Summary
The EphA4 tyrosine kinase cell surface receptor regulates an array of physiological processes and is the only currently known class A Eph receptor that binds both A and B class ephrins with high affinity. We have solved the crystal structure of the EphA4 ligand binding domain alone and in complex with (1) ephrinB2 and (2) ephrinA2. This set of structures shows that EphA4 has significant conformational plasticity in its ligand binding face. In vitro binding data demonstrate that it has a higher affinity for class A than class B ligands. Structural analyses, drawing on previously reported Eph receptor structures, show that EphA4 in isolation and in complex with ephrinA2 resembles other class A Eph receptors but on binding ephrinB2 assumes structural hallmarks of the class B Eph receptors. This interactive plasticity reveals EphA4 as a structural chameleon, able to adopt both A and B class Eph receptor conformations, and thus provides a molecular basis for EphA-type cross-class reactivity.
Keywords: SIGNALING, CELLBIO
Introduction
Eph (Erythropoietin-producing hepatoma) tyrosine kinase cell surface receptors comprise the largest group of receptor tyrosine kinases (Hirai et al., 1987). These receptors with their membrane-bound ligands, the ephrins (Eph receptor interacting proteins), contribute to fundamental developmental processes such as boundary formation and cell migration in multicellular organisms ranging from sponges to vertebrates (Drescher, 2002; Kullander and Klein, 2002). The signals resulting from Eph-ephrin complex formation can be bidirectional (forward: into the Eph-presenting cell, reverse: into the ephrin-presenting cell) (Cowan and Henkemeyer, 2002; Lim et al., 2008; Lu et al., 2001).
There are fourteen Eph receptors and eight ephrin ligands in the human genome, each family divided into two classes (A and B) (Eph Nomenclature Committee, 1997). The extracellular portions of ephrinA ligands are attached to the cell by a glycosylphosphatidylinositol anchor, while ephrinB ligands contain a transmembrane helix and a short intracellular region. Eph receptors are type 1 membrane proteins with an N-terminal extracellular region consisting of a ligand binding domain (LBD), a cysteine rich region, and two fibronectin type III domains. The extracellular region is followed by a single transmembrane span connecting to a cytoplasmic region comprising a protein tyrosine kinase domain that mediates autophosphorylation and phosphorylation of other proteins (Pasquale, 2005), a sterile α motif (SAM), and a PDZ-binding motif.
Generally, it has been found that high affinity binding (nM to low μM KD) of Eph receptors to ephrin ligands is restricted within classes (i.e., class A ligands bind class A receptors) (Gale et al., 1996). However, this rule only holds to an extent. EphA4, for example, has been found to bind both to A and B class ligands and EphB2 has been found to bind to ephrinA5 (Gale et al., 1996; Himanen et al., 2004).
EphA4 is associated with an extensive range of biological activity; for example, it is critical for the development of the nervous system (Canty et al., 2006; Coulthard et al., 2002; Dottori et al., 1998; Fu et al., 2007), is also highly expressed during glioma cell proliferation (Fukai et al., 2008), and has been linked with melanoma tumor suppression (Easty and Bennett, 2000). The potential to target Eph receptors for cancer therapeutics is of growing interest. Structural studies of the LBD of EphB2 and EphA4, for example, have shown that these receptors may be useful targets for the design of small molecule cancer therapeutics (Chrencik et al., 2006b; Noberini et al., 2008; Qin et al., 2008).
Knowledge of the molecular determinants that define Eph receptor cross-reactivity and specificity is a powerful prerequisite for the rational development of Eph receptor- and ephrin-based disease treatments. In this study, we sought to elucidate the mechanism by which the LBD of the EphA4 cell surface receptor binds to both A and B class ephrin ligands at the molecular level. We present binding affinity data of EphA4 with both A and B class ephrins and report the structure of the LBD of EphA4 alone and in complex with the receptor binding domain (RBD) of ephrinA2 and ephrinB2. From our analysis of these structures, we suggest that the LBD of EphA4 is structurally plastic and, as a consequence, has a bimorphic A and B class Eph receptor nature. These data provide a molecular basis for how differing patterns in Eph-ephrin binding define properties that influence class-dependent Eph receptor specificity and promiscuity.
Results
Crystal Structure of EphA4 Ligand Binding Domain
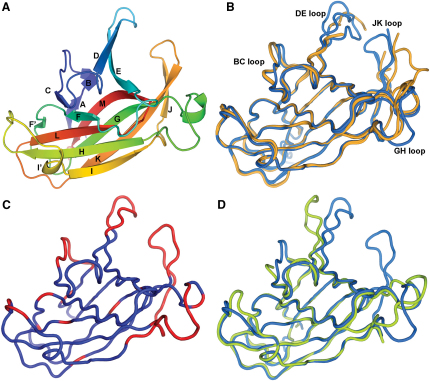
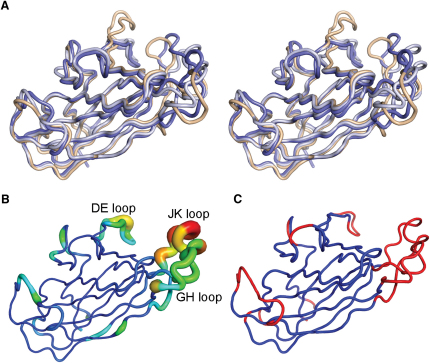
The crystal structure of the uncomplexed (apo) EphA4 cell surface receptor LBD was solved and refined to 1.85 Å resolution (Table 1) with two molecules in the asymmetric unit, using EphB2 (Protein Data Bank [PDB] code 1NUK) as a model for molecular replacement. Similar to the previously reported apo-EphA2 (PDB code 3C8X) LBD (55% sequence identity in the LBD), EphA4 consists of 12 anti-parallel β strands (designated A–M) with two disulfide bonds and is arranged as a β sandwich (Figure 1A; see Figure S1 available online). We note that during the preparation of this manuscript an independently determined apo structure of EphA4 (PDB code 3CKH; 2.8 Å resolution) has been published and released in the PDB (Qin et al., 2008). Superposition of both molecules in the asymmetric unit of our high resolution structure with the two molecules in the asymmetric unit of PDB deposition 3CKH (overall 1.0 Å rms average deviation over 163 equivalent Cα atoms) reveals that there is a high degree of conformational plasticity in the regions of the BC, DE, GH, and JK loops (Figures 1B and 1C). The flexibility inherent in these regions is consistent with their relatively high average crystallographic main chain B factor values in comparison with the rest of the protein (for our high resolution structures an average of 62 Å2 in BC, DE, GH, and JK loops versus 31 Å2 over the entire molecule). Given that the DE and JK loops map to regions in EphA2 (Figure 1D) and EphB2 crystal structures (Himanen et al., 2001), which move upon binding to their respective ephrin ligands, their flexibility may be of direct functional relevance.
Table 1.
Data Collection and Refinement Statistics
| Data Collection | EphA4 | EphA4-ephrinA2 | EphA4-ephrinB2 |
|---|---|---|---|
| Beamline | ESRF BM14 | Diamond I03 | ESRF BM14 |
| Resolution (Å) | 25.0–1.85 (1.92–1.85) | 50.0–2.35 (2.43–2.35) | 30.0–2.45 (2.54–2.45) |
| Spacegroup | P21 | P64 | P61 |
| Cell dimensions (Å) and angles (°) | a = 48.7, b = 73.2, | a = 115.4, b = 115.4, | a = 107.4, b = 107.4, |
| c = 54.4, | c = 59.3, | c = 47.6, | |
| β = 98.3 | γ = 120 | γ = 120 | |
| Wavelength (Å) | 0.977 | 1.810 | 0.977 |
| Unique reflections | 31875 (3127) | 18961 (1872) | 11645 (1084) |
| Completeness (%) | 98.2 (97.4) | 99.9 (99.4) | 99.3 (94.2) |
| Rmerge (%)a | 0.069 (0.344) | 0.091 (0.749) | 0.054 (0.531) |
| I/σI | 19.5 (4.0) | 30.0 (1.8) | 43.2 (2.6) |
| Average redundancy | 3.7 (3.7) | 16.0 (8.1) | 11.7 (6.6) |
| Refinement | |||
| Resolution range (Å) | 30.0–1.85 (1.90–1.85) | 50.0–2.35 (2.41–2.35) | 28.0–2.45 (2.51–2.45) |
| Number of reflections | 30277 | 17987 | 11062 |
| Rwork (%)b | 18.7 | 19.8 | 21.6 |
| Rfree (%)c | 23.4 | 24.5 | 26.0 |
| Rmsd bonds (Å) | 0.010 | 0.011 | 0.006 |
| Rmsd angles (°) | 1.2 | 1.3 | 1.0 |
| Rmsd main chain bond B (Å2) | 0.8 | 0.8 | 0.3 |
| Rmsd side chain bond B (Å2) | 1.8 | 1.6 | 0.7 |
| Rmsd between NCS related Cα atoms (Å) | 0.5 | NA | NA |
| Number of atoms per asymmetric unit (protein/water/sugar/ligandd) | 2936/296/NA/20 | 2576/136/28/NA | 2533 /49/14/NA |
| Average B factors (protein/water/sugar/ligandd) (Å2) | 31.4/27.0/NA/40.2 | 43.7 /25.1/47.9/NA | 49.9/22.2/57.2/NA |
| Model quality | |||
| Ramachandran (%) (favored, additionally allowed, outlier) | 94.2, 5.5, 0.3 | 97.5, 2.5, 0 | 96.4, 3.6, 0 |
Numbers in parentheses refer to the relevant outer resolution shell. Rmsd, root mean square deviation from ideal geometry.
Rmerge = Σhkl Σi|I(hkl;i) − < I(hkl) > |/Σhkl ΣiI(hkl;i), where I(hkl;i) is the intensity of an individual measurement and < I(hkl) > is the average intensity from multiple observations.
Rfactor = Σhkl‖Fobs| − k|Fcalc‖/Σhkl |Fobs|.
Rfree is calculated as for Rwork, but using only 5% of the data that was sequestered prior to refinement.
Isopropan-1-ol was observed in the crystal structure and was derived from the screening condition.
Figure 1.
The Structure of the EphA4 LBD
(A) Cartoon representation of the apo LBD of EphA4 colored as a rainbow with the N terminus in blue and the C terminus in red and with secondary structure labeled according to standard nomenclature.
(B–D) Cα traces.
(B) Superposition of all EphA4 molecules from the asymmetric unit of reported unbound crystal structures. Molecules in the asymmetric unit of the EphA4 structure we report here are colored blue, and molecules in the asymmetric unit of PDB code 3CKH are colored orange.
(C) Representative EphA4 molecule colored according to regions of flexibility as defined by ESSET (Schneider, 2002). Regions colored blue are conformationally invariant and regions colored red are flexible.
(D) Superposition of representative EphA4 (light blue) with crystal structure of unbound EphA2 (green; PDB code 3C8X).
EphA4 Displays a Broad Affinity Range for Ephrin Ligands
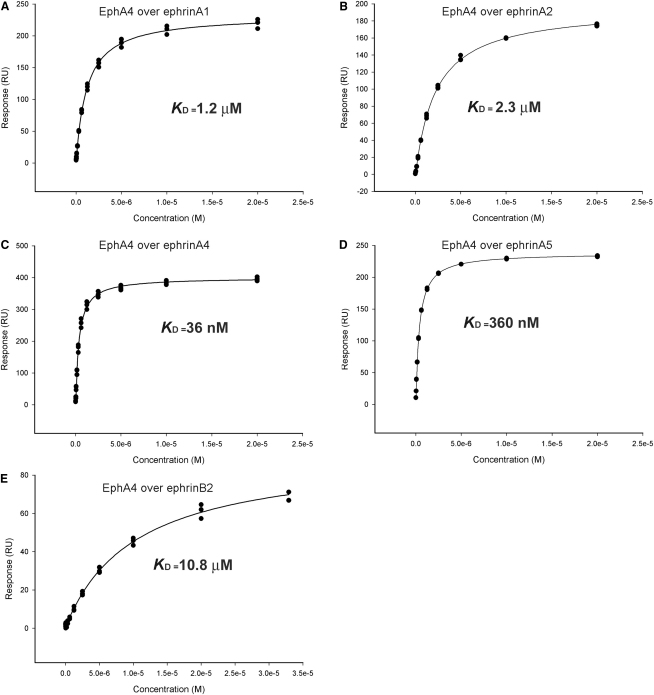
To ascertain the strength of EphA4 binding to a panel of ephrins, affinity measurements of EphA4 with ephrinA1, ephrinA2, ephrinA4, ephrinA5, ephrinB1, and ephrinB2 were performed using surface plasmon resonance. Monomeric ephrin ligands were immobilized onto carboxymethyl (CM5) chip surfaces via a biotin-streptavidin linkage while monomeric EphA4 was used as an analyte with concentrations ranging from 20 nM to 30 μM. Consistent with previously reported fluorescence titration spectroscopy, isothermal calorimetry, and surface plasmon resonance studies that measured monomeric Eph-ephrin interactions, our data yielded Eph-ephrin binding constants in the nanomolar to low micromolar range (Figure 2) (Day et al., 2005; Himanen et al., 2004; Nikolov et al., 2005; Pabbisetty et al., 2007).
Figure 2.
KD Determination of EphA4 LBD with Ephrin RBD
(A–E) Overlay association plots of surface plasmon resonance sensorgrams recording response of EphA4 (analyte) interaction with immobilized ephrinA1 (A), ephrinA2 (B), ephrinA4 (C), ephrinA5 (D), and ephrinB2 (E). Calculated affinity binding KD values are shown.
Our results show that the EphA4 receptor has a broad affinity range for different types of ephrin ligands with cross-class Eph receptor binding weaker (5–300 times) than EphA-ephrinA interactions. We find that EphA4 has greatest affinity for ephrinA4 (KD = 36 nM), intermediate affinities to ephrinA1, ephrinA5, and ephrinA2 (KD = 1.2 μM, KD = 360 nM, and KD = 2.3 μM, respectively), binds most weakly to ephrinB2 (KD = 10.8 μM), and shows no detectable binding to ephrinB1.
Crystal Structures of EphA4-ephrinB2 and EphA4-ephrinA2 Complexes
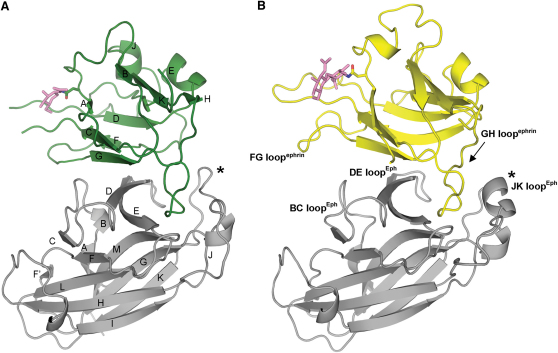
In order to investigate the molecular mechanisms underlying EphA4 cross-reactivity, we sought to determine representative complexes with A and B class ephrins. The EphA4-ephrinB2 and EphA4-ephrinA2 complex structures were solved and refined to 2.4 and 2.3 Å resolution, respectively (Table 1). Our determination of these two structures increases the number of known Eph-ephrin complexes to six. The EphA4-ephrinB2 structure is the first cross-class EphA type complex. Both the overall fold and architecture of complex binding in these two structures remain consistent with previously reported Eph-ephrin complexes (Figure S2). EphA4 maintains the same β sandwich fold that is present in the unliganded structures, and the ephrin ligands retain a Greek key β barrel fold consisting of eight β strands (designated A–K) with hydrophobic residues in the GHephrin loop buried with almost no solvent accessibility between the DEEphA4 and JKEphA4 loops on EphA4 (Figure 3 and Figure S3).
Figure 3.
Crystal Structures of EphA4-ephrinB2 and EphA4-ephrinA2
(A and B) Cartoon representation of EphA4-ephrinB2 (A) and EphA4-ephrinA2 (B). Secondary structure elements are labeled according to standard nomenclature in A and the main interaction loops are labeled in B. The JKEph loops are marked with an asterisk and undergo major conformational changes dependent on the class of ephrin ligand bound. Acetyl-glucosamine moieties observed at N-linked glycosylation sites are shown as pink sticks.
To facilitate crystallization of the EphA4-ephrinA2 complex, a site-directed mutation, N174Q, was incorporated into the sequence of ephrinA2 to remove a glycosylation site not conserved between ephrins (Figure S4). As a result, only electron density corresponding to two reducing-terminal N-acetylglucosamine residues of an N-linked sugar at Asn42ephrinA2 was observed in the EphA4-ephrinA2 structure (Figure S5). In the EphA4-ephrinB2 structure, electron density corresponding to a single N-acetylglucosamine moiety was observed at Asn36ephrinB2 (conserved with Asn42ephrinA2) but not Asn139ephrinB2 (Figure S5).
The first structure of an Eph receptor-ephrin complex (EphB2-ephrinB2) (Himanen et al., 2001) revealed a heterotetrameric arrangement of molecules suggestive of a clustering mode involved in signaling. However, two subsequent complexes (EphB2-ephrinA5 and EphB2-ephrinB2) (Chrencik et al., 2006a; Himanen et al., 2004) have not shown such higher order arrangements and we do not observe any evidence of heterotetramerization in our structures of EphA4-ephrinB2 and EphA4-ephrinA2.
The overall total buried surface areas for the receptor-ligand interface in EphA4-ephrinB2 and EphA4-ephrinA2 are 2590 and 1930 Å2, respectively (Krissinel and Henrick, 2007). These values are comparable to those observed in the EphA2-ephrinA1 (PDB code 3CZU), EphB2-ephrinB2 (PDB code 1KGY), and EphB4-ephrinB2 (PDB code 2HLE) structures (buried surface areas of 2260, 2380, and 2120 Å2, respectively), but are in contrast to EphB2-ephrinA5 (PDB code 1SHW; buried surface area of 1220 Å2), the only other cross-class complex crystallized. Our EphA4-ephrinB2, EphA4-ephrinA2, and the recently deposited EphA2-ephrinA1 structures also have relatively high surface complementarity scores (Lawrence and Colman, 1993) (0.71, 0.76, and 0.73, respectively), which distinguishes them from the previously reported EphB2-ephrinB2, EphB4-ephrinB2, and EphB2-ephrinA5 structures (surface complementarity scores of 0.67, 0.64, and 0.66, respectively).
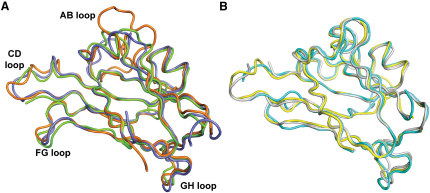
Structural Rigidity of the Ephrin Ligand
Comparison of ephrin ligands from multiple Eph-ephrin structures reveals that both A and B class ephrins are structurally invariant and independent of their Eph receptor pair (Figure 4). Upon detailed inspection, it is apparent, however, that the AB loop region is variable in A class ephrins. This is due to a six amino acid insertion in ephrinA2 that is not present in other ephrin ligands. As this region is remote from the Eph receptor binding face, it is unlikely to influence Eph binding.
Figure 4.
Structural Alignment of Ephrin Ligands in Their Bound States
Ephrin A class ligands (A) and ephrinB2 ligands (B) are shown in cartoon representation. EphrinA1 from EphA2-ephrinA1 (PDB code 3CZU) is colored green, ephrinA2 from EphA4-ephrinA2 is colored orange, and ephrinA5 from EphB2-ephrinA5 (PDB code 1SHW) is colored blue. EphrinB2 from EphA4-ephrinB2 is colored cyan, ephrinB2 from EphB2-ephrinB2 (PDB code 1KGY) is colored yellow, and ephrinB2 from EphB4-ephrinB2 (PDB code 2HLE) is colored gray.
Due to the high structural rigidity of ephrin ligand structures, we suggest that the overall binding conformation of an Eph receptor is molded by the identity of its cognate ligand. However, ephrinB2 can show structural plasticity in a different context; the conformation of the GHephrinB2 loop is altered upon binding to the Nipah virus and Hendra virus attachment glycoproteins (Bowden et al., 2008a, 2008b).
Plasticity of Eph Receptor A4 at the Ephrin Binding Interface
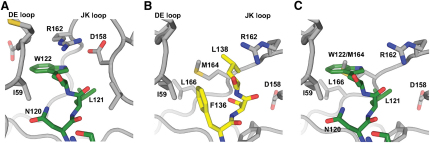
While ephrins generally exhibit a low degree of structural variance between different bound forms, structural analysis of EphA4 reveals distinct structural differences between unbound EphA4 and its ephrinA2 (1.7 Å rmsd over 166 equivalent Cα atoms)- and ephrinB2 (2.3 Å rmsd over 168 equivalent Cα atoms)-bound states (Figure 5A). Most notable are the structural changes that occur to the DEEphA4 and JKEphA4 loops (Figures 5B and 5C); regions which we identify above as having a high degree of flexibility in unbound EphA4. While the DEEphA4 loop moves appreciably between its unbound and bound states, its main chain conformation is maintained (0.5 Å rmsd over Cα atoms of residues Ser58EphA4-Arg68EphA4) between the ephrinA2- and ephrinB2-bound structures (Figure S6). This suggests that the DEEphA4 loop interacts in a conserved manner that is independent of the identity of the ephrin ligand. In contrast, the JKEphA4 loop is very different between its two bound states (2.8 Å rmsd over Cα atoms of residues Ala150EphA4-Leu166EphA4); forming an extended conformation when bound to ephrinB2 and an α helix (Gln156EphA4-Asp161EphA4) when bound to ephrinA2. The extended conformation of the JKEphA4 loop when bound to ephrinB2 appears to be stabilized by a hydrophobic stacking interaction between Arg162EphA4 and Trp122ephrinB2 (Figure 6A; Trp122 is equivalent to Trp125 in mouse). It is likely that this stacking interaction will be conserved in the interaction of EphA4 with ephrinB3 (Trp122 is conserved). These results also provide a rationale for why EphA4 does not bind to ephrinB1. Trp122ephrinB2 (equivalent to Met122ephrinB1) is not conserved and Tyr121ephrinB1 (equivalent to Leu121ephrinB2) is likely to clash with the JKEphA4 loop. The Arg162EphA4-Trp122ephrinB2 stacking interaction contrasts with that observed in the EphA4-ephrinA2 structure, where Arg162EphA4 is involved in hydrogen bonding interactions with the carbonyl oxygen of Leu138ephrinA2, while Met164EphA4and Leu166EphA4 participate in hydrophobic interactions with Leu138ephrinA2 and Phe136ephrinA2 (Figure 6B).
Figure 5.
Conformational Changes of EphA4 upon Binding
(A) Superposition of unbound EphA4 (orange), with its ephrinB2 (dark blue)- and ephrinA2 (gray)-bound states shown in stereo representation.
(B) Rms displacement of equivalent residues between unbound, ephrinB2-bound, and ephrinA2-bound EphA4 mapped onto the Cα trace structure of EphA4 receptor (from EphA4-ephrinA2). The tube radius and color of the trace represent the rms displacement (ramped from blue to red). Regions with high deviations between structures are thick and colored red, while regions with low deviation are thin and colored blue.
(C) Cα trace of EphA4 from EphA4-ephrinA2 colored according to regions of flexibility as defined by ESSET (Schneider, 2002). Regions colored blue are conformationally invariant and regions colored red are flexible.
Figure 6.
EphrinB2 Precludes the Formation of an α Helix in the JKEphA4 Loop upon Binding
(A) Residues from the GHephrinB2 loop (stick representation with nitrogen blue, oxygen red, and carbon green) bind between the DEEphA4 and JKEphA4 loops (stick representation with nitrogen blue, oxygen red, sulfur gold, and carbon gray).
(B) Residues from the GHephrinA2 loop (stick representation with nitrogen blue, oxygen red, and carbon yellow) bind between the DEEphA4 and JKEphA4 loops (gray van der Waals surface; stick representation with nitrogen blue, oxygen red, and carbon gray).
(C) W125ephrinB2 from the GHephrinB2 loop (green sticks, colored as in [A]) clashes with M164EphA4 when superposed onto EphA4 in its ephrinA2-bound state.
Although the JKEphA4 loop is α-helical in the EphA4-ephrinA2 structure, sequence-based secondary structure prediction programs (Bryson et al., 2005) predict the JKEphA4 loop region to be a coil or a turn. However, we note that this helix is not unprecedented and is analogous to that existing in the recently released EphA2-ephrinA1 structure (PDB code 3CZU; residues Ser153-Ala158EphA2). It is therefore likely that this helix is a structural motif specific to A class Eph receptor-ligand interactions and its formation is dependent on the nature of residues present in the GHephrin binding loop. This helix is not present in the EphA4-ephrinB2 structure due to the steric clash that would occur between Trp122ephrinB2 (rather than Leu138ephrinA2) and Met164EphA4 (Figure 6C). As a result, we suggest that the plasticity of the JKEphA4 loop region allows EphA4 to physically adapt to Trp122ephrinB2 and the resulting complementary stacking interactions with Arg162EphA4 are compensatory.
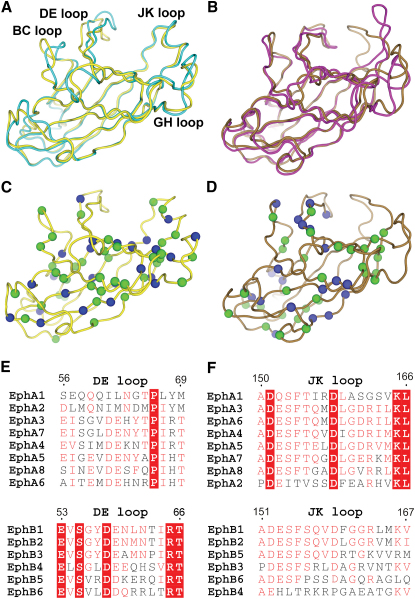
These observations prompted us to undertake a full comparison of EphA-ephrinA- and EphB-ephrinB-type complexes to detect any additional class-dependent trends (Figure 7). Superposition of EphA4 and EphA2 from their ephrinA2- and ephrinA1-bound structures, respectively, reveals that the JKEph α helix is well conserved both in structure and in amino acid sequence (Figures 7A, 7C, and 7F). This contrasts noticeably with the lack of sequence identity and varied extended coil conformations present in EphB2 and EphB4 when in complex with ephrinB2 (Figures 7B, 7D, and 7F). It is striking that the reverse observations can be made with the DEEph loop. EphB-ephrinB complexes demonstrate both a conserved structure and sequence (Figures 7B, 7D, and 7E), while EphA-ephrinA class complexes are structurally dissimilar and have relatively low sequence identity (Figures 7A, 7C, and 7E). On the basis of markedly different trends in sequence and structural conservation in these regions, we suggest that the DEEphA4 and JKEphA4 loops provide a mechanism by which A and B class Eph receptors, respectively, generate specificity and cross-class reactivity.
Figure 7.
Structure and Sequence Conservation of EphA-ephrinA and EphB-ephrinB Complexes
(A) Superposition of EphA4 (yellow) and EphA2 (cyan) from their ephrinA2- and ephrinA1-bound states, respectively.
(B) Superposition of EphB2 (brown; PDB code 1KGY) and EphB4 (magenta; PDB code 2HLE) from their ephrinB2-bound states.
(C and D) EphA4 from its ephrinA2-bound state (C; yellow) and EphB2 from its ephrinB2-bound state (D; brown; PDB code 1KGY) with sequence conservation mapped onto the structure as blue (residues completely conserved between EphA sequences) and green (residues at least 80% conserved across Eph sequences) spheres.
(E) Sequence alignment of the DEEph loop numbered according to EphA4.
(F) Sequence alignment of the JKEph loop numbered according to EphA4.
The Architecture of Complex Binding is Dependent on the Ephrin Ligand
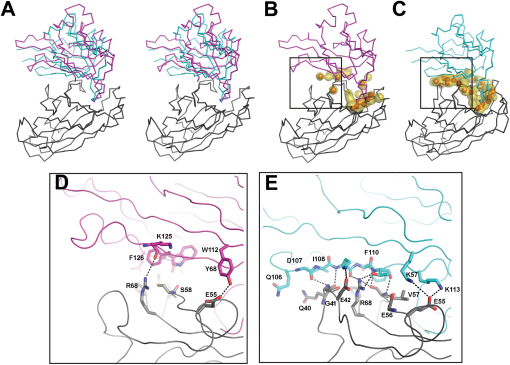
Superposition of our EphA4 complexes reveal that the relative orientation of ephrinA2 when bound to EphA4 differs by a tilt of 12° in comparison to the ligand in the EphA4-ephrinB2 structure (Figure 8A) and results in fewer intracomplex interactions beyond the DEEphA4 and JKEphA4 surface channel (Figures 8B and 8C). The observation that ephrinA-type ligands bind to their respective Eph receptor with a different tilt, which therefore results in a lack of an extended binding site, has been made by previous comparison of the EphB2-ephrinA5, EphB2-ephrinB2, and EphB4-ephrinB2 structures (Himanen et al., 2007) and holds true for the recently deposited EphA2-ephrinA1 structure (Figure S7). The presence or absence of an extended binding site correlates with the van der Waals and hydrogen bonding patterns (Figures 8D and 8E and Figure S7). While the EphA4-ephrinB2 interface is comprised of 15 hydrogen bonds and a total of 30 residues participating in van der Waals interactions, EphA4-ephrinA2 contains only 6 and 22, respectively. This ephrin class difference is also consistent with previously reported structures where EphB2-ephrinB2, EphB2-ephrinB4, EphA2-ephrinA1, and EphB2-ephrinA5 have 14, 12, 8, and 2 hydrogen bonds, respectively, which lie away from the hydrophobic surface channel formed by the DEEphA4 and JKEphA4 loops. Although these observations do not take into account differences in crystal packing, these patterns suggest a second characteristic dissimilarity between class A and B ephrin binding.
Figure 8.
The Extended Ephrin Binding Site
(A–E) Eph and ephrin Cα traces where EphA4 is colored gray, ephrinA2 is colored pink, and ephrinB2 is colored cyan.
(A) Comparison of ephrinA2 (pink)- and ephrinB2 (cyan)-bound to EphA4 (gray) based on superposition of the EphA4 component (shown in stereo representation) reveals a 12° relative tilt between ephrinA- and ephrinB-bound complexes.
(B and C) The presence or absence of an extended Eph-ephrin binding surface correlates with the degree of van der Waals and hydrogen bonding interactions. Midpoints between van der Waals interactions are shown as gold surfaces and intracomplex hydrogen bonds are shown as red spheres in the EphA4-ephrinA2 (B) and EphA4-ephrinB2 (C) complexes. Boxes in (B) and (C) refer to areas highlighted in (D) and (E). Colored as in (A).
(D and E) Van der Waals and hydrogen bonding interactions taking place in the extended binding surface. Residues involved in protein-protein contacts are shown as sticks where carbon atoms are colored as in (A)–(C), nitrogen atoms are colored blue, oxygen red, and carbon yellow. Dashed lines correspond to intracomplex hydrogen bonds.
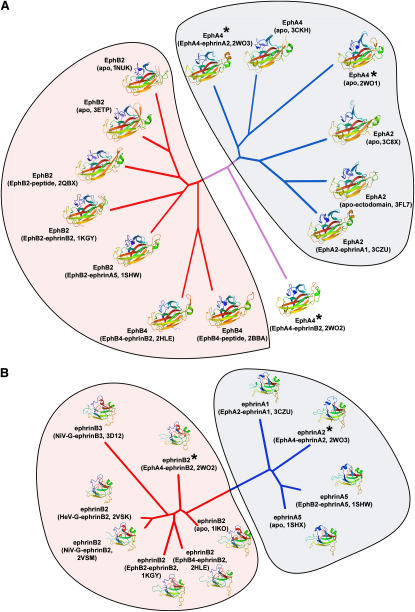
Structural Phylogenetic Analysis of Multiple Eph Receptors and Ephrin Ligands
Given the recent availability of multiple high resolution Eph and ephrin crystal structures, we performed structure-based phylogenetic analyses on the LBD of the Eph receptor and RBD of the ephrin ligand (Figure 9). A structural-based phylogenetic tree was derived with SHP (Stuart et al., 1979) and plotted with PHYLIP (Felsenstein, 1989) using all currently available Eph receptors and ephrin ligands (from 14 and 11 PDB entries, respectively). Similar to a sequence-based phylogenetic analysis (Figure S8), Eph receptors (Figure 9A) and ephrin ligands (Figure 9B) are structurally divided into two different branches dependent on their A or B type classification. Conspicuously, in the Eph receptor tree (Figure 9A), EphA4 in its ephrinB2-bound state is the most structurally similar out of all A class Eph receptors to the B class Eph receptors. Other EphA receptor structures, including the apo and ephrinA2-bound EphA4 structures, on the other hand, are more distantly related to B class Eph receptors. It appears that EphA4 maintains an element of conformational duality and flexibility that allows it to act as a pseudo class B Eph receptor, but, in corroboration with our surface plasmon resonance binding data, at an energetic cost with reduced binding affinity. The link between structure and cross-class promiscuity, however, is more difficult to establish with EphB2, the other known cross-class, high-affinity binding receptor. In contrast to the relatively large structural distance between different forms of the EphA4 LBD, EphB2 is much less structurally variant and, as a result, these structures cluster together much more closely.
Figure 9.
Structure-Based Phylogenetic Analysis of Eph Receptors and Ephrin Ligands
(A and B) Unrooted trees of the Eph receptor LBD (A) and ephrin ligand RBD (B). The structures were superposed and a pairwise distance matrix was constructed as previously described (Graham et al., 2008). PDB codes for each representative structure are labeled beneath a cartoon representation ramped from blue (N terminus) to red (C terminus). Branches colored blue correspond to A class Eph and ephrin families, branches colored red correspond to B class Eph and ephrin families, and the branch colored lilac (A) corresponds to EphA4 from its ephrinB2-bound state, which does not belong to either family. Structures marked with an asterisk are those presented within this manuscript.
Discussion
Here, we have presented equilibrium binding data and crystal structures of the apo, ephrinB2-bound, and ephrinA2-bound forms of the cross-class reactive EphA4 LBD. Our surface plasmon resonance data show that EphA4 interacts with ephrin ligands with a broad affinity range. Eph receptors and their ephrin ligands are known to be expressed in complementary gradients (Hafner et al., 2004), a characteristic that appears consistent with receptor binding spanning a wide range of ligand concentrations. We suggest that this wide affinity range reflects a mechanism of fine tuning the response to multiple ligands encountered and perhaps different physiological functions.
From our structural analysis, we do not detect any indication of higher order EphA-ephrin heterotetramers. The structural differences between the two available apo LBD structures of EphA4 and the divergence in conformational changes that occur upon ephrinB2 and ephrinA2 binding are consistent with the EphA4 receptor maintaining a high degree of structural plasticity in its DEEphA4 and JKEphA4 ephrin binding loops. These changes include the introduction of unpredicted α-helical secondary structure in the JKEphA4 loop of the EphA4-ephrinA2 complex. Based on the high degree of sequence conservation in the JK loop across all A class Eph receptors, it is likely that this α helix motif is common to all EphA-ephrinA complexes. In contrast, in EphB-ephrinB complexes, the DE loop is more conserved in both sequence and structure. Therefore, we suggest that these two structural features are class-specific characteristics that distinguish between EphA-ephrinA and EphB-ephrinB modes of interaction.
Based on their structures of complexed and uncomplexed EphA2, Himanen et al. (2009) have very recently proposed that A class Eph receptor-ephrin interactions can be described by a “lock-and-key” mechanism. However, inspection of the EphA2 unbound structures deposited by the Structural Genomics Consortium (Toronto) indicates that for these crystal structures the primary EphA2 ligand binding loops (JK and DE loops) are partially disordered or are in conformational states quite different to their bound forms. These properties are similar to those observed in our EphA4 structures and the previously reported unbound EphA4 structure (Qin et al., 2008), results that suggest that the ligand binding surface of the EphA receptor is molded by the ephrin ligand. In light of these observations, although we cannot discount a lock-and-key mechanism until a greater range of A class Eph receptor structures are available, the evidence that A class Eph receptors are more likely than B class Eph receptors to be structurally rigid between their bound and unbound states is still inconclusive.
The bidirectional downstream signaling processes that result from Eph receptor-ephrin ligand binding regulate a range of biological processes from axon guidance to vascular development. The specific biology is dependent on the identity of the Eph receptor-ephrin ligand pair (Pasquale, 2005). By decoding the molecular basis for specificity and promiscuity of binding, we can aim to understand the hierarchy of Eph-ephrin interactions.
Experimental Procedures
Cloning and Expression
Human ephrinA2 (residues 33–177; GenBank accession number NP_001396) and human ephrinB2 (residues 27–167; GenBank accession number NP_004084) were cloned into the pHLSec vector (Aricescu et al., 2006b) and human EphA4 (residues 30–202; GenBank accession number NP_004429) was cloned into the pOPING vector (Berrow et al., 2007). Both vectors encode an optimal signal peptide for HEK293T cell-based expression and a C-terminal Lys-His6 tag. Proteins were expressed transiently in HEK293T cells in the presence of the class I α-mannosidase inhibitor kifunensine (1 mg L−1 culture). Transfections were performed with polyethyeleneimine either with a single DNA construct or as cotransfections for complex formation.
Purification
EphA4 alone and in complex with ephrinA2 and ephrinB2 was purified by immobilized metal affinity chromatography using nickel-charged chelating Sepharose beads (GE Healthcare). EphA4-ephrinB2 complex was treated with endoglycosidase F1 (75 μg mg−1 protein, 12 hr, 21°C) to cleave glycosidic bonds of N-linked sugars within the di-N-acetylchitebiose core. Following deglycosylation, protein complexes were purified by size exclusion chromatography using a Superdex 75 10/30 column (Amersham) in 150 mM NaCl and 10 mM Tris (pH 7.5–8.0) buffer. The typical yield of purified protein (or protein complex) was approximately 2.5 mg complex L−1 culture, and cotransfection of EphA4 with ephrinB2 resulted in an approximately 50% increase in overall yield (4.0 mg complex L−1).
Mutagenesis of N-linked Glycosylation Sites on EphrinA2
Mutagenesis of the potential N-linked glycosylation site N174Q was achieved by incorporation of the desired mutation in the C-terminal primer used for PCR cloning (Pyrobest Polymerase, Takara). Following PCR, the product was cloned into the pHLsec vector.
Crystallization, Data Collection, Structure Determination, and Refinement
Protein crystals were grown using the sitting drop vapor diffusion method using 100 nL protein plus 100 nL precipitant according to previously described methods (Walter et al., 2005) (Table S1). Crystals were flash frozen by immersion of the crystal into a cryoprotectant containing glycerol, PFO-X125/03 (Lancaster Synthesis), or PEG (polyethylene glycol) 400 followed by rapid transfer to a gaseous nitrogen stream at 100 K. Data were collected at the European Synchrotron Radiation Facility (ESRF) beamline BM-14 and Diamond beamline I03. Images were integrated and scaled using the programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). Crystallographic statistics are presented in Table 1. For all structures, 5% of reflections were randomly set aside to calculate the Rfree. Structures were solved by molecular replacement using the program Phaser (Storoni et al., 2004). The structure of EphA4 was solved using EphB2 (PDB code 1NUK) as a molecular replacement model and the structures of EphA4 in complex with ephrinB2 and ephrinA2 were solved using the EphB2-ephrinB2 structure (PDB code 1KGY). Initial model building for the high resolution EphA4 structure was performed with the program ARP-wARP (Perrakis et al., 2001). Structure refinement included iterative model building using Refmac 5 (Murshudov et al., 1997). Refinement cycles included rigid body refinements and restrained refinement with TLS. The molecular graphics program COOT was used for manual rebuilding (Emsley and Cowtan, 2004). The program MolProbity was used to validate models (Davis et al., 2007).
Surface Plasmon Resonance Binding of EphA4 to Ephrin Ligands
The RBDs of human ephrinA1 (residues 16–158; GenBank accession number AAH32698), ephrinA2 (residues 33–174; GenBank accession number NP_001396), ephrinA4 (residues 23–162; GenBank accession number AAI07484), ephrinA5 (residues 27–166; GenBank accession number NP_001953), ephrinB1 (residues 26–170; GenBank accession number NP_004420.1), and ephrinB2 (residues 25–168; GenBank accession number NP_004429) were subcloned into the pHLsec-avitag-3 vector (Aricescu et al., 2006b). Ephrin ligands were immobilized on chelating Sepharose Fast Flow Ni2+-agarose beads (GE Healthcare) and biotinylated as described previously (Aricescu et al., 2006a). All ephrin ligands used for these binding studies had wild-type glycosylation. Affinity measurements between ephrin ligands and EphA4 receptor were performed using a BIAcore T100 (Biacore). Purified, biotinylated ephrin ligands were immobilized on a CM5 BIAcore sensor chip (Biacore) coated with streptavidin (Sigma). A surface with only streptavidin present on the chip was reserved to subtract background responses due to differences in refractive indices of running and sample buffers. For affinity measurements of EphA4 analyte binding to immobilized ephrin ligands, samples were injected at 20 μl min−1 (25°C) in HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.005% surfactant P20, and 3 mM EDTA). To calculate binding affinity, several cycles of association and dissociation at different concentrations of analyte were required. Analyte concentrations ranged from 20 nM to 30 μM. KD values were calculated using Biacore Evaluation Software Version 1.1.
Superpositions and Illustrations
All molecular superpositions were calculated using SHP (Stuart et al., 1979). All molecular representations were produced with Pymol (http://pymol.sourceforge.net). Sequence alignments were performed with MultAlin (Corpet, 1988) and formatted with ESPript (Gouet et al., 1999). Figures were prepared using Adobe Illustrator, Adobe Photoshop, CorelDraw, and Microsoft Publisher.
Acknowledgments
We thank W. Lu for technical assistance with tissue culture, K. Harlos for data collection, M. Crispin for helpful discussions, and the staff of beamlines IO3 and BM-14 at Diamond Light Source and the ESRF, respectively. This work was funded by Cancer Research UK (C375/A3964), the UK Medical Research Council (G0701506), the Wellcome Trust, and the European Commission as SPINE2 Complexes (FP6-RTD-031220). A.R.A., R.J.O., D.I.S., and E.Y.J. designed and supervised experiments. T.A.B., A.R.A., N.R.H., and J.E.N. performed cloning, protein preparation, mutagenesis, and crystallization. T.A.B., A.R.A., C.S., D.I.S., and E.Y.J. performed the structural analysis. T.A.B. performed the in vitro surface plasmon resonance experiments and analysis. All authors participated in writing the manuscript.
Published: October 13, 2009
Footnotes
Supplemental data include eight figures and one table and can be found with this article online at http://www.cell.com/structure/supplemental/S0969-2126(09)00339-6.
Accession Numbers
Atomic coordinates and structural factors have been deposited in the Protein Data Bank with accession codes 2WO1 (EphA4), 2WO2 (EphA4-ephrinB2), and 2WO3 (EphA4-ephrinA2).
Supplemental Data
References
- Aricescu A.R., Hon W.C., Siebold C., Lu W., van der Merwe P.A., Jones E.Y. Molecular analysis of receptor protein tyrosine phosphatase μ-mediated cell adhesion. EMBO J. 2006;25:701–712. doi: 10.1038/sj.emboj.7600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Berrow N.S., Alderton D., Sainsbury S., Nettleship J., Assenberg R., Rahman N., Stuart D.I., Owens R.J. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden T.A., Aricescu A.R., Gilbert R.J., Grimes J.M., Jones E.Y., Stuart D.I. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008;15:567–572. doi: 10.1038/nsmb.1435. [DOI] [PubMed] [Google Scholar]
- Bowden T.A., Crispin M., Harvey D.J., Aricescu A.R., Grimes J.M., Jones E.Y., Stuart D.I. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: a template for antiviral and vaccine design. J. Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K., McGuffin L.J., Marsden R.L., Ward J.J., Sodhi J.S., Jones D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty A.J., Greferath U., Turnley A.M., Murphy M. Eph tyrosine kinase receptor EphA4 is required for the topographic mapping of the corticospinal tract. Proc. Natl. Acad. Sci. USA. 2006;103:15629–15634. doi: 10.1073/pnas.0607350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrencik J.E., Brooun A., Kraus M.L., Recht M.I., Kolatkar A.R., Han G.W., Seifert J.M., Widmer H., Auer M., Kuhn P. Structural and biophysical characterization of the EphB4-ephrinB2 protein-protein interaction and receptor specificity. J. Biol. Chem. 2006;281:28185–28192. doi: 10.1074/jbc.M605766200. [DOI] [PubMed] [Google Scholar]
- Chrencik J.E., Brooun A., Recht M.I., Kraus M.L., Koolpe M., Kolatkar A.R., Bruce R.H., Martiny-Baron G., Widmer H., Pasquale E.B., Kuhn P. Structure and thermodynamic characterization of the EphB4/Ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure. 2006;14:321–330. doi: 10.1016/j.str.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard M.G., Duffy S., Down M., Evans B., Power M., Smith F., Stylianou C., Kleikamp S., Oates A., Lackmann M. The role of the Eph-ephrin signalling system in the regulation of developmental patterning. Int. J. Dev. Biol. 2002;46:375–384. [PubMed] [Google Scholar]
- Cowan C.A., Henkemeyer M. Ephrins in reverse, park and drive. Trends Cell Biol. 2002;12:339–346. doi: 10.1016/s0962-8924(02)02317-6. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., To C., Himanen J.P., Smith F.M., Nikolov D.B., Boyd A.W., Lackmann M. Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3. J. Biol. Chem. 2005;280:26526–26532. doi: 10.1074/jbc.M504972200. [DOI] [PubMed] [Google Scholar]
- Dottori M., Hartley L., Galea M., Paxinos G., Polizzotto M., Kilpatrick T., Bartlett P.F., Murphy M., Kontgen F., Boyd A.W. EphA4 (Sek1) receptor tyrosine kinase is required for the development of the corticospinal tract. Proc. Natl. Acad. Sci. USA. 1998;95:13248–13253. doi: 10.1073/pnas.95.22.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher U. Eph family functions from an evolutionary perspective. Curr. Opin. Genet. Dev. 2002;12:397–402. doi: 10.1016/s0959-437x(02)00316-7. [DOI] [PubMed] [Google Scholar]
- Easty D.J., Bennett D.C. Protein tyrosine kinases in malignant melanoma. Melanoma Res. 2000;10:401–411. doi: 10.1097/00008390-200010000-00001. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eph Nomenclature Committee Unified nomenclature for Eph family receptors and their ligands, the ephrins. Eph Nomenclature Committee. Cell. 1997;90:403–404. doi: 10.1016/s0092-8674(00)80500-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Fu W.Y., Chen Y., Sahin M., Zhao X.S., Shi L., Bikoff J.B., Lai K.O., Yung W.H., Fu A.K., Greenberg M.E., Ip N.Y. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat. Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- Fukai J., Yokote H., Yamanaka R., Arao T., Nishio K., Itakura T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer Ther. 2008;7:2768–2778. doi: 10.1158/1535-7163.MCT-07-2263. [DOI] [PubMed] [Google Scholar]
- Gale N.W., Holland S.J., Valenzuela D.M., Flenniken A., Pan L., Ryan T.E., Henkemeyer M., Strecker K., Hirai H., Wilkinson D.G. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Graham S.C., Bahar M.W., Cooray S., Chen R.A., Whalen D.M., Abrescia N.G., Alderton D., Owens R.J., Stuart D.I., Smith G.L., Grimes J.M. Vaccinia virus proteins A52 and B14 Share a Bcl-2-like fold but have evolved to inhibit NF-kappaB rather than apoptosis. PLoS Pathog. 2008;4:e1000128. doi: 10.1371/journal.ppat.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner C., Schmitz G., Meyer S., Bataille F., Hau P., Langmann T., Dietmaier W., Landthaler M., Vogt T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin. Chem. 2004;50:490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- Himanen J.P., Rajashankar K.R., Lackmann M., Cowan C.A., Henkemeyer M., Nikolov D.B. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- Himanen J.P., Chumley M.J., Lackmann M., Li C., Barton W.A., Jeffrey P.D., Vearing C., Geleick D., Feldheim D.A., Boyd A.W. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat. Neurosci. 2004;7:501–509. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- Himanen J.P., Saha N., Nikolov D.B. Cell-cell signaling via Eph receptors and ephrins. Curr. Opin. Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen J.P., Goldgur Y., Miao H., Myshkin E., Guo H., Buck M., Nguyen M., Rajashankar K.R., Wang B., Nikolov D.B. Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 2009;10:722–728. doi: 10.1038/embor.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Maru Y., Hagiwara K., Nishida J., Takaku F. A novel putative tyrosine kinase receptor encoded by the eph gene. Science. 1987;238:1717–1720. doi: 10.1126/science.2825356. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kullander K., Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- Lawrence M.C., Colman P.M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lim Y.S., McLaughlin T., Sung T.C., Santiago A., Lee K.F., O'Leary D.D. p75(NTR) mediates ephrin-A reverse signaling required for axon repulsion and mapping. Neuron. 2008;59:746–758. doi: 10.1016/j.neuron.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Sun E.E., Klein R.S., Flanagan J.G. Ephrin-B reverse signaling is mediated by a novel PDZ-RGS protein and selectively inhibits G protein-coupled chemoattraction. Cell. 2001;105:69–79. doi: 10.1016/s0092-8674(01)00297-5. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nikolov D.B., Li C., Barton W.A., Himanen J.P. Crystal structure of the ephrin-B1 ectodomain: implications for receptor recognition and signaling. Biochemistry. 2005;44:10947–10953. doi: 10.1021/bi050789w. [DOI] [PubMed] [Google Scholar]
- Noberini R., Koolpe M., Peddibhotla S., Dahl R., Su Y., Cosford N.D., Roth G.P., Pasquale E.B. Small molecules can selectively inhibit ephrin binding to the EphA4 and EphA2 receptors. J. Biol. Chem. 2008;283:29461–29472. doi: 10.1074/jbc.M804103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski A., Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr., Sweet R.M., editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pabbisetty K.B., Yue X., Li C., Himanen J.P., Zhou R., Nikolov D.B., Hu L. Kinetic analysis of the binding of monomeric and dimeric ephrins to Eph receptors: correlation to function in a growth cone collapse assay. Protein Sci. 2007;16:355–361. doi: 10.1110/ps.062608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Harkiolaki M., Wilson K.S., Lamzin V.S. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Qin H., Shi J., Noberini R., Pasquale E.B., Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high-affinity ephrin-binding channel of the EphA4 receptor. J. Biol. Chem. 2008;283:29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.R. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr. D Biol. Crystallogr. 2002;58:195–208. doi: 10.1107/s0907444901019291. [DOI] [PubMed] [Google Scholar]
- Storoni L.C., McCoy A.J., Read R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Stuart D.I., Levine M., Muirhead H., Stammers D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- Walter T.S., Diprose J.M., Mayo C.J., Siebold C., Pickford M.G., Carter L., Sutton G.C., Berrow N.S., Brown J., Berry I.M. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.