Abstract
The ability to design effective enzymes is one of the most fundamental challenges in biotechnology and in some respects in biochemistry. In fact, such ability would be one of the most convincing manifestations of a full understanding of the origin of enzyme catalysis. In this work we explore the reliability of different simulation approaches, in terms of their ability to rank different possible active site constructs. This validation is done by comparing the ability of different approaches to evaluate the catalytic contributions of various residues in chorismate mutase. It is demonstrated that the empirical valence bond (EVB) model can serve as a practical yet accurate tool in the final stages of computer aided enzyme design (CAED). Other approaches for fast screening are also examined and found to be less accurate and mainly useful for qualitative screening of ionized residues. It is pointed out that accurate ranking of different options for enzyme design cannot be accomplished by approaches that cannot capture the electrostatic preorganization effect. This is in particular true with regard to current design approaches that use gas phase or small cluster calculations and then estimate the interaction between the enzyme and the transition state (TS) model rather than the TS binding free energy or the relevant activation free energy. The ability of the EVB model to provide a tool for quantitative ranking in the final stage of CAED may help in progressing towards the design of enzymes whose catalytic power is closer to that of native enzymes than to that of the current generation of designer enzymes.
Enzyme design has become a subject of major attention and major activity in recent years (1–5). Effective design is expected to have a great potential in industrial application and eventually in medicine. Furthermore, one can argue that the ability to design efficient enzymes is the best manifestation of a true understanding of enzyme catalysis. However, at present there has been a limited success in most attempt of rational enzyme design (6, 7), and the resulting constructs have been significantly less effective than the corresponding natural enzymes (1, 8).
Many proposals for the reasons of the limited success have been brought forward, but most are not based on actual validation studies (for review see (1, 8)). In fact, it has been argued (9) that the problems are due to the incomplete modeling of the transition state (TS) and, in part, to the limited awareness to the key role of the reorganization energy. However only demonstration of better performance in enzyme design can tell us what is exactly missing in current approaches. In fact, the ultimate reliability of computer aided enzyme design (CAED) is determined by the reliability of the calculations of mutational effects on catalysis. In other words, at the end of the day a reliable CAED approach must be able to rank different design options by predicting their activation barriers and the corresponding catalytic effect. Thus, it is possible to ask what are the missing factors in current design strategies and in particular in the final screening stages, by examining the performance of different approaches in calculations of mutational effects. In this respect it is useful to note that semiquantiative computational studies of the effect of mutations of enzyme catalysis date back the empirical valence bond (EVB) simulations of the effect of mutations in the catalytic power of trypsin (10) and to more qualitative transition state description (11). Subsequent calculations of mutational effect include EVB studies (e.g. (9, 12–16) and more recent (molecular orbital) quantum mechanics/molecular mechanics (QM/MM) studies (e.g. (17–22)). Relevant approaches that may be used for fast initial screening included the linear response approximation (LRA) evaluations of the contributions of different residues to the TS energy (23) and less systematic attempts of evaluating group contributions. The above studies, and in particular the quantitative one have established that the effects of the mutations are associated with the changes in reorganization energy upon mutations (9, 13, 14, 24).
Although recent design studies have produced encouraging results they have not been based on methods capable of estimating the energetics of the final design constructs (the approaches used are unable to predict the activation barriers in the generated active sites). More specifically current CAED approaches are based on fast and elegant construction of active sites that can provide reasonable interaction with the gas phase TS model. Here the emphasis is on the generation of protein structures with reasonable interactions with the TS model. While such approaches are very effective in generating structural candidates, the scoring function used is problematic, as it cannot reproduce the TS binding free energy or the catalytic effect. This is true regardless of the attempts to identify the estimated interaction energies with the TS binding energy. Our point can be easily verified by comparing the calculated scoring function to the experimental TS binding energy. Even more sophisticated methods such as the linear response approximation (LRA) method, that will be considered here, cannot provide quantitative scoring function. Thus the current CAED approaches basically focus on generating reasonable active sites with good hydrogen bonding etc. and leave the final decision to the experimental screening.
Our philosophy is quite different than the above approaches, since we believe that it is hard to design a function when the relevant property is not reproduce by the calculations. Thus we demand that the scoring function will reproduce the correct catalytic effect (a task that cannot be accomplished by current design approaches). Of course, the more expensive is the evolution of the free energy, the less practical is our approach as compared to experimental mutational studies. Thus we will focus on the price per performance issues. Of course, the EVB cannot be used in the preliminary enzyme design since the number of options is overwhelming. This step is not the focus of the present work and will only be considered in a preliminary way.
The above discussion can be summarized by stating that the experience from quantitative computer simulations of mutational studies has not been translated to a general guideline in current enzyme design methods, and the present work is aimed at moving in such a direction. This is done by trying to establish what it takes to get reliable results in the final screening stage, in terms of the relationship between accuracy and computer time. It is found that the EVB approach can provide a reliable way of performing the final screening steps in CAED while other approaches can be useful in more preliminary steps.
II. Systems and Methods
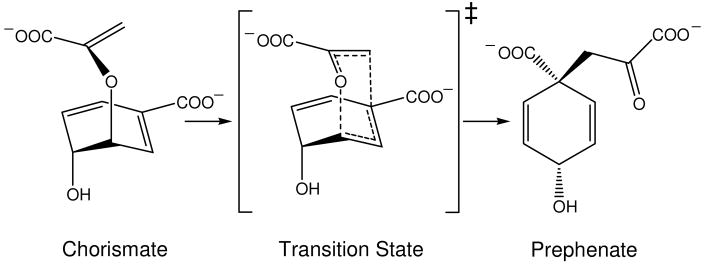
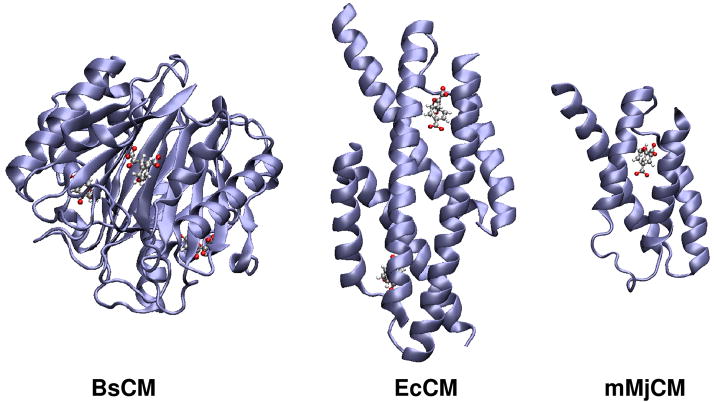
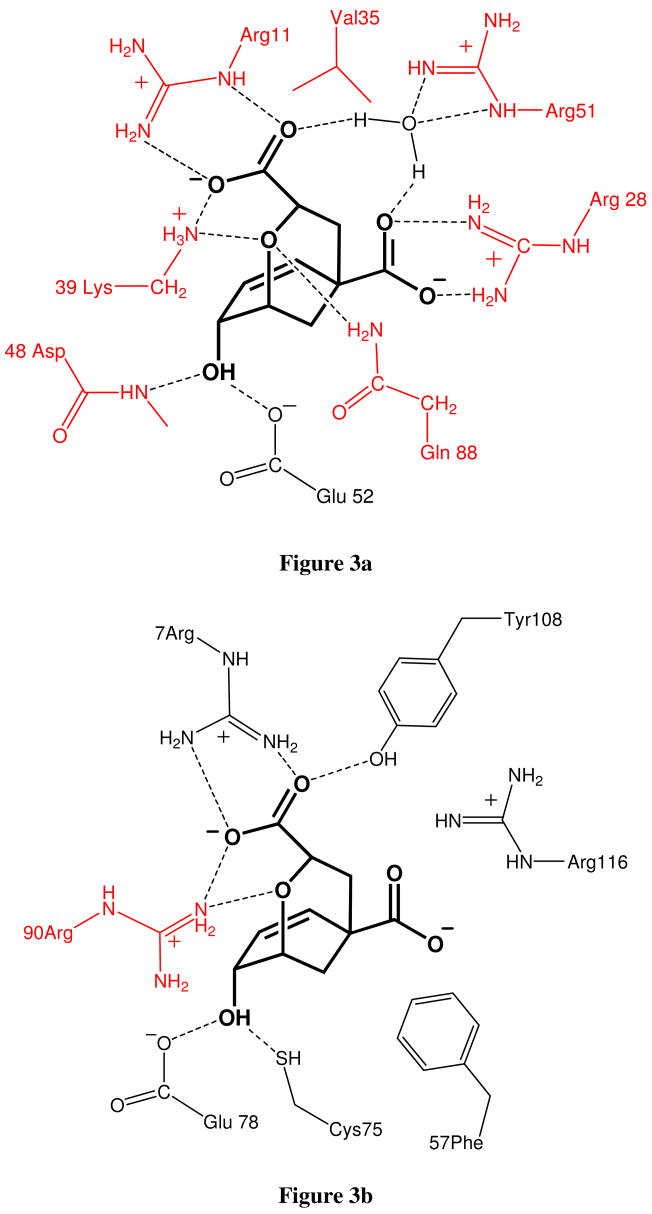
In this work we chose as a benchmark the ability to reproduce the catalytic activity of different mutants of different forms of enzyme chorismate mutase (CM) that catalyzes the reaction shown in Figure 1. The studied systems are the trimer Bacillus subtilis chorismate mutase (BsCM),(25) the homodimeric chorismate mutase from Escherichia coli (EcCM) (26) and the monomeric chorismate mutase mMjCM obtained by Hilvert and coworkers (27) by topological redesign of the thermostable EcCM homologue from M. jannaschii (MjCM). The coordinates for BsCM (X-Ray), EcCM (X-Ray) and mMjCM (NMR) structures were obtained from the Protein Data Bank, Brookhaven National Laboratory, with PDB access codes 1COM, 1ECM and 2GTV, respectively. The trimer resembles a beta-barrel structure in which a core beta-sheet is surrounded by helices whereas the dimer and monomer adopt a helix-bundle structure (see Figure 2). The active site residues that interact strongly with the transition state analogue (TSA) are depicted in Figure 3.
Figure 1.
Rearrangement of chorismate to prephenate
Figure 2.
Structures of the trimeric BsCM (left), dimeric EcCM (center) and monomeric mMjCM (right) proteins. The actives sites contain prepenate, TSA and TSA in the trimer, dimer and monomer, respectively. These active site ligands are represented by ball-and-stick models
Figure 3.
(a) Schematic description of the EcCM and mMjCM active sites, depicting key residues that are involved in the binding of the transition-state analogue (bold). (b) Schematic description of the BsCM active site, depicting key residues that are involved in the binding of the transition-state analogue (bold). The targeted residues for simulations of mutational effects are colored red.
CM has been the subject of intensive design studies (1) and was also considered in the examination of the origin of the difference between transition states (TSs) and transition state analogues (TSAs) (1, 28). The selection of this system allows us to examine three different enzymes that catalyze the same chemical reaction. Furthermore, since the protein topology and active site residues are different in the three CM forms they provide a diverse test set while keeping the same reference reaction. Thus we have a system that clearly presents a major challenge to CAED.
Below we will review the simulation approaches that will be used in this study. We start with the clarification that we do not like to use approaches of the type used recently in enzyme design studies (6, 7), where the TS features are determined in the gas phase since such approaches cannot be used to rank in a quantitative the different design constructs.
We will start with what we believe is the most reliable current approach for evaluating activation barriers of enzymatic reactions, namely the EVB. This method was used by our group and others in quantitative studies of the catalytic power of many enzymes (e.g. (9)). The EVB has been described in great details elsewhere (12, 13) and was used recently in a study of CM (29, 30). We give here only key relevant points.
The EVB is a QM/MM method that can be considered as a mixture of force fields of reactant and products (or intermediate) in a way that retains the correct change in structure and charge distribution along the reaction coordinate. The reason for the remarkable reliability of the EVB is that it is calibrated on the reference solution reaction and then the calculations in the enzyme active site only reflects (consistently) the change in the environment, exploiting the fact that the reacting system is the same in enzyme and solution. Thus, the EVB approach has to be calibrated only once per in a study of a given type of enzymatic reaction and this is done while considering the uncatalyzed reaction. More specifically the EVB begins with the resonance states (or more precisely, diabatic states) corresponding to classical valence-bond structures. These basis states are mixed to describe the reacting system. The potential energies of the diabatic states (H11 and H22) and the mixing term (H12) are represented by the Hamiltonian matrix elements,
| (1a) |
| (1b) |
Here R and Q represent the atomic coordinates and charges, respectively, of the reactants or products (“solute”) in the diabatic states, and r and q are the coordinates and charges of the surrounding water or protein (“solvent”). is the energy of the ith diabatic state in the gas-phase, where all the fragments are taken to be at infinity; is the intramolecular potential of the solute system (relative to its minimum) in this state; represents the interaction between the solute atoms and the surrounding solvent atoms; and represents the potential energy of the solvent.
The adiabatic ground-state energy (Eg) and the corresponding eigenvector (Cg) are obtained by solving the secular equation
| (2) |
The simplicity of the EVB formulation makes it relatively straightforward to obtain analytical derivatives of the potential surface by using the Hellmann-Feynman theorem for the first derivatives of Eg, and thus to sample the EVB energy surface by molecular-dynamics (MD) simulations. This is done by a combined free energy perturbation (FEP) umbrella sampling (US) procedure that provides the free energy function (Δg‡(x)) that is needed to calculate the activation free energy (Δg‡). The FEP/US mapping procedure used to evaluate the EVB free energy surface is described elsewhere (12), and here we review only essential points for the simple case of two diabatic states. In such a case we use a mapping potential of the form,
| (3) |
where θm changes from 0 to 1 in n+1 fixed increments (θm = 0/n, 1/n, 2/n, …, n/n). The free energy ΔGm associated with changing λ from 0 to m/n can be evaluated by a free-energy perturbation (FEP) procedure. The free energy functional that corresponds to the adiabatic ground state surface, Eg, is obtained by the FEP-umbrella sampling (FEP/US) method, which can be written as,
| (4) |
In this expression, εm is the mapping potential that keeps the reaction coordinate x in the region of x′, 〈⋯〉m denotes an average over an MD trajectory on this potential, β = (kBT)−1, kB is the Boltzmann constant, and T is the temperature. If the changes in εm are sufficiently gradual, the free energy functional Δg(x′) obtained with several values of m overlap over a range of x′, and patching together the full set of Δg(x′) gives a complete free energy curve for the reaction.
The FEP-US approach also can be used to obtain the free energy functional of the individual diabatic states. For example, the free energy of the reactant state (Δg1) is,
| (5) |
In order to relate the origin of the catalytic effect to the EVB results it is convenient to approximate the activation free energy by the modified Marcus equation (12),
| (6) |
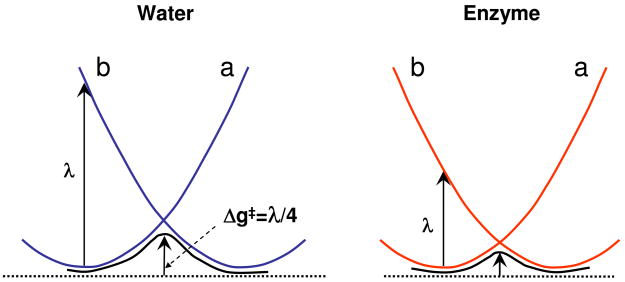
where w̄ is the so called “work term” that describes the free energy of bringing the donor and acceptor to the interaction distance (at the reactant state, λ is the reorganization energy, ΔG0 is the reaction free energy, Γ is the nuclear quantum mechanical correction and the H̄12 are the average values of H12 in the TS (x‡) and the reactant state (x0). The nature of this expression is defined schematically in Figure 4 (see also ref. (9)). The first two terms of Eq. 6 are those used in Marcus formulation for electron transfer reactions (31) and the rest of the expression represents the effect of the very strong mixing (H12) between the diabatic states.
Figure 4.
Relationship between the reaction barrier (Δg‡) and the reorganization energy (λ) in aqueous solution and in an enzyme. In aqueous solution λ is large and Δg‡ is large, whereas in enzyme environment λ and (Δg‡) are small.
The reorganization energy can also be obtained directly from the EVB diabatic free energies of Eq. 5, which follow the same trend as that shown in Figure 4. The work term w̄ is discussed in ref. (14, 32) and it is related to the potential of mean force (PMF) of bringing the donor and acceptor together. When the PMF is close to zero the work term it is similar to the cage effect discussed in many of our works (see (9)).
The reorganization energy can also be estimated by using the expression (9),
| (7) |
where Δε is the difference between εa and εb and Δε〉 designates average over trajectories on εa.
The EVB calculations were evaluated by using the MOLARIS simulation program (33, 34) using the ENZYMIX force field. The EVB activation barriers were calculated at the configurations selected by using the same free energy perturbation umbrella sampling (FEP/US) approach used in all of our EVB studies. The simulation systems were solvated by the surface constrained all atom solvent (SCAAS) model (33) using a water sphere of 18 Å radius centered on the substrate and surrounded by 3 Å grid of Langevin dipoles and then by a bulk solvent, while long-range electrostatic effects were treated by the local reaction field (LRF) method (33). The EVB region consisted of the entire substrate, a total of 24 atoms. The FEP mapping was evaluated by 21 frames of 20 ps each for moving along the reaction coordinate with our all atom surface constrained spherical model. All the simulations were done at 300 K with a time step of 1 fs. In order to obtain reliable results we repeated the simulations several times with different initial conditions (obtained from arbitrary points in the relaxation trajectory). The final results were obtained from the average of the different simulations. The different mutations were generated from the native enzymes by 100 ps relaxation runs.
The EVB approach is quite expensive (when one insists on converging results) and requires major computer time for a reasonable convergence when one deals with electrostatic effects in protein interiors. In some cases it is possible to obtain reasonable results by semimacroscopic models that focus on the TS electrostatic energy. This is true in particular with regard to the semimacroscopic version of the protein dipole Langevin dipole (PDLD/S) method in its linear response approximation (PDLD/S-LRA) version (35) that provides a direct link between the microscopic and macroscopic concepts. Since this method was reviewed extensively, we review here only its main features. The PDLD/S-LRA method evaluates the change in electrostatic free energies upon transfer of a given ligand (l) from water to the protein by using,
| (8) |
where the ΔG is the free energy of changing the ligand in the given environment (i.e., water (w) or protein (p)). Using the cycles described in ref. (35), we start with the effective PDLD potentials;
| (9) |
where ΔGsol denotes the electrostatic contribution to the solvation free energy of the indicated group in water (e.g., denotes the solvation of the protein-ligand complex in water). To be more precise, ΔGsol should be scaled by 1 / (1 − 1/εw) but this small correction is neglected here. The values of the ΔGsol’s are evaluated by the Langevin dipole solvent model. l and l′ denote the polar and nonpolar ligand respectively. is the electrostatic interaction between the charges of the ligand and the protein dipoles in vacuum (this is a standard PDLD notation) and is the intramolecular electrostatic interaction within the ligand. We also evaluate the non electrostatic contributions to the binding energy as described in ref. (35). Now the PDLD/S results obtained with a single protein-ligand configuration cannot capture properly the effect of the protein reorganization (see discussion in ref. (35)) and a more consistent treatment should involve the use of the LRA or related approaches (e.g., (35)). This approach provides a reasonable approximation for the corresponding electrostatic free energies:
| (10) |
where the effective potential Ū is defined in Eq. 9, and 〈 〉l and 〈 〉l′ designate an MD average over the coordinates of the ligand-complex in their polar and nonpolar forms respectively. It is important to realize that the average of Eq. 10 is always done where both contributions to the relevant Ūelec are evaluated at the same configurations. That is, the PDLD/S energies of the polar and nonpolar states are evaluated at each averaging step by using the same structure. However, we generate two set of structures one from MD runs on the polar state and one from MD runs on the nonpolar state. This is basically the same approach used in the microscopic LRA but now with the effective potential Ūelec.
Even the PDLD/S-LRA approach is computationally demanding and would require enormous computer time if one would likes to use this approach in the preliminary screening stages which involve exploration of many possible mutations. A promising strategy for this stage may be provided by the evaluation of the so called “electrostatic group contributions” (36, 37). These contributions are defined here as the effect of “mutating” all the residual charges of the given group to zero. In principle, we can perform such mutations and evaluate the PDLD/S-LRA binding energy for the given native and mutant. However, when we are dealing with charged and polar residues, it is reasonable to start with the faster screening approximation introduced by Muegge et al (36, 37). This approach evaluates the electrostatic group contributions to the binding energy by looking at the terms in Eq. 9, this leads to,
| (11) |
where εx is taken as εx ≈ 4 for polar residues and εx = εeff ≈ 40 for ionized residues. This approach was examined in several test cases (e.g. (37, 38)) and apparently provides a reasonable result for an initial screening.
The calculations of Eqs. 10 and 11 were done by using the MOLARIS simulation program (33, 34) The PDLD/S-LRA simulations involved the generation of 5 configurations in the charged and uncharged forms of the TS by MD runs of 10 ps with a 1 fs time step at 300 K. The calculations use the SCAAS spherical boundary condition (33) and the LRF long range treatment (33). The averages of Eq. 11 are evaluated on the 5 configurations with the charged forms.
III. Results
As stated above we took the CM system as our benchmark. More specifically we considered; a) the native EcCM and the V35I, V35A mutants that has been used recently by Mayo and coworkers (39), b) we also considered the monomer mMjCM and the F77W-mMjCM, Q88N-mMjCM, R51Q-mMjCM and D48G-mMjCM mutants that have been examined by Hilvert and coworkers (40). This system is of a particular interest since this enzyme behaves like a molten globule when it does not bind the substrate (41) and since this enzyme have a wider catalytic landscape than that of the native enzyme. We also considered c) the R11A-EcCM, R28A-EcCM and K39A-EcCM mutations of ref. (42) and d) the native Bs-CM and the R90G-BsCM and R90Cit-BsCM mutations of refs (43, 44), where the removal of an active site charge group leads to a major loss of catalytic power. The latter mutation consists of the substitution of a charged residue, Arg90, with a neutral hydrogen donor, citrulline (see ref. (44)).1 In all of these cases the observed values of kcat/Km are known and the observed values of kcat are also known except for the mutations mentioned in section c. Here we explore the performance of several screening approaches.
III.1 Qualitative approaches for fast screening
The difference between the activation barrier ΔΔg‡p that corresponds to kcat/KM (or more precisely to kcat/Kbind(RS)) and the activation barrier for the uncatalyzed reaction is related to the TS binding energy by (23),
| (12) |
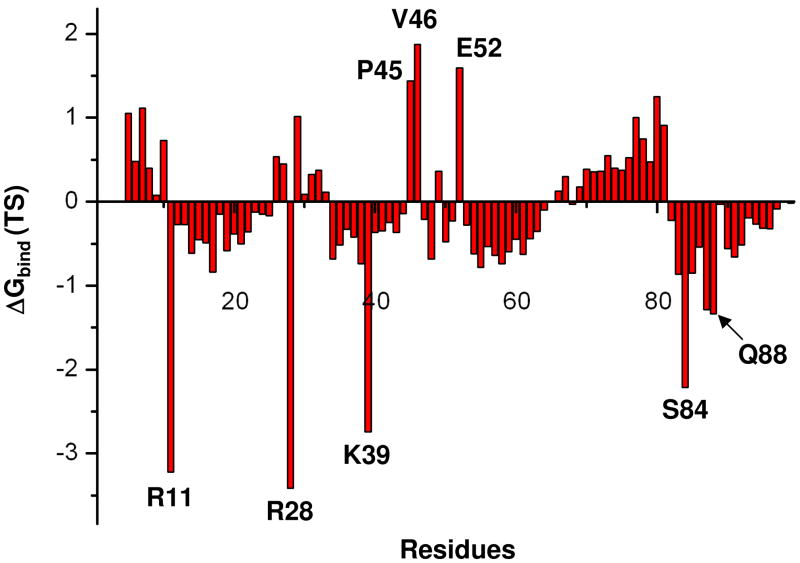
The electrostatic contributions to this binding free energy can be estimated by the PDLD/S approach and by Eq. 11. Here we started by exploring first the most qualitative approach, evaluating the group contributions to the TS binding by using Eq. 11. The corresponding results are shown if Figure 5, for the EcCM dimer. As seen from the figure, the group contributions captured the effect of the positively charged active site groups. However, for the other residues the performance is not encouraging.
Figure 5.
Electrostatic group contributions obtained by Eq. 11 for the TS binding in the native EcCM in kcal/mol. The contributions are for the residues of both subunits which are close to active site considered in our simulations.
Next we examined the performance of the PDLD/S-LRA in the evaluation of the TS binding free energy. The performance of this approach is summarized in Table 1. As seen from the table this approach is useful when we deal with strong direct electrostatic interactions (e.g. the interaction with Arg 11, Arg 28 and Lys 39 for EcCM system) but less effective when it comes to less direct effects. Overall it seems that at present both approaches can mainly be used in a screening of the effect of ionized residues and in some cases of polar residues (23).
Table 1.
The ΔΔg‡P and the electrostatic contribution, ΔΔg‡P,elec (or ), obtained by the PDLD/S-LRA calculations at different dielectric constants (εp=4 and 6) and the corresponding observed values.a
| (ΔΔg‡P,elec)εp=4 | (ΔΔg‡P)εp=4 | (ΔΔg‡P,elec)εp=6 | (ΔΔg‡P)εp=6 | (ΔΔg‡P)obs | |
|---|---|---|---|---|---|
| EcCM | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| V35I-EcCM | 1.8 | 0.4 | 1.2 | −0.2 | −0.1 |
| V35A-EcCM | 0.0 | 0.2 | 0.0 | 0.2 | 0.8 |
| R11A-EcCM | 9.5 | 8.6 | 6.5 | 5.6 | 3.7 |
| R28A-EcCM | 12.7 | 11.9 | 8.6 | 7.8 | 3.9 |
| K39A-EcCM | 11.0 | 9.6 | 7.5 | 6.0 | 6.1 |
| mMjCM | −1.4 | 0.4 | −0.9 | 0.0 | 1.2 |
| F77W-mMjCM | −3.2 | −1.6 | −2.1 | −0.6 | 2.1 |
| Q88N-mMjCM | −0.4 | −0.9 | −0.3 | −0.8 | 6.0 |
| R51Q-mMjCM | 3.1 | 2.9 | 2.1 | 1.9 | 2.5 |
| D48G-mMjCM | 5.6 | 4.0 | 3.7 | 2.2 | 4.4 |
| BsCM | −3.5 | −3.5 | −2.4 | −2.3 | −1.0 |
| R90Cit-BsCM* | 3.2 | 3.1 | 2.3 | 2.6 | 5.6 |
| R90G-BsCM | 6.9 | 6.1 | 4.7 | 4.9 | 6.6 |
Energies in kcal/mol. The observed value of ΔΔg‡p is given by ΔΔg‡p= −RT*ln((kcat/Km)mut/(kcat/Km)nat). The value of ΔΔg‡p for mMjCM and BsCM was obtained relative to that of EcCM. The calculated values are obtained directly from the TS binding energy through Eq. 12, where the TS binding energy is evaluated by the PDLD/S-LRA approach using Eq. 10. The observed value for kcat/Km for R90Cit-BsCM* was taken from the semisynthetic B. subtilis chorismate mutase containing the D102E mutation and control experiments confirmed that this mutation does not significantly alter the catalytic properties of the enzyme (44).
III.2 Accurate final screening
In order to move to a more accurate screening we had to move to the EVB approach. This approach can evaluate Δg‡cat (that corresponds to kcat) rather than the Δg‡p considered in the previous section. Here we explored the catalytic power of the trimer (BsCM), dimer (EcCM) and monomer (mMjCM) by calculating the EVB surfaces for different mutants and native proteins considered. The results are shown in Table 2. In each of the systems considered we started by generating the given sequence from the native protein proceeded to 100 ps MD simulations in order to relax the given structures. Five structures were saved during each relaxation process and then used to generate the EVB surface and obtain the activation free energies and reorganization energies.
Table 2.
Calculated and observed Δg‡cat(ΔΔg‡cat).a
| Δg‡cat(ΔΔg‡cat)calc | Δg‡cat(ΔΔg‡cat)obs | |
|---|---|---|
| EcCM | 15.3(0.0) | 15.3(0.0) |
| V35I-EcCM | 13.3(−2.0) | 15.0(−0.3) |
| V35A-EcCM | 15.2(−0.1) | 15.7(0.4) |
| mMjCM | 16.2(0.9) | 16.8(1.5) |
| F77W-mMjCM | 17.7(2.4) | 17.6(2.3) |
| Q88N-mMjCM | 20.5(5.2) | 20.2(4.9) |
| D48G-mMjCM | 20.1(4.8) | 18.3(3.0) |
| BsCM | 16.6(1.3) | 15.3(0.0) |
| R90Cit-BsCM | 23.7(8.4) | 21.1(5.8) |
| R90G-BsCM | 23.8(8.5) | 22.5(7.2) |
Energies in kcal/mol. The values between brackets designate the ΔΔg‡cat relative to EcCM. The RMSD of the calculated values are 1.5, 0.6, 0.9, 1.7, 1.1, 1.5, 1.4, 1.6, 2.5 and 2.2 kcal/mol for the systems studied, according to the order in the table.
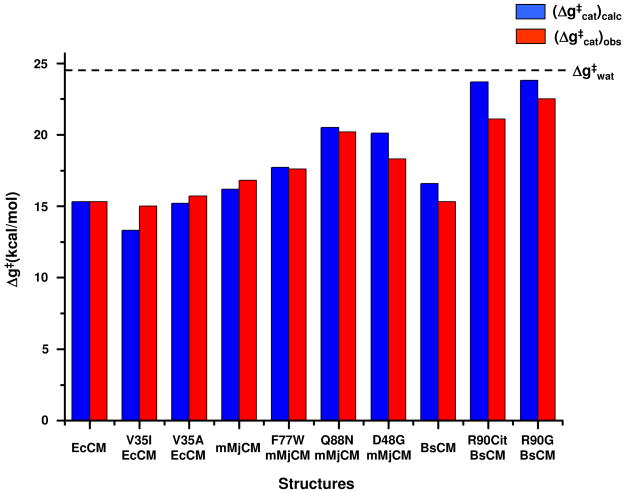
The calculated activation barriers of the different mutants are compared to the corresponding observed values in Table 2 and Figure 6. As seen from the table the agreement between the calculated and observed results is excellent and can be considered to be a quantitative agreement. It should be noted in this respect that the performance of our calculations may not be fully appreciated by those who note the small deviations between the calculated an observed in Figure 6 while overlooking the fact that we actually reproduced quantitatively the absolute values of the activation free energies without any parameterization on the reaction in the enzyme. Obtaining such a quantitative prediction is very encouraging and cannot be expected from current design approaches that use gas phase calculations.
Figure 6.
Comparing the calculated (in blue) and observed (in red) activation energies for the indicated systems. The dash line designates the activation free energy of the reaction in aqueous solution.
Thus the EVB approach can be used in the final screening stage of CAED approaches and perhaps can be defined as the “gold standard” of our approaches. However, this approach is quite expensive (see below) and it is important to explore less quantitative approaches. Since the activation barrier is correlated with the reorganization energy (see Figure 4 and ref 9), we started the search for a faster screening approach by examining the relationship between the reorganization energy obtained from the full EVB free energy functional, λ1 (see Figure 4) and from the LRA approach Eq.7, λ2. The results are shown in Table 3 and in both cases we have significantly less good agreement between the calculated and observed results than in Table 2. The situation can be improved in screening other enzymes, since in the case of CM the reorganization energy involves a large intermolecular contribution, which requires longer simulations for full convergence. Also in the case of CM we have a very large charge-charge interaction in the solute system and this makes the outer sphere reorganization energy a less stable quantity.
III.3 Efficient Prescreening
The approaches examined above used the native structure as the starting point for the screening process. Obviously, a more general treatment should start from configurations where at lease the initial orientation of the active site side chains is randomized. Although we have not focused on this issue in the present work we have developed a potentially very effective approach for this stage of the screening. That is, our approach of using a simplified folding model as a reference potential for explicit folding calculations (24, 45), which can be called Folding with Coarse Grained Reference (CGR) simulations, has been used recently in exploring the catalytic landscape of CM (30). This approach or its variants can be used in prescreening. For example, we can start by taking the structure of the native main chain and sample the positions of the simplified side chains with an explicit model of the substrate (see ref. (30)). Next one can convert the simplified model to a “simplified explicit model” where the solvent is treated implicitly, and use a Monte Carlo (MC) procedure to generate the explicit side chains. The difference between the explicit and the simplified potential can be used in the MC procedure. The simplified model can also be used for fast screening of the optimal residues and thus to suggest which residues will be included in the explicit treatment. Furthermore, we can also generate optimized simplified sequences and structures and then monitor the energy difference between the simplified and explicit model for the TS binding of different mutants. Those residues that would give lowest energy can then be screened by the EVB approach. In fact we can use this MC procedure for calculating the TS binding free energy.
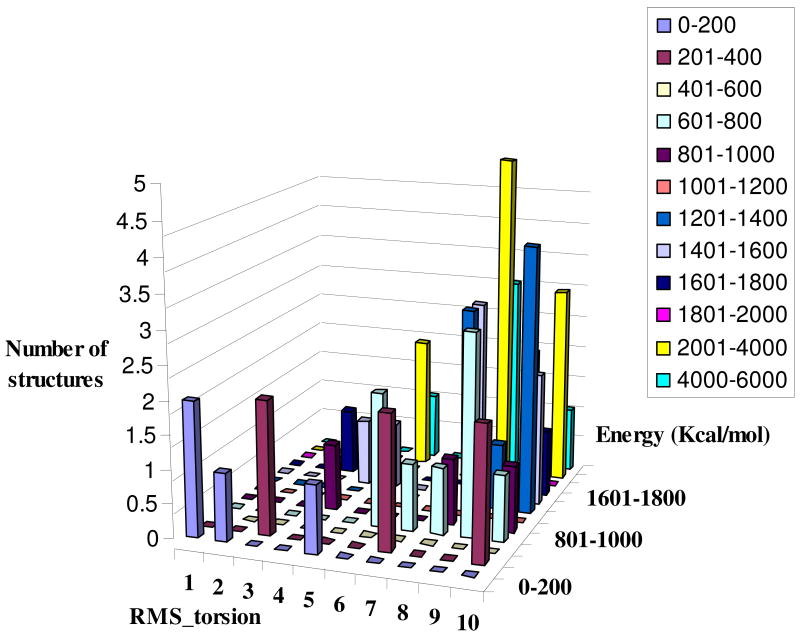
In the present work we decided to leave the exploration of the full CGR approach to a subsequent study and to use a simpler related treatment, where the main chain was fixed and the configurations of several side chains were sampled by using first a rotamer library and then a MC optimization in the torsional space of the side chains of the simplified explicit model The lowest energy configurations found by the above treatment were then used as starting points in EVB calculations (with the explicitly solvated full microscopic model) by taking both the mutant Q88N-mMjCM and the native enzyme and trying to reproduce their observed activation energies. This was done while starting from randomized arrangements of the configurations of the mutated residue and several other residues. The point in this validation study is that we can view the native enzyme and the mutant as the targets in a design experiment (“pretending” that we do not know the actual structure). At any rate, our study considered different configurations of the mutated residue and neighboring residues while keeping the main chain fixed and generating optimized structures with low energy. The performance of this approach, for the case where we explore the configurational space of residues 84, 88 and 91, is considered in Figure 7 for the Q88N mutant. This is done in terms of the rapidly obtained energies of the simplified explicit model rather that in terms of the EVB free energy, which will be considered below. The figure indicates that the structures with a low RMS from the “correct” structure (the one generated by a single mutation of the native structure) have low energy (a similar pattern was obtained for the native enzyme). Thus the low energy structures correspond frequently to structures that are close to the actual active site structure and the use of the rapidly obtained low energy structures as starting point for the extensive EVB simulations may provide powerful screening approach.
Figure 7.
Energy versus RMS from the correct structure for a case where we consider the rotamer library for the residues Asn84, Gln88 and Tyr91. Each number in RMS_torsion axis represents a 15 degrees range of torsional RMS. For example: 1 and 2 and 3 correspond to RMS of (1–15) and (16–30) and (31–45), respectively. The energies reported correspond to the explicit model with implicit solvent rather that to the EVB free energies. The point is that the configurations with low energy are not far from the correct active site structure and thus can be used as reasonable starting points for the EVB calculations.
The above procedure was examined first in the case of the native enzyme, where we explored the configurational space of residues 84, 88 and 91 and took the four lowest energy configurations and used them in EVB studies. It was found that starting with two of the low energy configurations reproduces the observed activation barrier (calculated barrier of about 15 kcal/mol).
The situation became more complicated with the Q88N mutant. That is, using the above procedure while randomizing only the structure of residue 88 and then picking the lowest energy configuration, gave EVB barriers of about 20 kcal/mol (close to the corresponding observed barrier). Similar result was obtained for mutant structures that were generated from the native enzyme. However, taking the mutant structures generated by using the rotamer library of three residues gave EVB barriers that were significantly lower than the observed barrier. Exploring the origin of this effect, by calculating the binding energy of the substrate, indicated that the system was locked in an unstable ground state that led to a reduction in Δg‡cat, while still having reasonable Δg‡p (similar problem occurs in our study of DNA polymerase β (46)). In order to deal with this problem we used an alternative approach where we calculated the binding free energy of the EVB transition state. These calculations were performed by building a TS from the combination of the two EVB states with a mapping parameter (the θm of Eq. 3) that corresponds to the TS (see (15) for a related approach). The charging free energy of the TS was then calculated by a FEP charging approach (for the water and enzyme systems) and taken as the approximated TS binding free energy. The non-electrostatic contribution was assumed to be similar for the native and mutant enzymes and it was also assumed that the solvation free energy of the free enzyme is similar for the native and mutant enzymes. Furthermore, estimating the non-electrostatic contribution by the LIE approximation (47) gave similar contributions for the native and mutant enzyme. We also imposed a weak constraint on the protein to prevent a major rearrangement at the state where the substrate is fully uncharged (in this state the repulsion between the protein positively charged groups leads to major reorganization in the absence of the substrate negative charges). This constraint is justified since the rigorous final state should be the state where the substrate is replaced by water. At any rate, now the TS binding energies of the native and mutant enzyme appeared to follow now the observed trend. That is, the TS of the mutant generated by using the rotamer library of three residues had about 2 kcal/mol less negative binding energy than the mutant generated starting from the native structure and 4 kcal/mol less negative than the binding energy of the native enzyme. This result is in a reasonable agreement with the observed difference in Δg‡p although the error range of these calculations is larger than in the EVB calculations. Nevertheless in cases where the active site is constructed by extensive conformational search it may be beneficial to evaluate the TS binding energy rather that to calculate Δg‡cat. Here we believe that the above FEP charging approach can be very effective. However, more studies along this line are clearly needed before selecting the most effective screening strategy.
III. 4 Performance assessment
In order to assess the effectiveness of a given CAED approach it is important to have a clear idea on the computational resources needed to obtain the given result. Here we report the computer time needed for each of the approaches considered above, or in other words the price per performance ratio. The relevant estimates are summarized in Table 4. As seen from the table we can screen 28 mutants by the EVB approach, while using 200 processors. Of course, it is possible to use much more massive power and to screen more mutants by the EVB approach. We can also screen 1000 mutants on 200 processors by using the PDLD/S-LRA approach but these calculations are significantly less accurate than the EVB calculations. Thus although the EVB screening may look like a major investment in computational resources it might provide at present the most practical way of getting accurate screening. In other words, while the PDLD/S-LRA may help in the preliminary screening stage, particularly for charged mutants, its relatively small price does not solve the fact that the results are not sufficiently accurate. Here the fact that the price of computer power is rapidly declining suggests that, at least in the final screening stage, we have to use the EVB approach.
Table 4.
The performance times of the different models.a
| Computation time per mutant (runs, processor) | Number of mutants per 24 hours/per 200 processors | Number of mutants per 24 hours/per 1000 processors | |
|---|---|---|---|
| Δg ‡cat using EVBb | 7.5 hours (5, 21) | 28 | 142 |
| λ1 based on mapping energy profilesb | 7.5 hours (5, 21) | 28 | 142 |
| λ 2 based on LRA calulationsb | 1.5 hours (5, 10) | 280 | 1,600 |
| Δg ‡p using PDLD/S- LRAc | 4.8 hours (1,1) | 1,000 | 5,000 |
| Group Contributionc, d | 1 hour (1,1)/100 | 480,000 | 2,500,000 |
The calculations are done on the USC HPCC (High Performance Computing and Communication) Linux computer, using the Dual Intel P4 3.0 GHz 2GB Memory nodes.
Five runs per mutant.
These approximations are effective mainly for charge residues and to lesser extent for polar residues.
In each run we can calculate mutations of all the protein residues to fully neutral analogues.
IV. Conclusions
The ability to design effective enzymes can be considered as the Holy Grail of biotechnology and for some as the ultimate proof of understanding of enzyme catalysis. As much as computer aided enzyme design is concerned the challenge is not different than that addressed in our early 1986 study of computer aided mutations (10). In fact the simulation method has not changed (the same EVB) but the available computer power increases by four orders of magnitude.
At present the EVB is probably the most effective tool for quantitative screening 28 active site mutants in 24 hours with 100 nodes. Focusing on a few types of typical mutants can further optimize this procedure. Furthermore, a qualitative PDLD/S-LRA screening of 1000 mutations with significant electrostatic effect can be accomplished in 24 hours with 100 nodes and 500,000 can be screened by the same computer power using the group contribution approach.
One of the obvious questions about the performance of our method is the difference between our EVB calculations and the approaches used in other enzyme design studies. The difference is that current enzyme design approaches focus on effective an elegant initial screen but do not use methods that can provide a quantitative scoring. For example, refs. (6, 7) used gas phase calculations to estimated the TS structure and charges and then generate in a rapid screening approach protein configurations that accommodated this TS. Although this approach provided impressive results, it cannot provide quantitative information about the relative energy of different design options. The best way to realize this point is to try to see whether the method used can reproduce the absolute activation free energies, and in our experience it will result with very large errors. Furthermore, the use of gas phase QM calculations for enzyme studies is notoriously problematic (see discussion in e.g. (48)). In other words, an approach that does not treat the protein TS electrostatic free energy in a consistent way cannot capture the preorganization effect and the corresponding catalytic effect. Here our strategy is based on the philosophy that a computational approach for studies of enzymes must be able to reproduce the free energy of the TS in the enzyme active site and that approaches that ignore this requirement will be unable to really reach highly catalytic enzymes unless this is done by experimental trial and error.
It might be instructive to respond to a referee question about the reason for the quantitative performance of the EVB with the ENZYMIX force field as compare to that of some other computer programs. Here we must start by stating that very few research groups specialized in the extensive averaging approach used here and with the use of powerful long-range treatment and reliable polarization boundary conditions. Also most QM/M approaches do not involve the EVB model. Furthermore, the EVB force field has been calibrated on observed solvation free energies rather than on less relevant properties, and has been validated repeatedly on highly relevant properties such as pKas (see a review in (49)). Overall we believe that the accuracy of MOLARIS in reproducing reorganization free energy is a well-established fact (e.g. see recent study ketosteroid isomerase (50) and B12 enzymes (51)) as well as the results of the present study. Nevertheless, other modern simulation programs with properly parameterized force fields would probably give similar results once they implement the full EVB treatment and repeat the procedure used here.
This study has not focused on the early screening steps but, nevertheless, we did consider some key elements that can be used in the early screening stage. For example, we can benefit from the use of the PDLD/S-LRA or the group contribution approach in identifying residues that can contribute to electrostatic stabilization of the TS. Furthermore, in subsequent study we intend to exploit the power of the simplified model and the coarse-grained-reference approach in the early stages of the screening process. This approach can be quite effective in minimizing the energy in the sequence and configurational space.
Imposing relevant constraints in addition to the results of the actual final screening can help the general design significantly. For example, it is useful to have an estimate of the protein stability in the proposed design. Here we consider our recent electrostatic approach (52) as an extremely powerful approach. In fact, the major elements of this approach are already incorporated in the simplified model. Of course, one can use bioinformatics information on similar active sites (53, 54) but this would not solve the problem of designing a new activity.
This study has not explored our ability to design an enzyme in a blind way but in a posteriori way, focusing on the last steps of the screening process. Thus one may argue that our approach is not necessarily a predictive approach, as the results were known before the calculations. While we agree with this assessment, as much as the full design is concerned, we like to point out that as much as the final screening is concerned, the MOLARIS-EVB calculations can be considered as a relatively robust black box which contains no prior information about the enzyme catalytic power. Since this program can be used by anyone it is easy to verify that the reported results do not reflect our bias and that they are fully reproducible Thus we contend that if the program continues to produce similar agreement between calculated and observed mutational effects when applied to other enzymes, it may be justified to consider it as a predictive tool. We are fully aware of the philosophy that a prediction must be something that is done before the results are known, and we have our share of correct predictions (e.g. the primary events in vision (55) and photosynthesis (56) and even predictions of mutational effects (see discussion of Fig 4 in (15)). However, we believe that a systematic validation of enough test cases is, in principle, equivalent to an assessment of a predictive power.
We would also like to clarify that the EVB has been found to be effective only when we are dealing with a reasonable active site constructs and that generating a reasonable active site structure is a challenge that was only addressed here in a preliminary way and where we clearly cannot claim that we have a predictive power.
Recent years witnessed an impressive advances in enzyme design (e.g. (6, 7)). However, the enzymes generated by current design efforts are still far less efficient that naturally evolved enzymes. Although we are yet to see further advances, it seems to us that a part of the problem in previous CAED has been the limited focus on modeling of the actual chemical step in the actual enzyme active site, and in fact using approaches that cannot capture the critical electrostatic preorganization effect. That is, the main problem in designing enzymes with native activity is related to the ability to predict the proper TS stabilization. This difficulty is due to a large part to the difficulty of predicting the preorganization effect. Attempts to evaluate the catalytic effect by using gas phase models or by looking at the electrostatic interaction between different residues and the TS are unlikely to reproduce the correct catalytic effect since (among other problems considered elsewhere (57)) it is impossible to assess the preorganization effect without considering the protein reorganization in the simulations. Here we overcome this challenge by actually calculating the activation barrier by the EVB approach. Thus the present work is not so much about effective early screening but mainly about the requirements of the final stage in the screening process. In this respect we believe that the EVB provides a very effective way for performing the final stage in the screening process. We also provide some promising directions for the initial screening steps.
Finally, it is useful to clarify that our approach can be used to augment the initial screening steps of current CAED approaches. Furthermore, while it is hard at present to compete with the enormous power of directed evolution, the EVB can clearly help in understanding why the evolved enzymes are not perfect and in offering clues on how they can be improved.
Table 3.
Calculated reorganization energies and the corresponding observed ΔΔg‡cat.a
| ΔΔλ1 | ΔΔλ2 | (ΔΔg‡cat)obs | |
|---|---|---|---|
| EcCM | 0.0 | 0.0 | 0.0 |
| V35I-EcCM | −0.9 | 2.3 | −0.3 |
| V35A-EcCM | −1.2 | −0.1 | 0.4 |
| mMjCM | 2.9 | 2.1 | 1.5 |
| F77W-mMjCM | 2.5 | 4.2 | 2.3 |
| Q88N-mMjCM | 5.9 | 4.6 | 4.9 |
| D48G-mMjCM | 4.1 | 4.8 | 3.0 |
| BsCM | 0.0 | 0.0 | 0.0 |
| R90Cit-BsCM | 3.3 | 3.1 | 5.8 |
| R90G-BsCM | 2.4 | 2.9 | 7.2 |
Energies in kcal/mol. The reorganization energy λ1 was calculated from EVB mapping energy profiles shifting the parabolas so that ΔG0=0 and getting λ1 from the intersection using 4Δg‡=λ (ΔG0=0). The reorganization energy λ2 was calculated based on linear response estimate for EVB states: λ2=0.5*(〈Δε〉b−〈Δε 〉a). The reorganization energy of the native BsCM is taken as zero rather than relative to EcCM since with the very large λs values the error range in the absolute value can be significant.
Acknowledgments
We gratefully acknowledge the University of Southern California’s High Performance Computing and Communication Center (HPCC) for computer time. We would like to thank Professor Don Hilvert for insightful discussions and for letting us have his results prior to publication. M.R. thanks the Generalitat Valenciana from Spain for the postdoctoral fellowship.
Abbreviations
- EVB
empirical valence bond
- CAED
computer aided enzyme design
- FEP/US
free energy perturbation/umbrella sampling
- CM
chorismate mutase
- PDLD/s-LRA
protein dipole Langevin dipole/semimacroscopic with the linear response approximation
Footnotes
This work was supported by Grant GM024492 from the National Institutes of Health (NIH).
The R90Cit mutation was performed on the native BsCM rather than with BsCM* (44) that contains a D102E mutation because control experiments with this mutation confirms that the mutation does not alter significantly the catalytic power of BsCM.
References
- 1.Toscano MD, Woycechowsky KJ, Hilvert D. Minimalist active-site redesign: Teaching old enzymes new tricks. Angew Chem Int Ed. 2007;46:4468–4470. doi: 10.1002/anie.200604205. [DOI] [PubMed] [Google Scholar]
- 2.Seeling B, Szostak JW. Selection and evolution of enzyme from a partially randomized non-catalytic scaffold. Nature. 2007;448:828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varadarajan N, Gam J, Olsen MJ, Georgiou G, Iverson B. Proc Natl Acad Sci USA. 2005;102:6855–6860. doi: 10.1073/pnas.0500063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc Natl Acad Sci USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang J, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DSDB. Kemp Elimination Catalysts by Computational Enzyme Design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 7.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanka F, Barbas CFI, Hilvert D, Houk KN, Stoddard B, Baker D. De Novo Computational Design of Retro-Aldol Enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippow SM, Tidor B. Progress in computational protein design. Curr Opin Biotechnol. 2007;18:305–311. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warshel A, Sharma PK, Kato M, Xiang Y, Liu HB, Olsson MHM. Electrostatic basis for enzyme catalysis. Chem Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 10.Warshel A, Sussman F. Toward computer-aided site-directed mutagenesis of enzymes. Proc Natl Acad Sci USA. 1986;83:3806. doi: 10.1073/pnas.83.11.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao SN, Singh UC, Bash PA, Kollman PA. Free Energy perturbation calculations on binding and catalysis after mutating Asn 155 in subtilisin. Nature. 1987;328:551–554. doi: 10.1038/328551a0. [DOI] [PubMed] [Google Scholar]
- 12.Warshel A. Computer Modeling of Chemical Reactions in Enzymes and Solutions. Wiley Interscience; New York: 1991. [Google Scholar]
- 13.Aqvist J, Warshel A. Simulation of Enzyme-Reactions Using Valence-Bond Force-Fields and Other Hybrid Quantum-Classical Approaches. Chem Rev. 1993;93:2523–2544. [Google Scholar]
- 14.Liu H, Warshel A. The Catalytic Effect od Dihydrofolate Reductase and Its Mutants Is Determined by Reorganization Energies. Biochemistry. 2007;46:6011–6025. doi: 10.1021/bi700201w. [DOI] [PubMed] [Google Scholar]
- 15.Hwang JK, Warshel A. Semiquantitative calculations of catalytic free energies in genetically modified enzymes. Biochemistry. 1987;26:2669–2673. doi: 10.1021/bi00384a003. [DOI] [PubMed] [Google Scholar]
- 16.Hansson T, Nordlund P, Aqvist J. Energetics of Nucleophile Activation in a Protein Tyrosine Phosphatase. J Mol Biol. 1997;265:118–127. doi: 10.1006/jmbi.1996.0716. [DOI] [PubMed] [Google Scholar]
- 17.Mulholland AJ. Computational enzymology: modelling the mechanisms of biological catalysts. Biochem Soc Trans. 2008;36:22–26. doi: 10.1042/BST0360022. [DOI] [PubMed] [Google Scholar]
- 18.Marti S, Andres J, Moliner V, Silla E, Tunon I, Bertran J. Predicting an Improvement of Secondary Catalytic Activity of Promiscuos Isochorismate Pyruvate Lyase by Computational Design. J Am Chem Soc. 2008;130:2894–2895. doi: 10.1021/ja078334c. [DOI] [PubMed] [Google Scholar]
- 19.Marti S, Andres J, Moliner V, Silla E, Tunon I, Bertran J. Computer-Aided Rational Design of Catalytic Antibodies: The 1F7 Case. Angew Chem Int Ed. 2007;46:286–290. doi: 10.1002/anie.200603293. [DOI] [PubMed] [Google Scholar]
- 20.Ferrer S, Tunon I, Moliner V, Williams IH. Theoretical site-directed mutagenesis: Asp168Ala mutant of lactate dehydrogenase. J R Soc Interface. 2008;5:217–224. doi: 10.1098/rsif.2008.0211.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grigorenko BL, Nemukhin AV, Topol IA, Cachau RE, Burt SK. QM/MM Modeling the Ras-GAP Catalyzed Hydrolysis of Guanosine Triphosphate. Proteins. 2005;60:495–503. doi: 10.1002/prot.20472. [DOI] [PubMed] [Google Scholar]
- 22.Gherman BF, Goldberg SD, Cornish VW, Friesner RA. Mixed Qunatum Mechanical/Molecular Mechanical (QM/MM) Study of the Deacylation Reaction in a Penicillin Binding Protein (PBP) versus in a Class C β– Lactamase. J Am Chem Soc. 2004;126:7652–7664. doi: 10.1021/ja036879a. [DOI] [PubMed] [Google Scholar]
- 23.Ishikita H, Warshel A. Predicting drug-resistant mutations of HIV protease. Angew Chem Int Ed. 2008;47:697–700. doi: 10.1002/anie.200704178. [DOI] [PubMed] [Google Scholar]
- 24.Roca M, Liu H, Messer B, Warshel A. On the relationship between thermal stability and catalytic power of enzymes. Biochemistry. 2007;46:15076–15088. doi: 10.1021/bi701732a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chook YM, Gray JV, Ke H, Lipscomb WN. The monofunctional chorismate mutase from Bacillus subtilis. Structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction. J Mol Biol. 1994;240:476–500. doi: 10.1006/jmbi.1994.1462. [DOI] [PubMed] [Google Scholar]
- 26.Lee AY, Karplus PA, Ganem B, Clardy J. Atomic-Structure of the Buried Catalytic Pocket of Escherichia-Coli Chorismate Mutase. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 27.Vamvaca K, Vögeli B, Kast P, Pervushin K, Hilvert D. An enzymatic molten globule: Efficient coupling of folding and catalysis. Proc Natl Acad Sci USA. 2004;101:12860–12864. doi: 10.1073/pnas.0404109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbany M, Gutierrez-de-Teran H, Sanz F, Villa J, Warshel A. On the Generation of Catalytic Antibodies by Transition State Analogues. ChemBioChem. 2003;4:277–285. doi: 10.1002/cbic.200390048. [DOI] [PubMed] [Google Scholar]
- 29.Strajbl M, Shurki A, Kato M, Warshel A. Apparent NAC effect in chorismate mutase reflects electrostatic transition state stabilization. J Am Chem Soc. 2003;125:10228–10237. doi: 10.1021/ja0356481. [DOI] [PubMed] [Google Scholar]
- 30.Roca M, Messer B, Hilvert D, Warshel A. On the relationship between folding and chemical landscapes in enzyme catalysis. Proc Natl Acad Sci USA. 2008;105:13877–13882. doi: 10.1073/pnas.0803405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcus RA. Electron-Transfer Reactions In Chemistry - Theory And Experiment (Nobel Lecture) Angew Chem Int Ed. 1993;32:1111–1121. [Google Scholar]
- 32.Liu H, Warshel A. Origin of the Temperature Dependence of Isotope Effects in Enzymatic Reactions: The Case of Dihydrofolate Reductase. J Phys Chem B. 2007;111:7852–7861. doi: 10.1021/jp070938f. [DOI] [PubMed] [Google Scholar]
- 33.Lee FS, Chu ZT, Warshel A. Microscopic and Semimicroscopic Calculations of Electrostatic Energies in Proteins by the Polaris and Enzymix Programs. J Comput Chem. 1993;14:161–185. [Google Scholar]
- 34.Chu ZT, Villa J, Strajbl M, Schutz CN, Shurki A, Warshel A. MOLARIS version α9.06. University of Southern California; Los Angeles: 2006. [Google Scholar]
- 35.Sham YY, Chu ZT, Tao H, Warshel A. Examining methods for calculations of binding free energies: LRA, LIE, PDLD-LRA, and PDLD/S-LRA calculations of ligands binding to an HIV protease. Proteins. 2000;39:393–407. [PubMed] [Google Scholar]
- 36.Muegge I, Tao H, Warshel A. A fast estimate of electrostatic group contributions to the free energy of protein-inhibitor binding. Prot Eng. 1997;10:1363–1372. doi: 10.1093/protein/10.12.1363. [DOI] [PubMed] [Google Scholar]
- 37.Muegge I, Schweins T, Warshel A. Electrostatic Contributions to Protein-Protein Binding Affinities: Application to Rap/Raf Interaction. Proteins. 1998;30:407–423. doi: 10.1002/(sici)1097-0134(19980301)30:4<407::aid-prot8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 38.Schutz CN, Warshel A. What are the dielectric constants of proteins and how to validate electrostatic models? Proteins. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 39.Lassila JK, Keeffe JR, Kast P, Mayo SL. Exhaustive Mutagenesis of Six Secondary Active-Site Residues in Escherichia coli Chorismate Mutase Shows the Importance of Hydrophobic Side Chains and a Helix N-Capping Position for Stability and Catalysis. Biochemistry. 2007;46:6883–6891. doi: 10.1021/bi700215x. [DOI] [PubMed] [Google Scholar]
- 40.Woycechowsky KJ, Choutko A, Vamvaca K, Hilvert D. Relative Tolerance of an Enzymatic Molten Globule and Its Thermostable Counterpart to Point Mutation. Biochemistry. 2008;47:13489–13496. doi: 10.1021/bi801108a. [DOI] [PubMed] [Google Scholar]
- 41.Pervushin K, Vamvaca K, Vogeli B, Hilvert D. Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol. 2007;14:1202–1206. doi: 10.1038/nsmb1325. [DOI] [PubMed] [Google Scholar]
- 42.Liu DR, Cload ST, Pastor RM, Schultz PG. Analysis of Active Site Residues in Escherichia Coli Chorismate Mutase by Site-Directed Mutagenesis. J Am Chem Soc. 1996;118:1789–1790. [Google Scholar]
- 43.Kast P, Grisostomi C, Chen IA, Li S, Krengel U, Xue Y, Hilvert D. A Strategically Positioned Cation Is Crucial for Efficient Catalysis by Chorismate Mutase. J Biol Chem. 2000;275:36832–36838. doi: 10.1074/jbc.M006351200. [DOI] [PubMed] [Google Scholar]
- 44.Kienhofer A, Kast P, Hilvert D. Selective Stabilization of the Chorismate Mutase Transition State by a Positively Charged Hydrogen Bond Donor. J Am Chem Soc. 2003;125:3206–3207. doi: 10.1021/ja0341992. [DOI] [PubMed] [Google Scholar]
- 45.Fan ZZ, Hwang JK, Warshel A. Using simplified protein representation as a reference potential for all-atom calculations of folding free energy. Theor Chem Acc. 1999;103:77–80. [Google Scholar]
- 46.Xiang Y, Oelschlaeger P, Florian J, Goodman MF, Warshel A. Simulating the effect of DNA polymerase mutations on transition-state energetics and fidelity: Evaluating amino acid group contribution and allosteric coupling for ionized residues in human pol beta. Biochemistry. 2006;45:7036–7048. doi: 10.1021/bi060147o. [DOI] [PubMed] [Google Scholar]
- 47.Aqvist J, Hansson T. Analysis of Electrostatic Potential Truncation Schemes in Simulations of Polar Solvents. J Phys Chem B. 1998;102:3837–3840. [Google Scholar]
- 48.Karmelin SCL, Haranczyk M, Warshel A. Progress in Ab Initio QM/MM Free-Energy Simulations of Electrostatic Energies in Proteins: Accelarated QM/MM Studies of pk, Redox Reactions and Solvation Free Energies. J Phys Chem B ASAP. 2009 doi: 10.1021/jp8071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochim Biophys Acta, Proteins Proteomics. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 50.Warshel A, Sharma PK, Chu ZT, Aqvist J. Electrostatic Contributions to Binding of Transition State Analogues ca be very different from the corresponding Contributions to Catalysis: Phenolates Binding to the Oxyanion Hole of Ketosteroid Isomerase. Biochemistry. 2007;46:1466–1476. doi: 10.1021/bi061752u. [DOI] [PubMed] [Google Scholar]
- 51.Sharma PK, Chu ZT, Olsson MHM, Warshel A. A new paradigm for electrostatic catalysis of radical reactions in vitamin B12 enzymes. Proc Natl Acad Sci USA. 2007;104:9661–9666. doi: 10.1073/pnas.0702238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roca M, Messer B, Warshel A. Electrostatic contributions to protein stability and folding energy. FEBS Letters. 2007;581:2065–2071. doi: 10.1016/j.febslet.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 53.Poole AM, Ranganathan R. Knowledge-based potentials in protein design. Curr Opin Struct Biol. 2006;16:508–513. doi: 10.1016/j.sbi.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Pegg SCH, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging Enzyme Structure-Function Relationships for Functional Interference and Experimental Design: The Structure-Function Linkage Database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 55.Warshel A. Bicycle-pedal Model for the First Step in Vision Process. Nature. 1976 doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- 56.Creighton S, Hwang JK, Warshel A, Parson WW, Norris J. Simulating the dynamics of the primary charge separation process in bacterial photosynthesis. Biochemistry. 1988;27:774–781. [Google Scholar]
- 57.Shurki A, Warshel A. Structure/Function Correlations of Proteins using MM, QM/MM, and Related Approaches: Methods, Concepts, Pitfalls, and Current Progress. Adv Prot Chem. 2003;66:249–313. doi: 10.1016/s0065-3233(03)66007-9. [DOI] [PubMed] [Google Scholar]