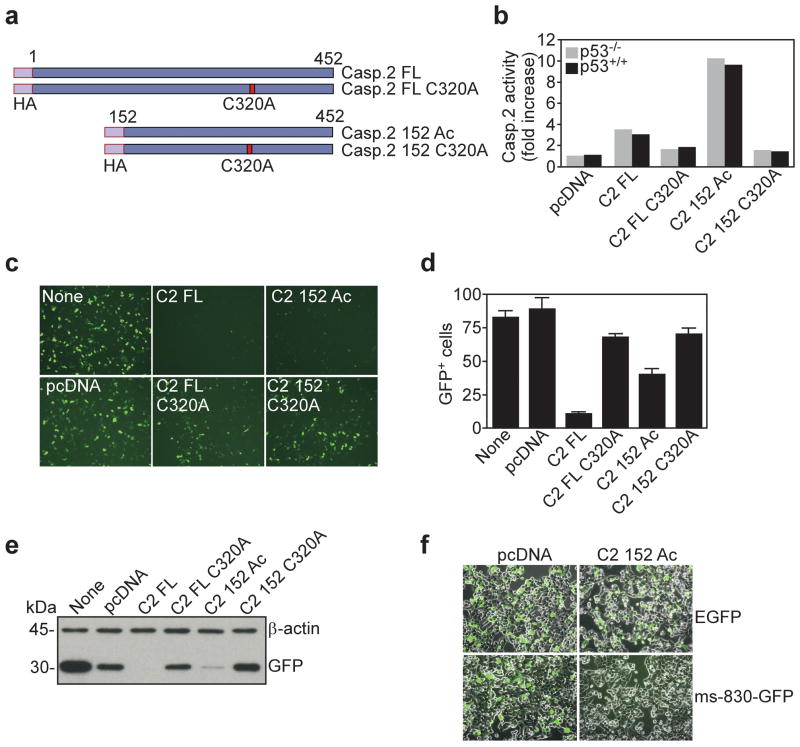
Requirement of caspase 2 catalytic activity for modulation of survivin gene expression
Forced expression of caspase 2 in cells has been associated with the generation of an active enzyme, potentially via autoproteolytic processing (Troy and Shelanski, 2003), or induced proximity (Bouchier-Hayes et al., 2009). To formally test whether the enzymatic activity of caspase 2 was required for survivin gene regulation, we next generated caspase 2 variants lacking the prodomain, thus mimicking a constitutively active enzyme (Casp.2 152 Ac) (Li et al., 1997), or carrying an Ala substitution of the active site Cys320 (C320A), which produces a catalytically dead enzyme (Figure 3a). Consistent with these predictions, p53+/+ or p53−/− HCT116 cells transfected with full length caspase 2 exhibited a 2-fold increase in enzymatic activity, whereas prodomain-deleted caspase 2 was considerably more active, and a C320A caspase 2 mutant had no activity (Figure 3b). Under these conditions, wild type caspase 2 or constitutively active caspase 2 suppressed survivin promoter-directed GFP expression in HCT116 cells, by fluorescence microscopy (Figure 3c, d), and Western blotting (Figure 3e). In contrast, active site-mutant caspase 2 did not affect survivin promoter activity in HCT116 cells (Figure 3c–e). Confirming the specificity of this response, transfection of HCT116 cells with active caspase 2 did not affect the expression of an unrelated plasmid, i.e. pEGFP (Figure 3f). Altogether, these data indicate that the catalytic activity of caspase 2 is required to suppress survivin gene transcription.
Figure 3.
Requirement of caspase-2 catalytic activity for survivin gene repression. (a) Schematic diagram of caspase 2 constructs. Casp. 2 FL, full length; Casp. 2 152 Ac, prodomain deleted, constitutively active caspase 2, Casp. 2 C320A, catalytically inactive caspase 2. The position of an HA tag is indicated. (b) Analysis of caspase 2 activity in transfected p53+/+ or p53−/− HCT116 cells. Data are representative of at least two independent experiments. (c) Fluorescence microscopy of ms-830-GFP expression in HCT116 cells transfected with the indicated caspase 2 constructs. Representative fields are shown. (d) Quantification of GFP+ cells. The experimental conditions are as in (c). Data are the mean±SD of two independent experiments. (e) Western blotting of GFP expression in transfected HCT116 cells. The experimental conditions are as in (c). (f) Fluorescence microscopy of GFP expression in HCT116 cells transfected with ms-830-GFP or pEGFP plus pcDNA or active caspase 2 (C2 152 Ac).
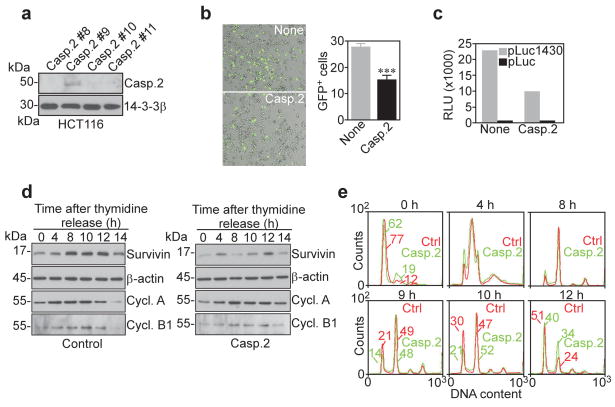
Mitotic defects and apoptosis induced by caspase 2 silencing of survivin
To investigate the consequence(s) of caspase 2 downregulation of survivin, we next generated clones of HCT116 cells that stably express caspase 2, or control plasmid. Generation of stable caspase 2 transfectants was feasible (Figure 4a), and resulted in significant repression of survivin gene transcription, by fluorescence microscopy of ms-830-GFP expression (Figure 4b), and luciferase reporter assay (Figure 4c). Cell cycle-synchronized HCT116 cells transfected with control plasmid exhibited a periodic increase in survivin expression, coinciding with entry into the G2/M phase of the cell cycle 8 h after thymidine release, and persisting throughout the completion of mitosis, 10 to 12 h after release (Figure 4d), in agreement with previous observations (Altieri, 2008). In contrast, HCT116 caspase 2 transfectants showed no increase in survivin expression at mitosis (Figure 4d), and this was associated with slower mitotic transitions, with 21% and 40% of these cells re-entering G1 10 and 12 h after thymidine release, respectively, as opposed to 30% and 51% of control HCT116 cells (Figure 4e). Accordingly, caspase 2-expressing cells exhibited persistence of cyclin A expression, as compared with control transfectants (Figure 4d).
Figure 4.
Characterization of caspase 2 HCT116 transfectants. (a) Western blotting of HCT116 stably transfected with caspase 2. Clone #9 was used in subesquent experiments. (b) Fluorescence microscopy of ms-830-GFP expression in stably transfected HCT116 cells. Right, Quantification of GFP+ cells, ***p<0.0001. (c)β-galactosidase-normalized survivin promoter (pLuc 1430) luciferase activity in stably transfected HCT116 cells. Data are representative of at least two independent experiments. RLU, relative luciferase units. (d) Western blotting of cell cycle-synchronized HCT116 transfectants expressing pcDNA (Control) or caspase 2. (e) Cell cycle profile of synchronized HCT116 cells. The percentage of cells in the G1 or G2/M phase of the cell cycle is indicated per each clone. Ctrl, Control; Cycl., Cyclin.
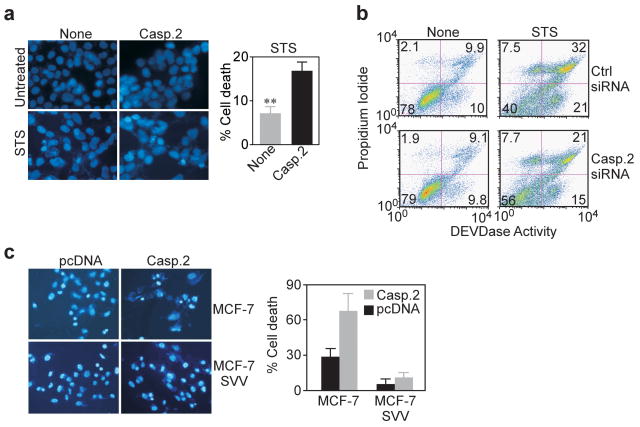
In the absence of cell death stimuli, stable expression of caspase 2 in synchronized HCT116 transfectants did not result in detectable apoptosis at any cell cycle phase tested, as compared with control cultures, by hypodiploid DNA content and flow cytometry (Figure 4e), or DAPI staining of nuclear apoptosis, by fluorescence microscopy (Figure 5a). In contrast, exposure of these cells to the cell death stimulus, staurosporine (STS), resulted in increased sensitivity to apoptosis, as compared with control HCT116 transfectants (Figure 5a). This cell death response had the hallmarks of apoptosis with increased caspase 3 activity, and loss of plasma membrane integrity, by multiparametric flow cytometry of DEVDase activity and propidium iodide staining (Figure 5b). In addition, knockdown of caspase 2 reversed STS-induced cell death in HCT116 cells, as compared with control siRNA (Figure 5b), whereas background cell death in the absence of STS was indistinguishable in control or caspase 2-silenced cultures (Figure 5b). In complementary experiments, stable expression of survivin in breast adenocarcinoma MCF-7 cells (MCF-7 SVV) completely reversed caspase 2-induced cell death to background levels of untreated cultures (Figure 5c). Conversely, acute expression of caspase 2 induced extensive nuclear apoptosis in parental MCF-7 cells (Figure 5c).
Figure 5.
Regulation of caspase 2-induced apoptosis. (a) Fluorescence microscopy of nuclear apoptosis in HCT116 cells stably transfected with caspase 2. Representative images are shown. STS, staurosporine. Right, quantification of apoptotic cells, **p=0.003. (b) Multiparametric flow cytometry analysis of DEVDase (caspase) activity and DNA content (propidium iodide) of transfected HCT116 cells treated with vehicle (None) or staurosporine (STS). The percentage of cells in each quadrant is indicated. (c) Nuclear morphology of apoptosis in parental MCF-7 cells or MCF-7 cells stably expressing survivin (MCF-7 SVV) after transfection of caspase 2 or pcDNA. Left, DAPI staining of representative microscopy fields. Magnification, ×200. Right, Quantification of cell death. Data are the mean±SD of replicates from at least two independent experiments.
Caspase 2 suppression of tumorigenesis
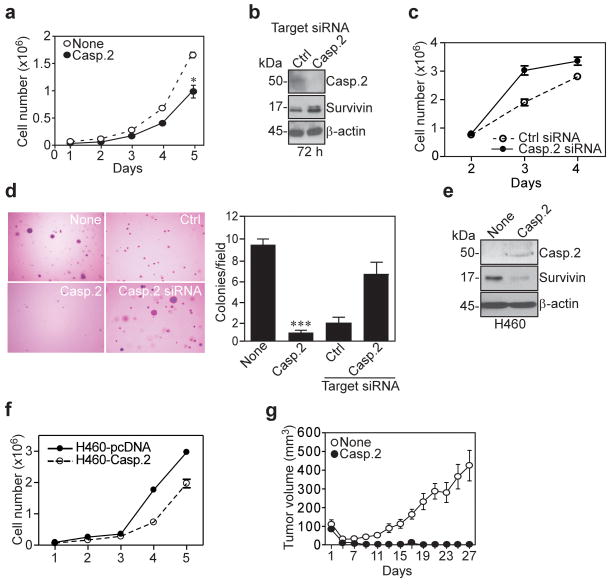
We next asked whether caspase 2 silencing of survivin affected tumorigenicity. First, HCT116 cells stably expressing caspase 2 exhibited significantly reduced cell proliferation, compared to control transfectants over a 5-d interval (Figure 6a). siRNA silencing of caspase 2 in stable HCT116 transfectants (Figure 6b) reversed this phenotype, and restored cell proliferation to levels comparable or exceeding those of control siRNA transfected cells (Figure 6c). Second, expression of caspase 2 in HCT116 cells completely abolished colony formation in soft agar, compared to control transfectants, in a reaction similarly reversed by siRNA silencing of caspase 2 in HCT116 transfectants, but not control siRNA (Figure 6d). To further test the specificity of caspase 2 inhibition of tumorigenicity, we next generated stable clones of caspase 2 transfectants in an unrelated tumor cell type, lung cancer H460 cells. Stable expression of caspase 2 in this cell type was also feasible (Figure 6e), and, similarly to the results with HCT116 cells, was associated with decreased rate of tumor cell proliferation (Figure 6f). Finally, HCT116 cells stably expressing caspase 2 completely failed to grow as superficial xenograft tumors in immunocompromised mice, whereas control transfectants gave rise to rapidly growing tumors in SCID/beige mice (Figure 6g).
Figure 6.
Caspase 2 suppression of tumorigenesis. (a) Proliferation of HCT116 caspase 2 stable transfectants; *, p=0.034. (b) Western blot of siRNA silencing of caspase 2 in HCT116 stable caspase 2 transfectants. (c) Cell proliferation of HCT116 stably expressing caspase 2 after transfection with control (Ctrl)- or caspase 2-directed siRNA. (d) Soft agar colony formation of HCT116 caspase 2 transfectants with or without caspase 2 silencing by siRNA. Left, representative microscopy fields per condition. Right, quantification of colony formation in soft agar; ***, p<0.0001. (e) Western blot of stable expression of caspase 2 in H460 cells. (f) Proliferation of H460 cells stably transfected with caspase 2 or pcDNA. (g) Kinetics of xenograft tumor growth of HCT116 caspase 2 stable transfectants. Mean tumor volume in mm3 is shown for each time point. Data are representative of two independent experiments.
Specificity of caspase 2 regulation of survivin expression and tumorigenicity
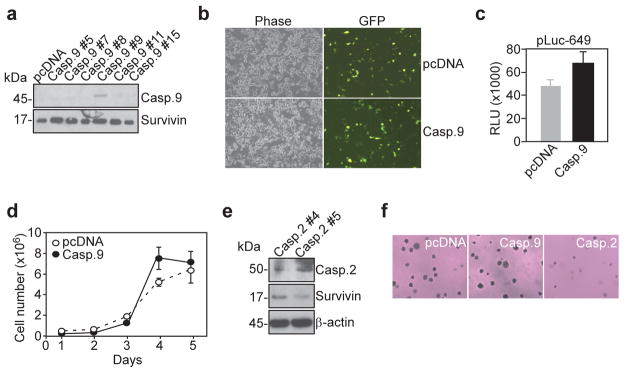
To validate the specificity of these findings, we next generated stable clones of HCT116 cells that expressed comparable levels of caspase 9 (Figure 7a), another long prodomain-containing apical caspase (Shi, 2002), which can also be activated by induced proximity and overexpression in cells. In sharp contrast with caspase 2, HCT116 caspase 9 transfectants exhibited no modulation of endogenous survivin expression, by Western blotting (Figure 7a), and no reduction in survivin promoter-dependent GFP expression, by fluorescence analysis of ms-830-GFP (Fig. 7b), or survivin promoter luciferase activity (Figure 7c). Functionally, caspase 9-expressing HCT116 cells exhibited kinetics of cell proliferation indistinguishable from those of pcDNA transfectants (Fig. 7d), and formed extensive colonies in soft agar (Figure 7f). For comparison, another independently established clone of HCT116 cells stably expressing caspase 2 exhibited downregulation of endogenous survivin (Figure 7e), and complete ablation of anchorage-independent cell growth (Figure 7f).
Figure 7.
Characterization of HCT116 caspase 9 stable transfectants. (a) Western blot of HCT116 cells stably transfected with caspase 9. Clone #9 expressing caspase 9 was selected for subsequent experiments. (b) Fluorescence microscopy of ms-830-GFP expression in HCT116 caspase 9 transfectants. (c) Analysis of survivin promoter luciferase activity in HCT116 caspase 9 transfectants. RLU, relative luciferase units. (d) Colony formation of HCT116 caspase 9 transfectants. (e) Western blot of an independent clone of HCT116 cells stably expressing caspase 2 (clone #5). (f) Soft agar colony formation assay of HCT116 cells stably transfected with pcDNA, caspase 9 (clone #9) or caspase 2 (clone #5). For panels c and e, data are the mean±SD of at least two independent experiments.
Caspase 2 targeting of NFκB activity
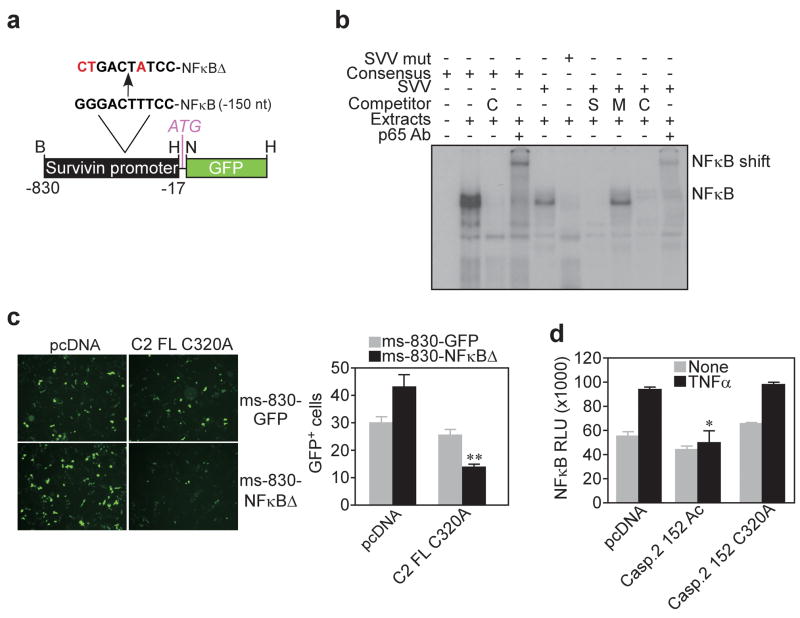
We next asked whether caspase 2 signaling, particularly NFκB regulation (Tinel et al., 2007), contributed to survivin gene silencing and tumor suppression. First, a radiolabeled DNA probe containing a consensus NFκB site in the proximal survivin promoter, but not a mutant NFκB DNA probe (Figure 8a) bound nuclear extracts of tumor cells, in a reaction supershifted by an antibody to the p65 subunit of NFκB (Figure 8b). Accordingly, a wild type, but not mutant unlabeled NFκB consensus sequence inhibited the formation of a survivin promoter-NFκB complex, by EMSA (Figure 8b).
Figure 8.
Caspase 2 targets NFκB-mediated survivin gene expression. (a) Position and mutagenesis of an NFκB consensus binding site in the survivin promoter (ms-830-NFκBΔ). (b) EMSA of 32P-labeled survivin (SVV) or NFκB consensus probe. Unlabeled excess competitor of a generic NFκB consensus sequence (C), or wild type (S), or mutant (M) survivin was used. (c) Fluorescence microscopy of ms-830-GFP expression in transfected HCT116 cells. Right, quantification of GFP+ cells. **, p=0.0028. (d) NFκB luciferase reporter activity in HCT116 transfectants in the presence or absence of TNFα. RLU, relative luciferase units; *, p=0.04.
Next, we mutated one of the NFκB consensus sites in the survivin promoter of ms-830-GFP (Figure 8a), and tested a potential effect on survivin gene expression. Consistent with the data presented above, transfection of these cells with catalytically dead caspase 2 had no effect on GFP expression driven by the wild type survivin promoter (Figure 8c). In contrast, GFP expression was partially reduced in HCT116 cells transfected with the mutant NFκB ms-830-GFP promoter and catalytically dead caspase 2 (Figure 8c). Altogether, these data suggest that caspase 2 functions as an upstream negative regulator of NFκB, in a pathway that requires at least one NFκB site in the proximal survivin promoter. Consistent with this model, active caspase 2 abolished NFκB reporter activity in TNFα-stimulated HCT116 cells, whereas catalytically dead caspase 2 had no effect (Figure 8d).
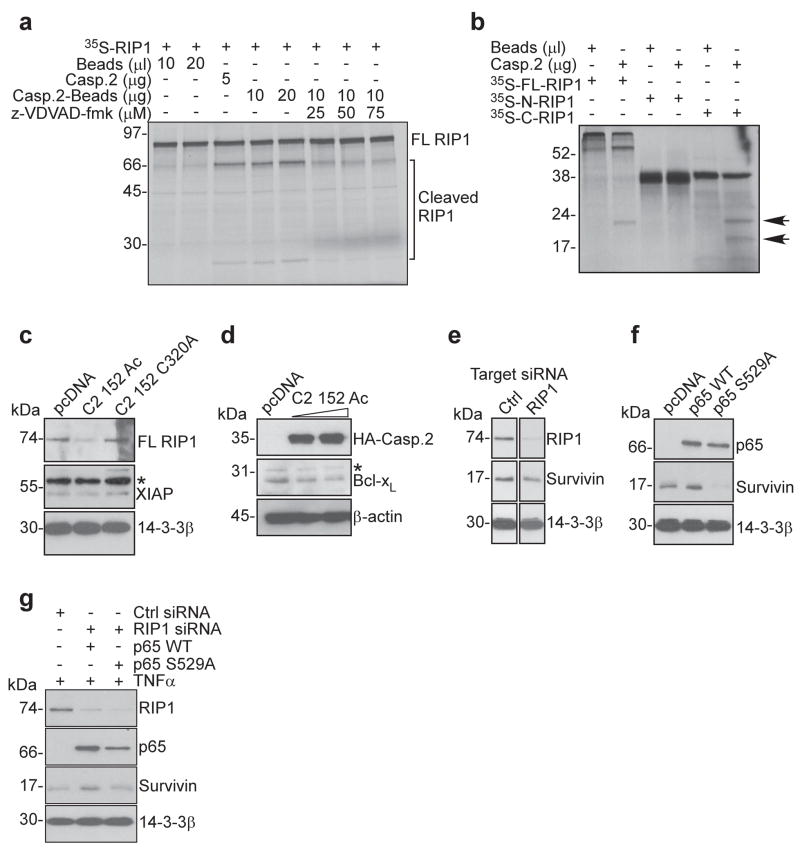
Caspase 2 cleavage of RIP1
We next searched for upstream activator(s) of NFκB that may be potentially cleaved by caspase 2, and we focused on RIP1 (Festjens et al., 2007) for its role in TNFα (Micheau and Tschopp, 2003)-, and DNA damage (Janssens et al., 2005)-induced NFκB activation. Unconjugated or bead-conjugated recombinant caspase 2 cleaved 35S-labeled recombinant RIP1, in vitro, with appearance of proteolytic fragments of approximate molecular weight of 56 and 20 kDa, respectively (Figure 9a). Addition of the caspase 2 inhibitor, z-VDVAD-fmk prevented RIP1 cleavage by caspase 2 (Figure 9a). In addition, caspase 2 cleaved a 35S-labeled–COOH terminus RIP1 fragment comprising residues 351-672, whereas a RIP1 NH2 fragment comprising residues 1-350 was not affected (Figure 9b). Consistent with these data, transfection of constitutively active caspase 2 in HCT116 cells decreased the expression of full length RIP1, and reduced the levels of Bcl-xL, another NFκB target gene (Figure 9d). In contrast, XIAP levels were not affected (Figure 9c), and transfection of catalytically dead caspase 2 did not modulate RIP1 expression (Figure 9c).
Figure 9.
Caspase 2 cleavage of RIP1. (a) Autoradiography of caspase 2 cleavage of 35S-labeled RIP1. z-VDVAD-fmk was used as a caspase 2 inhibitor. FL, full length. (b) Caspase 2 cleavage of 35S-labeled RIP1 constructs, including full length (FL), NH2-terminus (residues 1-350) or –COOH terminus (residues 351-672) fragments. Arrows, cleavage products visualized by autoradiography. (c) Western blot of HCT116 cells transfected with active (C2 152Ac), or mutant (C2 152 C320A) caspase 2. *, nonspecific. (d) Western blot of modulation of endogenous Bcl-xL expression in HCT116 cells transfected with constitutively active caspase 2. (e) Western blot of TNFα-stimulated HCT116 cells transfected with control (Ctrl) or RIP1-directed siRNA. (f) Western blot of HCT116 cells transfected with wild type or mutant (S529A) p65-NFκB. (g) Western blot of TNFα-stimulated HCT116 cells transfected with control (Ctrl) or RIP1-directed siRNA, in the presence of wild type or mutant p65-NFκB.
Consistent with a role of RIP1 in NFκB regulation of survivin, siRNA silencing of RIP1 in TNFα-stimulated HCT116 cells resulted in decreased expression of endogenous survivin (Figure 9e). Similarly, transfection of HCT116 cells with a p65 subunit of NFκB (p65-NFκB) increased endogenous survivin (Figure 9f), whereas a S529A p65-NFκB dominant negative mutant (Wang and Baldwin Jr, 1998) caused loss of survivin expression (Figure 9f). Finally, siRNA silencing of RIP1 in HCT116 cells inhibited the induction of survivin mediated by TNFα (Figure 9g), in a response reversed by transfection of wild type, but not S529A mutant p65-NFκB (Figure 9g).
DISCUSSION
In this study, we have shown that caspase 2, an apical caspase with still largely elusive functions (Krumschnabel et al., 2009), actively represses survivin gene transcription in tumor cells. In turn, acute loss of survivin causes mitotic defects, increased sensitivity to apoptosis and complete loss of tumorigenicity, in vivo. Mechanistically, this pathway involves caspase 2 cleavage of the NFκB activator RIP1 (Festjens et al., 2007), which shuts off transcription of NFκB-responsive antiapoptotic genes, including survivin (Kawakami et al., 2005).
Apart from a well-characterized mechanism of caspase 2 activation in response to DNA damage (Tinel and Tschopp, 2004), other pathophysiological activation platform(s) for this caspase have been far less clear (Krumschnabel et al., 2009). Here, conventional (CDDP or taxol), or molecularly targeted (17-AAG) anticancer agents produced concentration- and time-dependent caspase 2 activity in tumor cells, in agreement with the effect of other chemotherapeutic agents (Mhaidat et al., 2007), or stress response modifiers (Yeung et al., 2006). Caspase 2 activation under these conditions was not uniformly preceded by generation of caspase 3 activity, which activates caspase 2 by removing its prodomain (Van de Craen et al., 1999), suggesting that at least certain anticancer regimens may activate caspase 2 upstream of effector caspase(s) (Yeung et al., 2006).
Recent evidence suggests that survivin functions as a “nodal” IAP protein (Srinivasula and Ashwell, 2008), orchestrating cell division mechanisms, cytoprotection, and modulation of the cellular stress response, especially in cancer (Altieri, 2008). In this context, a role of caspase 2 as a novel repressor of the survivin gene adds to the multiple pathways that fine-tune survivin expression in tumor cells, including transcriptional (Altieri, 2008), translational (Vaira et al., 2007), and post-translational (O’Connor et al., 2000; Wheatley et al., 2007) modifications. Similar to the phenotype induced by blocking transcriptional activators of survivin, for instance Wnt/β catenin (You et al., 2004), Stat3 (Zhou et al., 2009), or Notch (Lee et al., 2008), heightened caspase 2 activity resulted in loss of endogenous survivin, causing mitotic defects, reduced cell proliferation, and sensitization to apoptotic stimuli (Altieri, 2008). Functionally, this resulted in complete loss of tumorigenicity, in vivo, a phenotype that mirrors the effect of genetic deletion of caspase 2 of delayed apoptosis (Ho et al., 2008), increased cell proliferation, enhanced cellular transformation, and accelerated tumor growth, in vivo (Ho et al., 2009). Similar to the data observed here, a recent study implicates a role for nuclear caspase 2 in the maintenance of a G2/M checkpoint in response to DNA damage (Shi et al., 2009). Although these results collectively suggest a role of caspase 2 in cell cycle progression, more work is needed to ascertain whether changes in survivin levels also contribute to this pathway. Taken together, these data support a model of caspase 2 as a novel tumor suppressor, in vivo, and identify acute survivin gene silencing as one of the pivotal effectors of this response.
Although the overexpression of survivin seen in most human cancers likely reflects activation of multiple oncogenic pathways (Altieri, 2008), complementary pathways have also been identified that keep survivin levels low in normal cells. Accordingly, tumor suppressors, including p53 (Hoffman et al., 2002; Mirza et al., 2002), Rb (Jiang et al., 2004), BRCA1 (Wang et al., 2008), or PTEN (Guha et al., 2009) have all been shown, similarly to caspase 2 (this study), to acutely silence the survivin gene by different mechanisms. Whether this pathway is responsible for the low to undetectable levels of survivin observed in most normal adult tissues (Altieri, 2008) remains to be elucidated. However, it is possible that transcriptional silencing of survivin by various effectors may provide a mandatory requirement for efficient tumor suppression (Lowe et al., 2004), whereas unrestrained survivin gene expression may constitute an obligatory step in the establishment of the transformed clone(s).
The mechanistic underpinning of how caspase 2 antagonizes tumor growth was not elucidated in a recent study (Ho et al., 2009). Here, we show that this pathway involves inhibition of NFκB signaling, via direct cleavage of its upstream activator, RIP1 (Festjens et al., 2007). Consistent with this model, the catalytic activity of caspase 2 was required for survivin gene silencing, and differential expression of RIP1 or NFκB was sufficient to modulate endogenous survivin levels in tumor cells. This is in agreement with other data of NFκB induction of survivin gene expression (Kawakami et al., 2005) in different cell types (Anand et al., 2008; Makishi et al., 2008), and the preliminary identification reported here of at least one functional NFκB-responsive site in the proximal survivin promoter. Clearly, other pathways for NFκB activation could participate in modulating survivin gene expression, utilizing potential additional NFκB sites in the survivin promoter. In this context, RIP1 functions as an ubiquitin-regulated component of a TNFα-induced multimolecular protein complex that participates in cell survival via NFκB and MAPK signaling (Festjens et al., 2007).
Although a non-catalytic mechanism of caspase 2-mediated NFκB activation has been proposed (Lamkanfi et al., 2005), our findings are at variance with this model (Lamkanfi et al., 2005), and suggest that caspase 2 functions as a pro-apoptotic effector antagonizing NFκB-mediated cell survival (Tinel et al., 2007). There is precedent for a role of caspase(s) executing a similar response via RIP1 cleavage, and caspase 8 protelysis of RIP1 at Asp324 has been implicated in suppression of NFκB-dependent survival in tumor cells (Lin et al., 1999), and regulation of macrophage differentiation (Rebe et al., 2007). Caspase 2 cleavage of RIP1 was not observed in an earlier study (Lamkanfi et al., 2005). However, we showed here that this response was largely reversed by a caspase 2 inhibitor, z-VDVAD-fmk (Krumschnabel et al., 2009), and involved a –COOH terminus fragment of RIP1 comprising residues 351-672. Although several Asp residues are present in this region as candidate cleavage site(s), none conforms to a canonical VDVAD sequence, in agreement with the promiscuous substrate recognition of caspase 2.
In summary, we have found that caspase 2 functions in a broad tumor suppression network, lowering a general anti-apoptotic threshold via interruption of NFκB signaling (Karin, 2006), and acute silencing of survivin gene expression (Altieri, 2008). Among the portfolio of apoptosis modifiers, strategies to restore caspase activity in tumors are being pursued for therapeutic opportunities (Fesik, 2005). Based on the data presented here, restoring caspase 2 activity may be beneficial in tumors with elevated NFκB activity and high levels of survivin, thus potentially “addicted” to this general cytoprotective pathway (Karin, 2006).
Acknowledgments
We thank Drs. Bert Vogelstein, Michelle Kelliher and Neal Silverman for reagents. This work was supported by National Institutes of Health grants CA78810, CA90917 and CA118005.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–6. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- Anand P, Kunnumakkara AB, Harikumar KB, Ahn KS, Badmaev V, Aggarwal BB. Modification of cysteine residue in p65 subunit of nuclear factor-kappaB (NF-kappaB) by picroliv suppresses NF-kappaB-regulated gene products and potentiates apoptosis. Cancer Res. 2008;68:8861–70. doi: 10.1158/0008-5472.CAN-08-1902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Baptiste-Okoh N, Barsotti AM, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci U S A. 2008;105:1937–42. doi: 10.1073/pnas.0711800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–14. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–40. doi: 10.1016/j.molcel.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Valent A, Raslova H, Yakushijin K, et al. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 2004;23:4362–70. doi: 10.1038/sj.onc.1207572. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–56. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–85. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death Differ. 2007;14:400–10. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–94. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- Guha M, Plescia J, Leav I, Li J, Languino LR, Altieri DC. Endogenous tumor suppression mediated by PTEN involves survivin gene silencing. Cancer Res. 2009;69:4954–8. doi: 10.1158/0008-5472.CAN-09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Read SH, Dorstyn L, Lambrusco L, Kumar S. Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene. 2008;27:3393–404. doi: 10.1038/sj.onc.1211005. [DOI] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci U S A. 2009;106:5336–41. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–92. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA. Aberrant regulation of survivin by the RB/E2F family of proteins. J Biol Chem. 2004;279:40511–20. doi: 10.1074/jbc.M404496200. [DOI] [PubMed] [Google Scholar]
- Kang BH, Altieri DC. Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J Biol Chem. 2006;281:24721–7. doi: 10.1074/jbc.M603175200. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-[kappa]B in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kawakami H, Tomita M, Matsuda T, Ohta T, Tanaka Y, Fujii M, et al. Transcriptional activation of survivin through the NF-kappaB pathway by human T-cell leukemia virus type I tax. Int J Cancer. 2005 doi: 10.1002/ijc.20954. [DOI] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen’s view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, D’Hondt K, Vande Walle L, van Gurp M, Denecker G, Demeulemeester J, et al. A novel caspase-2 complex containing TRAF2 and RIP1. J Biol Chem. 2005;280:6923–32. doi: 10.1074/jbc.M411180200. [DOI] [PubMed] [Google Scholar]
- Lee CW, Raskett CM, Prudovsky I, Altieri DC. Molecular Dependence of Estrogen Receptor-Negative Breast Cancer on a Notch-Survivin Signaling Axis. Cancer Res. 2008;68:5273–5281. doi: 10.1158/0008-5472.CAN-07-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J 344 Pt. 1999;2:305–11. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Bergeron L, Cryns V, Pasternack MS, Zhu H, Shi L, et al. Activation of caspase-2 in apoptosis. J Biol Chem. 1997;272:21010–7. doi: 10.1074/jbc.272.34.21010. [DOI] [PubMed] [Google Scholar]
- Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–26. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishi S, Okudaira T, Ishikawa C, Sawada S, Watanabe T, Hirashima M, et al. A modified version of galectin-9 induces cell cycle arrest and apoptosis of Burkitt and Hodgkin lymphoma cells. Br J Haematol. 2008;142:583–94. doi: 10.1111/j.1365-2141.2008.07229.x. [DOI] [PubMed] [Google Scholar]
- Mhaidat NM, Wang Y, Kiejda KA, Zhang XD, Hersey P. Docetaxel-induced apoptosis in melanoma cells is dependent on activation of caspase-2. Mol Cancer Ther. 2007;6:752–61. doi: 10.1158/1535-7163.MCT-06-0564. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–90. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, et al. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci U S A. 2000;97:13103–13107. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebe C, Cathelin S, Launay S, Filomenko R, Prevotat L, L’Ollivier C, et al. Caspase-8 prevents sustained activation of NF-kappaB in monocytes undergoing macrophagic differentiation. Blood. 2007;109:1442–50. doi: 10.1182/blood-2006-03-011585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, et al. DNA-PKcs-PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell. 2009;136:508–20. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- Shin MS, Kim HS, Kang CS, Park WS, Kim SY, Lee SN, et al. Inactivating mutations of CASP10 gene in non-Hodgkin lymphomas. Blood. 2002;99:4094–9. doi: 10.1182/blood.v99.11.4094. [DOI] [PubMed] [Google Scholar]
- Soung YH, Lee JW, Kim SY, Park WS, Nam SW, Lee JY, et al. Somatic mutations of CASP3 gene in human cancers. Hum Genet. 2004;115:112–5. doi: 10.1007/s00439-004-1129-3. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ashwell JD. IAPs: what’s in a name? Mol Cell. 2008;30:123–35. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupack DG, Teitz T, Potter MD, Mikolon D, Houghton PJ, Kidd VJ, et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature. 2006;439:95–9. doi: 10.1038/nature04323. [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Troy CM, Shelanski ML. Caspase-2 redux. Cell Death Differ. 2003;10:101–7. doi: 10.1038/sj.cdd.4401175. [DOI] [PubMed] [Google Scholar]
- Upton JP, Austgen K, Nishino M, Coakley KM, Hagen A, Han D, et al. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol Cell Biol. 2008;28:3943–51. doi: 10.1128/MCB.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira V, Lee CW, Goel HL, Bosari S, Languino LR, Altieri DC. Regulation of survivin expression by IGF-1/mTOR signaling. Oncogene. 2007;26:2678–84. doi: 10.1038/sj.onc.1210094. [DOI] [PubMed] [Google Scholar]
- Van de Craen M, Declercq W, Van den brande I, Fiers W, Vandenabeele P. The proteolytic procaspase activation network: an in vitro analysis. Cell Death Differ. 1999;6:1117–24. doi: 10.1038/sj.cdd.4400589. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wang D, Baldwin AS., Jr Activation of Nuclear Factor-kappa B-dependent Transcription by Tumor Necrosis Factor-alpha Is Mediated through Phosphorylation of RelA/p65 on Serine 529. J Biol Chem. 1998;273:29411–29416. doi: 10.1074/jbc.273.45.29411. [DOI] [PubMed] [Google Scholar]
- Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Barrett RM, Andrews PD, Medema RH, Morley SJ, Swedlow JR, et al. Phosphorylation by Aurora-B Negatively Regulates Survivin Function During Mitosis. Cell Cycle. 2007:6. doi: 10.4161/cc.6.10.4179. [DOI] [PubMed] [Google Scholar]
- Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Res. 2006;66:3392–5. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- Yeung BH, Huang DC, Sinicrope FA. PS-341 (bortezomib) induces lysosomal cathepsin B release and a caspase-2-dependent mitochondrial permeabilization and apoptosis in human pancreatic cancer cells. J Biol Chem. 2006;281:11923–32. doi: 10.1074/jbc.M508533200. [DOI] [PubMed] [Google Scholar]
- You L, He B, Xu Z, Uematsu K, Mazieres J, Fujii N, et al. An anti-Wnt-2 monoclonal antibody induces apoptosis in malignant melanoma cells and inhibits tumor growth. Cancer Res. 2004;64:5385–9. doi: 10.1158/0008-5472.CAN-04-1227. [DOI] [PubMed] [Google Scholar]
- Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052–62. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]