Abstract
Mantle cell lymphoma (MCL) is a B-cell lymphoma characterized by overexpression of cyclin D1 due to the t(11;14) chromosomal translocation. While expression of cyclin D1 is correlates with MCL development, expression of wild type cyclin D1 transgene in murine lymphocytes is unable to drive B-cell lymphoma. Because cyclin D1 mutants that are refractory to nuclear export display heighten oncogenicity in vitro compared with wild type D1, we generated mice expressing FLAG-D1/T286A, a constitutively nuclear mutant, under the control of the immunoglobulin enhancer, Eµ. D1/T286A transgenic mice universally develop a mature B-cell lymphoma. Expression of D1/T286A in B lymphocytes results in promiscuous S-phase entry and increased apoptosis in spleens of young pre-malignant mice. Lymphoma onset correlates with loss of p53 suggesting that inactivation of the p53 signaling axis precedes lymphoma development. Our results describe a cyclin D1-driven model of B-cell lymphomagenesis and provide evidence that nuclear-retention of cyclin D1 is oncogenic in vivo.
Keywords: CDK4, cyclin D1, mantle cell lymphoma, apoptosis
Introduction
Cyclin D1 functions as a critical mitogenic sensor that serves to integrate growth factor signals with cell cycle progression. Growth factor stimulation triggers an increase in cyclin D1 transcription and translation as well as its assembly into an active cyclin D1/CDK4 complex. The active kinase triggers phosphorylation of the retinoblastoma protein (RB) relieving its transcriptional repressive activities and its capacity to regulate components of the DNA replication machinery (Gladden & Diehl, 2003).
Cyclin D1 activity is also controlled through regulated changes in its sub-cellular localization (Diehl et al., 1998; Diehl et al., 1997). During early G1 cyclin D1 assembly with CDK4 occurs in the cytoplasm prior to nuclear translocation (Diehl & Sherr, 1997; Matsushime et al., 1994). At the onset of S-phase, the GSK-3β kinase enters the nucleus and phosphorylates cyclin D1 at a distinct threonine residue, Thr-286, targeting cyclin D1 for CRM1 dependent nuclear export and subsequent degradation by the 26S proteasome (Alt et al., 2000; Diehl et al., 1998). Abolition of Thr-286 phosphorylation via site specific mutation of the phospho-acceptor site (D1/T286A) or through an alternative splicing mechanism that removes sequences directing cyclin phosphorylation results in the constitutive nuclear localization of cyclin D1 (Alt et al., 2000; Lu et al., 2003).
The subversion of normal growth signaling pathways is a hallmark of neoplasia, and transformed cells generally exhibit reduced growth factor requirements. Cell cycle regulators, particularly those governing mitogen-dependent G1 phase progression such as cyclin D1, make natural targets during oncogenesis as this provides cells with a clear growth advantage. Overexpression of cyclin D1 and its ensuing nuclear accumulation is frequently observed in cancer cells due to gene amplification, chromosomal translocation, or protein stabilization (Hall & Peters, 1996; Sherr, 1996). Cyclin D1 has been associated with a number of solid tumors and additionally was identified as the BcL-1 gene associated with the chromosomal translocation t(11;14)(q13;q32) in mantle cell lymphoma (MCL) (Akiyama et al., 1994; Seto et al., 1992). Of particular note, the initial attempts to establish cyclin D1 as a dominant B-cell oncogene were unsuccessful (Bodrug et al., 1994; Lovec et al., 1994a) implying that the participation of additional regulatory mechanisms in the regulation of cyclin D1/CDK4 activity. In contrast, attempts to model genetic changes associated with other human B-cell malignancies, such as those associated with Burkitt (c-myc) and follicular lymphoma (BcL-2), have met with considerable success (Adams et al., 1985; Strasser et al., 1993).
In addition to MCL, cyclin D1 is overexpressed in a variety of solid tumors implicating cyclin D1 in tumor initiation or progression. The inability of overexpressed wild type cyclin D1 to promote oncogenic transformation even in in vitro systems (Alt et al., 2000; Quelle et al., 1993) is thus paradoxical. We have previously found that cyclin D1 mutants that remain constitutively nuclear due to impaired Thr-286 phosphorylation, can indeed induce a transformed phenotype in cultured murine fibroblasts (Alt et al., 2000); (Lu et al., 2003) providing a potential resolution to this paradox. In addition, our laboratory and others have recently identified a constitutively nuclear cyclin D1 isoform that arises due to alternative splicing (Lu et al., 2003; Solomon et al., 2003). This alternative transcript lacks sequences directing phosphorylation and nuclear export and its expression is restricted to cancer cells. These results imply that the inability of overexpressed wild type cyclin D1 to promote cell transformation in vitro and perhaps induce B-cell lymphomas in mice reflects the capacity of the targeted cells to maintain proper temporal regulation of cyclin D1 activity through regulated nuclear export. Given the oncogenic potential of constitutively nuclear cyclin D1/T286A in vitro, we hypothesized that expression of this nuclear mutant would drive B-cell malignancies in mice.
Results
D1/T286A associates with CDK4 in transgenic lymphocytes
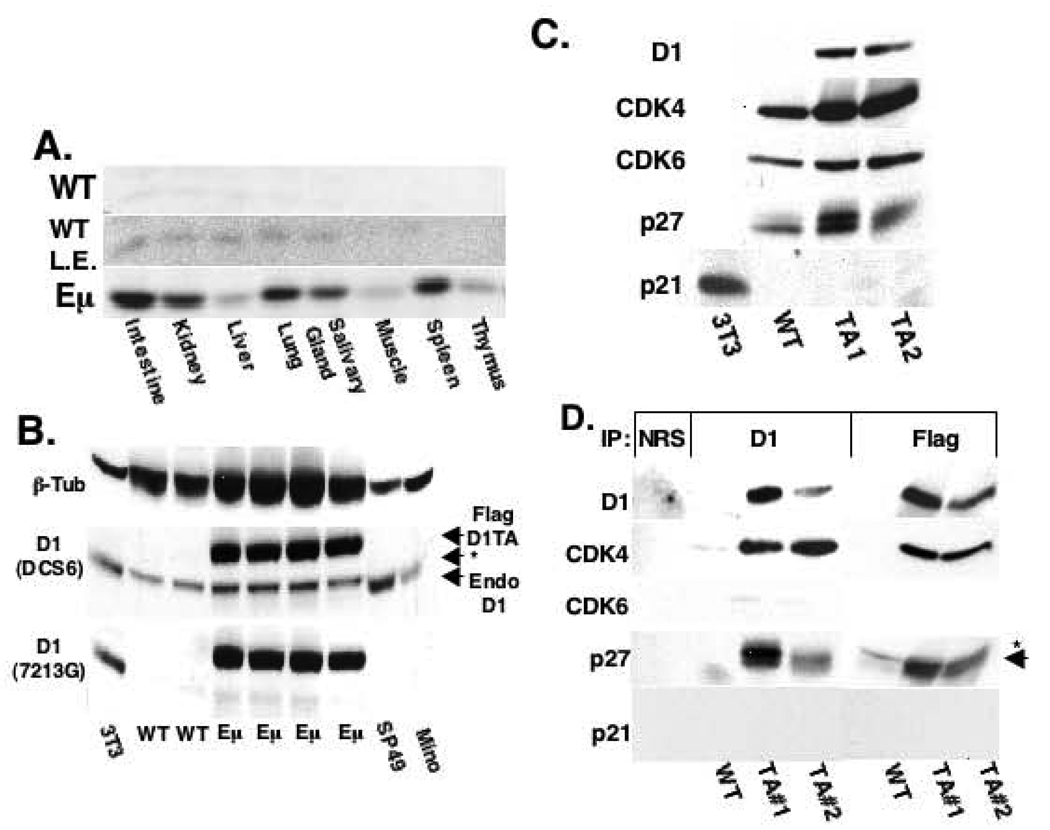
To determine whether a constitutively nuclear cyclin D1 mutant, D1/T286A, can promote neoplastic transformation in vivo, we generated transgenic mice wherein expression of FLAG tagged cyclin D1/T286A was controlled by the lymphoid specific Eµ enhancer. Four transgenic founder mice were identified; two were high copy carriers and were not useful in establishing lines due to morbidity (lymphoma) prior to breeding age. In contrast, one low copy and one single copy transgenic line was established and germ-line transmission confirmed (data not shown). Both lines exhibited similar phenotypes, and thus the single copy line was subjected to detailed analysis. The expression profile of FLAG-D1/T286A (D1/T286A) was evaluated by Western analysis of lysates prepared from the indicated tissues of six-week-old mice (Figure 1A). High levels of D1/T286A were present in the spleen, intestine, and lung while lower levels were observed in the thymus, kidney, and salivary gland (lower panel). Expression in non-lymphoid tissue reflects in part constitutive expression driven by the Pim-1 promoter and also some contribution of lymphoid infiltrates (data not shown). Endogenous cyclin D1 expression was not detectable in the non-transgenic spleen but was observed in the expected tissue compartments upon longer exposure (1A, upper and middle panels). To assess D1/T286A expression levels in transgenic lymphoid cells, we compared it with that of cyclin D1 in human MCL cell lines containing the t(11;14) translocation. Total protein from two established human MCL cell lines, SP49 and Mino along with total protein from wild type and transgenic spleens was analyzed for total cyclin D1 (Figure 1B). Antibodies specific for β-Tubulin (top panel), murine specific D1 antibody (72–13G) and an antibody that recognizes both mouse and human cyclin D1 (DCS6) (middle panel) were utilized. Normalization to β-Tubulin, revealed that levels of D1/T286A from transgenic spleens was similar to levels of cyclin D1 observed in the SP49 and Mino cell lines demonstrating the single copy transgenic mouse line did not express supra-physiological levels of cyclin D1.
Figure 1. D1/T286A associates with CDK4 and p27 in transgenic splenocytes.
a) Cyclin D1 direct Western of whole tissue protein lysates from either 6-week-old non-transgenic wild type (WT) or Eµ-D1/T286A (Eµ) mice. L.E. denotes longer exposure of the Western blot. b) Western analysis of protein lysates from NIH-3T3 cells (3T3), wild type spleens (WT), D1/T286A spleens (Eµ) or two human MCL cell lines performed with antibodies specific for β- Tubulin, human and mouse cyclin D1 (DCS6) or mouse D1 only (7213G). A non-specific background band recognized by the DCS6 antibody is noted by the (*). c) Western analysis with the indicated antibodies was performed on splenic extracts prepared from non-transgenic wild type (WT), D1/T286A (Eµ) mice, or NIH-3T3 fibroblasts. d) Protein lysates from (c) were precipitated using either a cyclin D1 antibody (D1), the M2 antibody (FLAG), or with normal rabbit serum (NRS). Proteins were separated by electrophoresis and probed with the indicated antibodies.
A majority of cyclin D1 is assembled into high order complexes composed of cyclin D1, CDK4/CDK6, and p21Cip1/p27Kip1 (LaBaer et al., 1997; Zhang et al., 1994). We initially assessed expression levels of the potential components of this complex by Western analysis with antibodies directed towards cyclin D1, CDK4, CDK6, p21Cip1 and p27Kip1 (Figure 1C). Relative to non-transgenic controls, expression of D1/T286A in transgenic spleens corresponded with increased levels of CDK4 and p27Kip1 while CDK6 levels remained constant (Figure 1C). To determine whether cyclin D1/T286A was incorporated into complexes containing CDK4/6 and p27Kip1, splenic lysates were subjected to precipitation with antibodies directed towards either cyclin D1 or the FLAG epitope expressed by the D1/T286A transgenic protein. While CDK4 and p27Kip1 co-precipitated with D1/T286A, CDK6 was not detected in precipitates (Figure 1D). This data demonstrates that CDK4 is the primary catalytic partner for cyclin D1/T286A in splenic lymphocytes.
D1/T286A mice develop disseminated IgM positive B-cell lymphoma
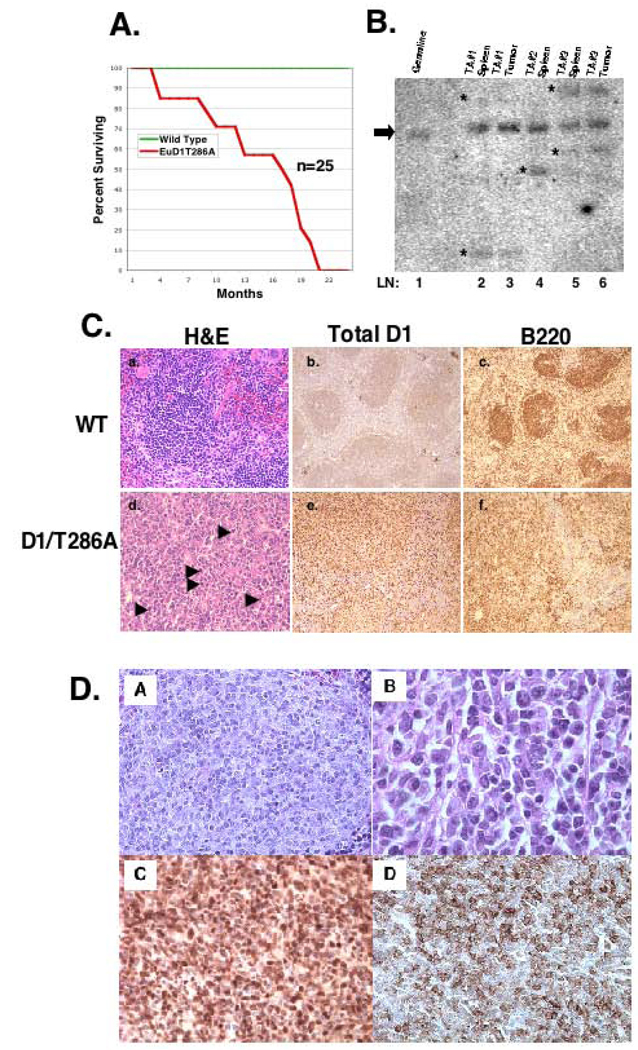
Expression of wild type cyclin D1 in murine fibroblasts or murine lymphocytes does not promote neoplastic transformation without co-expression of a cooperating oncogene such as Myc or Ras (Bodrug et al., 1994; Lovec et al., 1994a; Lovec et al., 1994b; Uchimaru et al., 1996). To ascertain whether expression of cyclin D1/T286A could drive lymphoid malignancies in vivo, a cohort of 25 non-transgenic and Eµ-D1/T286A transgenic mice were observed over a 24-month period. D1/T286A mice exhibited a significant reduction in lifespan relative to the non-transgenic controls (Figure 2A). The decreased survival rate of the transgenic mice was associated with the development of a widely disseminated malignant process that involved the spleen and mesenteric lymph nodes as well as non-lymphoid organs such as the lungs, intestine and liver. Southern blot analysis with the immunoglobulin heavy-chain JH probe (Alt et al., 1984; Borzillo & Sherr, 1989) performed on DNA isolated from the involved spleens, revealed clonal rearrangement of the IgH gene (Figure 2B, lanes 2, 4, 5) in contrast to the germ line pattern observed in the wild type mouse (lane 1). Where available, peripheral tumors from the same mouse were examined and found to exhibit identical IgH rearrangement as splenic tumors (lanes 3, 6). Tumor histology revealed diffuse infiltrates of lymphoid-appearing cells typically with large nuclei and with occasional cells exhibiting immunoblastic and plasmacytoid differentiation (Figure 2D, panel B). Tumors were of high mitotic indices, stained positive for cyclin D1 (Figure 2C panel E; 2D panel C) B220 (2C panel f; 2D panel D), negative for T-cell markers (data not shown), and negative for immature lymphoid markers (data not shown) confirming the malignant cells were of a mature B-cell origin.
Figure 2. D1/T286A mice have a decreased lifespan due to the onset of B-cell lymphoma.
a) Survival curve representing mean survival of either wild type (WT) (Green Line) or D1/T286A (Eµ) (Red Line) cohort over a 24-month period. b) Southern blot analysis of immunoglobulin heavy chain gene rearrangements in transgenic splenic tumors and available peripheral tumors from the identical mouse. Bold line represents the germline size of the IgH gene and (*) notes rearranged gene products. c) Representative histology of either wild type spleen (a, b, c) or a tumor burden D1/T286A spleen (d, e, f) stained with hematoxylin and eosin (a, d) with arrowheads denoting mitotic figures. Immunohistochemistry for either total cyclin D1 (b, e) or B220 (c, f). d) Higher magnification of D1/T286A tumor burden spleens stained with hematoxylin and eosin (A,B), for cyclin D1 (C) or B220 (D).
We also assessed potential tumor dissemination in non-lymphoid organs. Sizeable infiltrates of lymphoid cells were identified within the intestinal wall and lung parenchyma. Immunohistochemical analysis with antibodies specific for either B220 or cyclin D1 highlighted the infiltrate confirming their B-cell phenotype and transgenic derivation (data not shown).
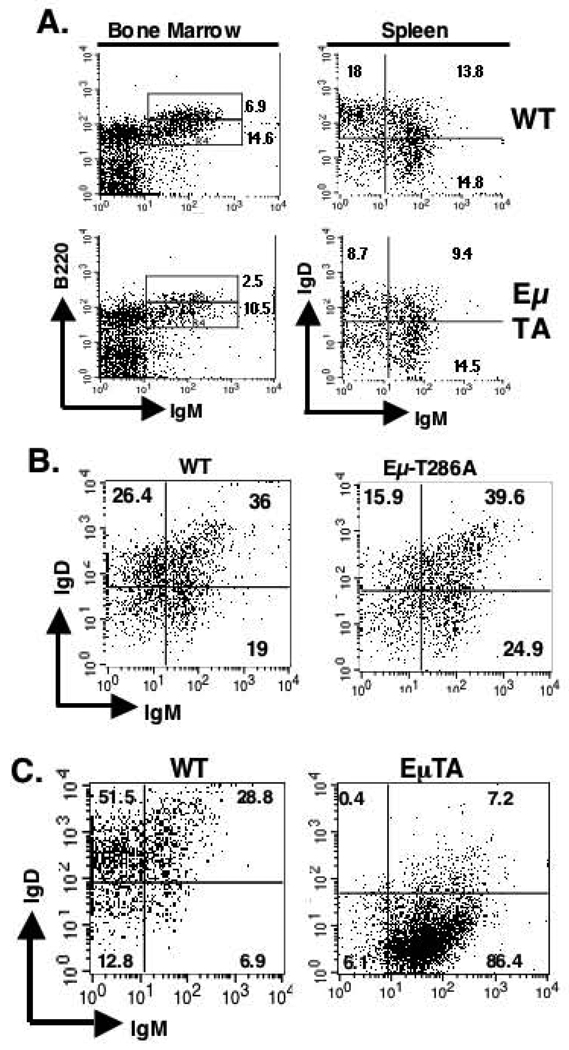
To ascertain the B-cell targeted during cancer genesis, D1/T286A and age matched non-transgenic mice were sacrificed and surface expression of B220 and IgM were assessed in primary bone marrow derived B-cells by flow cytometry. Approximately two-fold fewer recirculating B220high, IgM+ B-cells were present in non-transgenic controls compared to newly derived B220low, IgM+ B-cells (Figure 3A). In contrast, D1/T286A transgenics reproducibly contained four-fold fewer recirculating B-cells (Figure 3A). Once B-cells enter the spleen from the bone marrow, they progress from an IgMhigh, IgDlow to an IgMhigh, IgDhigh state and finally to an IgMlow, IgDhigh status as exemplified by the nearly 1:1:1 ratio detected in non-transgenic splenic B-cells (Figure 4A and 4B). Bone marrow isolated from Eµ-D1/T286A splenic B-cells have over two fold fewer IgMlow, IgDhigh and a lower proportion of IgMhigh, IgDhigh B-cells compared with that from non-transgenic mice (Figure 3A and 3B). To establish whether this change in surface immunoglobulin expression is similar in the fully transformed cells, splenocytes from transgenic, tumor burden mice reproducibly exhibited a significant enrichment of IgMhigh, IgDlow B-cells (Figure 3C). These results demonstrate the prime target cell is an early mature B-cell.
Figure 3. D1/T286A splenic B-cells display an altered surface immunoglobulin profile.
a) Bone marrow cells from either non-transgenic wild type (WT) or D1/T286A (Eµ-TA) six-week old mice were analyzed by flow cytometry with antibodies specific for either B220 or surface IgM. Likewise total splenic lymphocytes from either wild type or Eµ-D1/T286A mice were stained with B220, surface IgD and surface IgM. B220 positive cells were gated on and surface IgM and IgD expression was analyzed. b) Splenic cells from six-week-old pre-malignant mice were stained with B220, surface IgM and IgD, B220 cells were gated on and surface immunoglobulin levels were analyzed. c) Cells from a tumor burden spleen (EµTA) or an age-matched mouse were stained with B220, surface IgM or IgD. B220 positive cells were gated on and surface immunoglobulin levels were analyzed by flow cytometry.
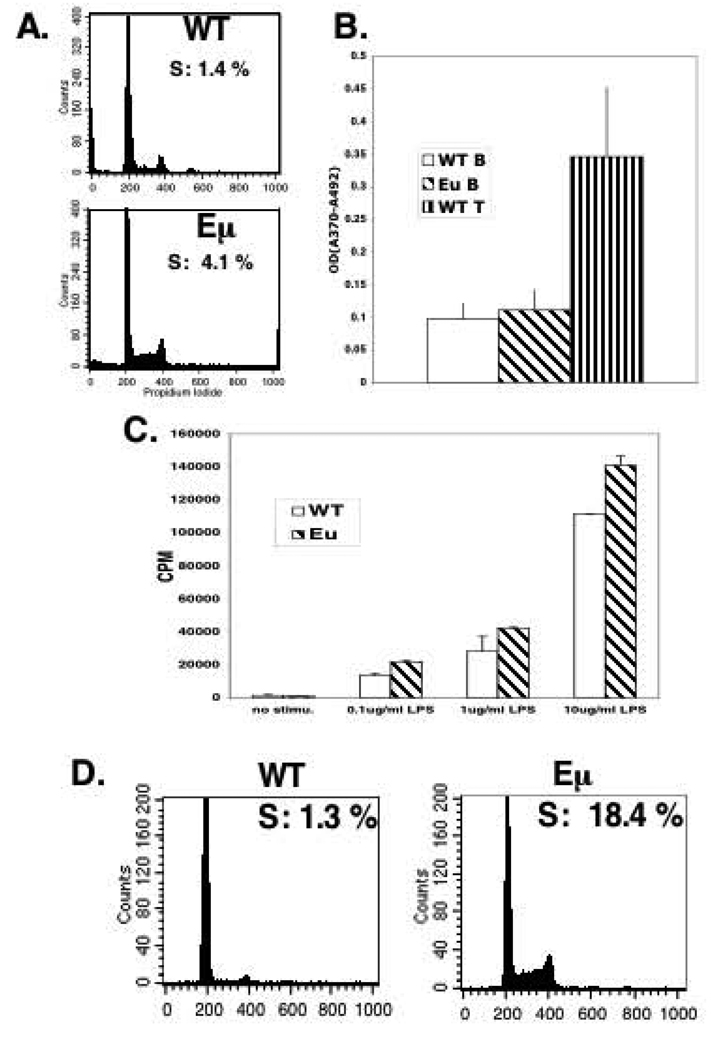
Figure 4. D1/T286A B-cells have a decreased dependence on mitogen stimulation.
a) Purified splenic B-cells from six-week old wild type or D1/T286A mice were stained with propidium iodide and DNA content was determined by flow cytometry. b) Six-week-old wild type or D1/T286A mice were injected with BrdU 24 hours prior to sacrifice. BrdU incorporation was quantitated by ELISA from purified splenic B or thymic T-cells. c) Purified splenic B-cells from pre-malignant transgenic (Eµ) and age matched wild type (WT) mice were stimulated with the indicated concentrations of LPS for 48 hrs. Proliferation was determined by incorporation of 3H-Thymidine. d) Cells from a tumor burden spleen (Eµ) or an age-matched mouse were stained with propidium iodide and DNA content was determined by flow cytometry.
Promiscuous S-phase entry induced by D1/T286A is countered by apoptosis in young transgenic mice
A majority of B-cells are quiescent until exposed to antigen (Noelle & Snow, 1990). Given that cyclin D1 overexpression results in reduced growth factor requirements and premature S-phase entry in cultured cells (Quelle et al., 1993), we considered the possibility that D1/T286A might trigger the promiscuous cell cycle entry of B lymphocytes. Total splenic B-cells were isolated from 6-week-old non-transgenic and transgenic mice and cell cycle profiles were assessed by FACS analysis. Essentially all (99%) of wild type splenic B-cells had a 2N DNA content consistent with G0 arrest (Figure 4A). In contrast, B-cells from D1/T286A mice reproducibly had 4 times (4%) more cells in S-phase and an accompanying increase in G2/M phase cells (Figure 4A). To corroborate this finding, asymptomatic six-week-old mice were injected with BrdU 24 hours prior to sacrifice. B-cells were purified from the spleens of both non-transgenic and transgenic mice and BrdU incorporation assessed by ELISA. B-cells expressing D1/T286A incorporated similar amounts of BrdU as wild type B-cells (Figure 4B). Our inability to detect BrdU incorporation in the transgenic B-cells did not reflect relative insensitivity of the assay, as we readily detected BrdU uptake in T-cells (Figure 4B). Thus, while the increase in cells with >2N DNA content was reproducibly noted, the data suggest that a majority of the cells remain quiescent or alternatively are rapidly eliminated following promiscuous S-phase entry in the absence of appropriate activating signals.
The small but reproducible fraction of S-phase cells suggested that while D1/T286A expression increases proliferative potential, a majority of the lymphocytes are initially able to counter this and maintain growth arrest. In fibroblasts, overexpression of the D-type cyclins reduces mitogen dependence during cell cycle reentry and accelerates G1 progression (Quelle et al., 1993). To determine whether transgenic B-cells exhibit increased proliferative potential following mitogen stimulation, young D1/T286A splenic B-cells and age matched non-transgenic controls were stimulated for 48hrs with increasing concentrations of LPS and proliferation evaluated by 3H-Thymidine incorporation (Figure 4C). At all doses of stimulation, the transgenic B-cells displayed a significant increase in 3H-Thymidine incorporation relative to the non-transgenic controls suggesting that D1/T286A expression does confer a proliferative advantage.
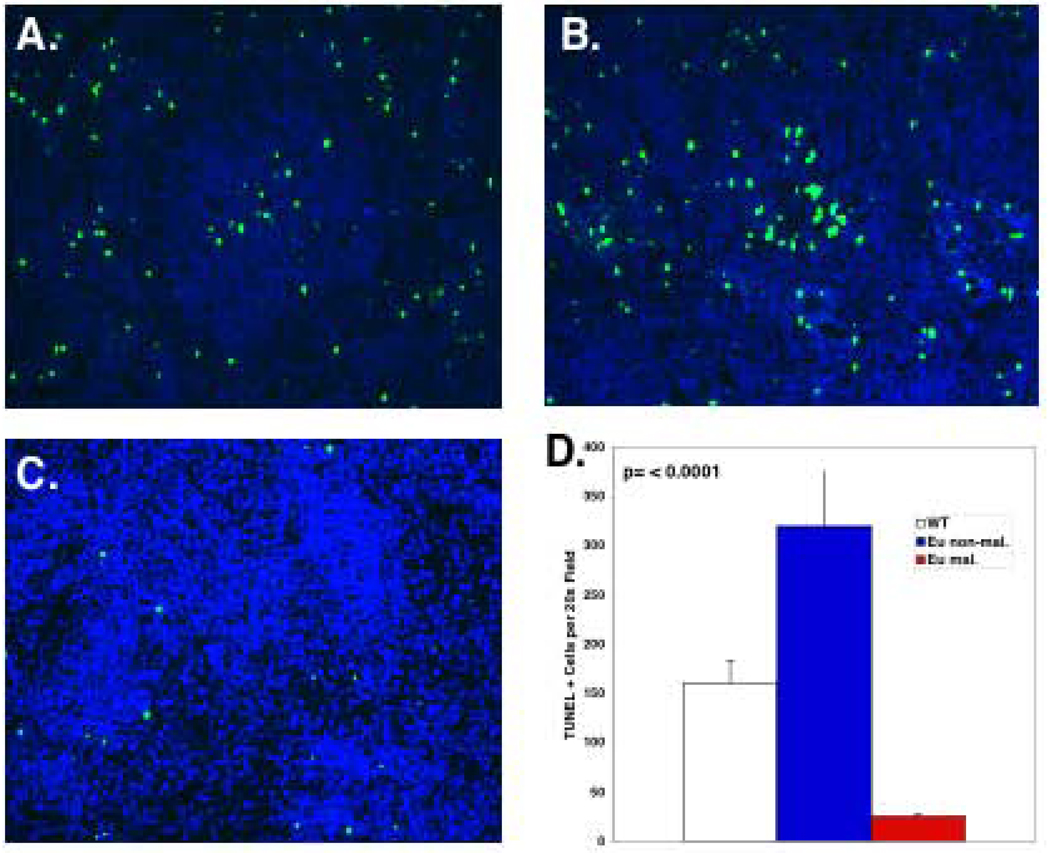
Given that D1/T286A expression provides B lymphocytes with a proliferative advantage, we were surprised at the relatively small increase in the fraction of transgenic B-cells in S-phase as assessed by FACS. One potential mechanism underlying this phenotype is that the promiscuous S-phase entry triggered by D1/T286A expression is countered by a high rate of apoptosis. Consistent with this notion, we noted increased accumulation of apoptotic cells (<2N) in transgenic B-cells (Figure 4A). To confirm this, spleens from 8-week old non-transgenic or D1/T286A transgenic mice were assessed by TUNEL assay. Transgenic spleens (Figure 5B) exhibited a significant increase in TUNEL positive cells relative to non-transgenic spleens (Figure 5A). If apoptosis counters proliferation in young mice, outgrowth of a fully transformed clone would only occur following a “second hit” that would counter or eliminate apoptosis. Consistent with this hypothesis tumorigenic spleens from transgenic mice (Figure 5C; quantification in 5D) exhibited almost no TUNEL positive cells.
Figure 5. D1/T286A transgenic spleens display an increased proportion of apoptotic cells.
Representative TUNEL analysis performed on a) non-transgenic spleens, b) age matched pre-malignant D1/T286A spleens, and c) tumors from malignant transgenic mice. d) Quantitation of TUNEL analysis.
The inability of D1/T286A expressing B-cells in young mice to survive following entry into S-phase must be overcome for malignant outgrowth. This should be reflected in a significant increase in the percentage of cycling B-lymphocytes in tumor burden spleens. To test this notion, splenic B-cells derived from either tumor burden transgenic mice or from age matched non-transgenic mice were fixed and stained with propidium iodide and their cell cycle profile analyzed by FACS. While the non-transgenic B-cells maintained a similar percentage of S-phase cells as the young wild type mice, B-cells from the tumor burden spleen of an D1/T286A mouse had 14.5 times more S-phase cells (18%). These results clearly demonstrate that the malignant B-cells had overcome the high apoptotic rate thereby permitting cell cycle progression (Figure 4D).
D1/T286A lymphomas exhibit alterations in pro-survival pathways
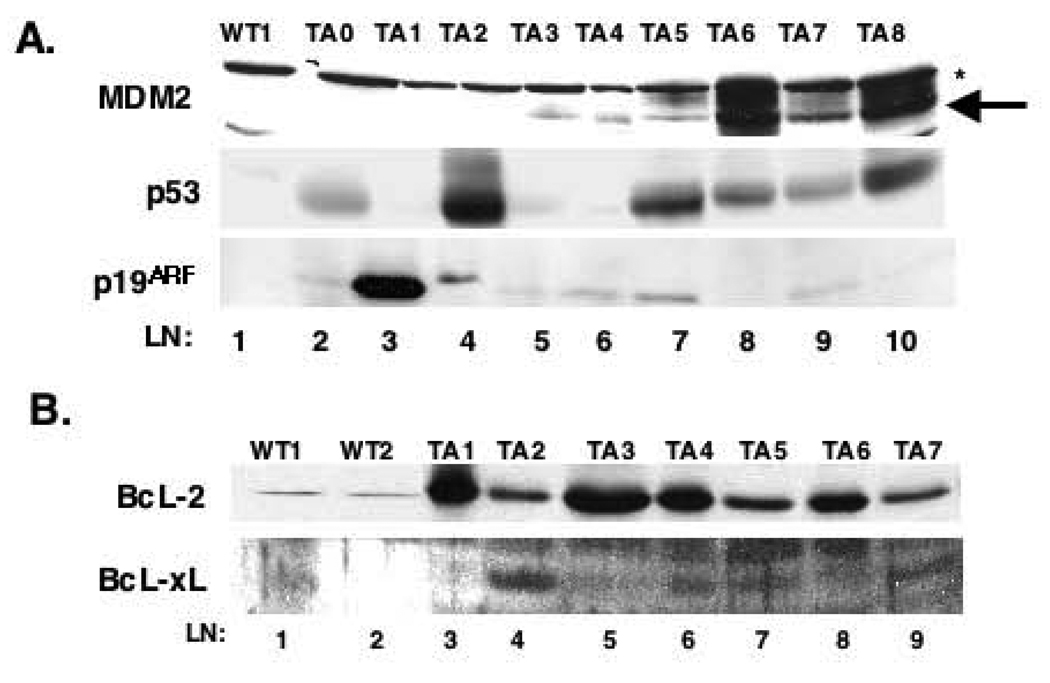
The latency in tumor development coupled with the proliferative block and increased apoptosis in B-cells of young D1/T286A mice suggests the need for co-operating mutations during the course of cancer genesis. Inactivation of the p19ARF-MDM2-p53 pathway accelerates lymphoma onset in Eµ-Myc mice (Eischen et al., 2001a; Eischen et al., 1999; Eischen et al., 2001b). While the p53 axis is rarely inactivated in human lymphoma, it is thought to be targeted as a late event in human MCL (Gronbaek et al., 2002; Lai et al., 2002; Martinez et al., 2003; Strasser et al., 1993). Inactivation of p53 predominantly occurs via point mutation of a single allele resulting in hyperaccumulation of dominant negative p53 (Eischen et al., 1999; Levine, 1997). Accordingly, we assessed p53 accumulation by Western analysis (Eischen et al., 1999; Zindy et al., 1998). Protein lysates from a cohort of B-cell tumors derived from Eµ-D1/T268A mice and control splenic lysates from non-transgenic mice were subjected to Western analysis using antibodies specific for MDM2, p53 or p19ARF. Lysates from Eµ-D1/T286A tumors displayed increased levels of p53 (Figure 6A, lane 2, 4, 7–10). One tumor sustained a loss of p53 (lane 3) resulting in the uncoupling of p53-p19ARF regulation and subsequent overexpression of p19ARF. Although expression of MDM2 did not fit a pattern, hyper-accumulation was noted in a majority of tumors examined (Eischen et al., 1999). In two tumors, MDM2 accumulation was accompanied by hypo-accumulation of p53 as expected (lanes 5–6). In other tumors, we noted accumulation of p53 and MDM2, suggesting that in these tumors, the p53 pathway may remain intact, and tumor progression may not be dependent upon p53 inactivation.
Figure 6. The p53 pathway is targeted D1/T286A tumors.
a) Direct western analysis was performed on protein lysates from either wild type spleen (WT) or D1/T286A (TA) tumors using antibodies specific for MDM2 (top panel) (*) denotes cross reacting band, p53 (middle panel) or p19ARF (bottom panel). b) Total protein lysates from wild type spleens or D1/T286A tumors were separated electrophoretically, transferred to nitrocellulose and probed with antibodies for either BcL-2 (top panel) or BcL-XL (bottom panel).
Overexpression of BcL-2, is also associated with B-cell lymphoma (Zhou et al., 2001). Overexpression of BcL2 and BcL-XL inhibits Myc induced apoptosis and accelerates lymphoma (Bissonnette et al., 1992; Eischen et al., 2001b; Fanidi et al., 1992). As expression of D1/T286A is associated with apoptosis, we considered the possibility that lymphoma onset might be associated with overexpression of BcL-2 or BcL-XL. Levels of BcL-2 and BcL-XL in D1/T286A tumors were determined by direct Westerns analysis of primary tumors. A majority of the tumors examined expressed elevated BcL-2 (Figure 6B; lanes 3, 5, 6, 8) relative to that found in non-transgenic spleen (lane 1, 2). We also noted elevated expression of BcL-XL in a smaller subset of tumor samples (lanes 4, 6, 7, 9); these higher levels of BcL-XL correlated with lower levels of BcL-2 suggesting that selective overexpression of one of the anti-apoptotic BcL-2 family members occurs in the tumors from D1/T286A mice. These data suggest that secondary mutations in either the p53 pathway and/or overexpression of anti-apoptotic BcL-2 family members contribute to lymphoma genesis in D1/T286A transgenic mice.
Discussion
While evidence suggests a role for cyclin D1 in either the onset or progression of human cancers, experimental evidence demonstrating a role for cyclin D1 as a bone fide oncogene in vitro or in mouse models is scarce. In an early attempt to model human MCL, wherein cyclin D1 is expressed due to the t(11;14) translocation, wild type cyclin D1 was placed under the control of the Eµ enhancer (Bodrug et al., 1994; Lovec et al., 1994a). Remarkably, the Eµ-wtD1 mice did not exhibit detectable lympho-proliferative disorder and showed little to no alterations in B or T-cell development. In contrast, we now demonstrated that expression of the nuclear export deficient and thus constitutively nuclear cyclin D1 mutant, D1/T286A, predisposes mice to B-cell lymphoma. The lymphoma-prone phenotype of the D1/T286A mice is consistent with our hypothesis that cyclin D1 oncogenicity is manifested upon deregulation of nuclear export and the resulting nuclear retention of the kinase during S-phase. Based on these findings we expect that in MCL and other malignancies wherein cyclin D1 is implicated, further analysis will reveal aberrations in the regulation of cyclin D1 nuclear export. Indeed we have recently identified a nuclear export deficient cyclin D1 isoform, cyclin D1b, in primary esophageal carcinoma (Lu et al., 2003), providing correlative support for our hypothesis. In addition, the cyclin D1b alternative transcript, which phenocopies the murine D1/T286A (Lu et al., 2003), has previously been identified in primary human MCL (Hosokawa et al., 1999; Howe & Lynas, 2001). Consistent with these findings, we have observed the cyclin D1b protein in human MCL-derived cell lines (data not shown) providing further evidence that nuclear accumulation of cyclin D1 is likely the contributing factor in cyclin D1-driven neoplasia.
While the D1/T286A mice develop lymphoma with complete penetrance, there is a relatively long incubation prior to overt lymphoma development. Given the proliferative advantage provided by D1/T286A, why don’t the transgenic mice exhibit a more distinct hyper-proliferative disorder or early tumorigenesis? Like other cancer promoting proteins, most notably c-Myc, cyclin D1 overexpression in certain contexts is associated with apoptosis (Sofer-Levi & Resnitzky, 1996). Indeed, we found that expression of the D1/T286A transgene in spleens of young pre-malignant mice was associated with a significant increase in apoptosis. Thus, the initial proliferative response to D1/T286A is countered by cell death such that development of lymphoma only results from secondary mutations that disrupt this response (e.g. overexpression of BcL-2 or loss of p53). Indeed, analysis of D1/T286A tumors revealed the frequent inactivation of the p53 pathway and overexpression of BcL-2. This suggests that p53 and or BcL-2 play a pivotal role in preventing D1/T286A-dependent cell growth.
Our analysis of the p53 pathway and BcL-2 family members is consistent with lymphomagenesis being a multi-step process wherein expression of D1/T286A provides a constant proliferative stress that in turn sensitizes mice to secondary genetic alterations. Similarly, while Eµ-Myc mice develop B-cell lymphoma with an earlier onset than D1/T286A mice, overt lymphoma in this model also reflects acquisition of secondary mutations. Extensive analysis of this model has revealed the frequent inactivation of p53 signaling components and the overexpression of BcL-2 and BcL-XL (Eischen et al., 1999; Eischen et al., 2001b). Similar to the Eµ-Myc mice, lymphomas arising in the D1/T286A mice have selected for inactivation of p53 and/or overexpression of the anti-apoptotic proteins BcL-2 or BcL-XL. Noteworthy in this context, micro-array analysis of human MCL indicates that both the p53 pathway and the BcL-2 proteins are targeted in a subset of high-grade mantle cell lymphomas (Rosenwald et al., 2003) suggesting these pathways may be commonly targeted during lymphoma progression wherein cyclin D1 represents the initiating step.
Using microarray-based gene expression profiling of purified B-lymphocytes from young asymptomatic mice, we identified Myc as highly expressed in transgenic mice (data not shown). Myc overexpression was in a subset of tumors by western analysis (data not shown). Myc is commonly overexpressed in MCL and was previously shown to cooperate with wild type cyclin D1 in doubly transgenic mice (Bodrug et al., 1994; Lovec et al., 1994a). Indeed, the overexpression of BcL-2 in a majority of Eµ-D1/T286A tumors may reflect deregulation of Myc-dependent transcriptional control as has been suggested for Eµ-Myc lymphomas (Eischen et al., 2001b; Hernandez et al., 1999; Nagy et al., 2003). In human MCL, expression of Myc is generally observed in the advanced, blastic type. Similarly, based on morphology, the analogous presence of the additional genetic changes in the p53 pathway and their disseminated nature, the B-cell lymphomas arising in D1/T286A mice resembled the blastic variant of MCL. D1/T286A transgenic mice exhibit a reduced percentage of IgMlow, IgDhigh splenic B-cells in pre-malignant stage. This imbalance is even more accentuated in lymphoma-bearing mice and is associated with an accumulation of IgM, IgD double positive B-cells that closely resemble the mature B-cell phenotype of human MCL.
The cyclin D1 translocation t(11;14) is a defining characteristic of human MCL. Considering the relatively poor prognosis and lack of durable response to current treatment modalities (Argatoff et al., 1997; Bosch et al., 1998), new therapies are needed for MCL. Accordingly, targeting the cyclin D1-dependent kinase appears therapeutically efficacious in vitro, where CDK4/6 inhibition reduced proliferation of a variety of tumor derived cell lines, whereas CDK2 inhibition was ineffective (Strasser et al., 1993). The recent development of a highly specific small molecule CDK4/6 inhibitor exhibited striking efficacy in both cell culture and subcutaneous tumor models (Fry et al., 2004). The development of a mouse model, that is dependent upon the cyclin D1/CDK4 kinase, should provide an excellent opportunity to evaluate the efficacy of targeting the cyclin D-dependent kinase in vivo and also provides a platform to further study the role that nuclear retention of cyclin D1 has on S-phase progression and cellular proliferation.
Materials and Methods
Transgenic Mouse Generation
FLAG tagged D1/T286A was sub-cloned into the HpaI site of TDK-TG90 and the subsequent 6.5kb HindIII fragment of TDK D1/T286A was injected into C57/BL6 pronuclei and implanted into C57/BL6 host mice. Founder mice were identified by Southern blot of BamHI digested genomic DNA and by PCR using primers specific for the FLAG tag (5’-CTGCTCCTCAGTGGATGTTGCC-3’) and murine cyclin D1 (5’-CTCACAGACCTCCAGCAT-3’), positive mice were subsequently bred to C57/BL6 mice to generate the Eµ-D1/T286A colony.
Cell Purification and Cell Culture conditions
Splenic B-cells from wild type or transgenic mice were purified by disassociating whole spleen in cold DMEM and ensuing purification using StemSep™ murine B-cell enrichment cocktail and negative selection columns per manufacturer’s directions (StemCell Technologies). Purified B-cells were cultured in RPMI 1640, 2mM L-glutamine (BioWhitaker), antibiotics and 5% heat inactivated fetal calf serum (FCS) (Gemini Bioproducts), stimulated with indicated concentrations of lipopolysaccharide (Sigma), and pulsed with 1 µCi of 3H-Thymidine.
Western Blotting and Immunoprecipitation
Fresh tissue was isolated and dissociated in Tween 20 buffer (50 mM HEPES (pH 8), 150 mM NaCl, 2.5 mM EGTA, 1 mM EDTA, 0.1% Tween 20, 0.1mM PMSF, 1 µg/ml aprotinin, 1 µg/ml leupeptin, 25 mM glycerol phosphate, 1 mM NaF and 1 mM dithiothreitol), sonicated two times and remaining debris cleared by centrifugation. Protein complexes were collected with the cyclin D1 monoclonal antibody (D1-17-13G) or the M2 monoclonal antibody (Sigma) and protein A Sepharose (Amersham Pharmacia); bound complexes were washed four times in Tween 20 buffer. Immunoprecipitated protein complexes and total protein lysates were resolved by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Millipore). Membranes were blotted with antibodies specific for cyclin D1 (D1-17-13G, Oncogene Research Products, Ab-3, or Dako, DCS-6), CDK4 (Santa Cruz Biotechnologies, C-22 or H-22), CDK6 (Ab3, Neomarkers), p21 (Santa Cruz, C-19), p27 (BD Transduction Laboratory, clone 57), MDM2 (Santa Cruz, C-18), p19 ARF (Abcam, ab80), p53 (PAB421), BcL-2 (3F11), and BcL-X. Antibody binding was visualized using protein A-conjugated horseradish peroxidase (PA-HRP, EY Laboratories), anti-mouse conjugated HRP, or anti-rabbit conjugated HRP (Cell Signaling).
BrdU analysis and Flow Cytometry
Mice were injected i.p. with 30 µg of BrdU per gram of body weight 24 hours prior to sacrifice. Splenic B-cells were purified and BrdU ELISA was performed according to the manufacturers instructions (Roche Applied Science). For cell surface marker analysis, the specified tissues were dissociated and washed in PBS containing 2% FCS. Staining was performed in PBS/2% FCS using antibodies specific for B220-APC, IgM-PE and IgD-FITC (BD Bioscience). For DNA analysis lymphoid cells were washed in PBS 2 times and resuspended in PI staining buffer (50 µg/ml propidium iodide, 1 µg/ml sodium citrate and 0.1% Triton-X). Analysis was performed with a BD FacsCalibur using CellquestPro software.
Southern Analysis
Genomic DNA from wild type or transgenic tissue was isolated, subjected to digestion with XbaI and separated on a 1% agarose gel. DNA fragments were then transferred to nylon membrane (Millipore) and probed with the JH probe made by random-prime labeling of a 1.2kb fragment released from the pJR34 JHIg plasmid by BamHI-XbaI double digestion (generous gift from M. F. Roussel and C. J. Sherr) (Borzillo & Sherr, 1989).
Immunohistochemistry
Tissue was fixed in 10% buffered formalin and subsequently dehydrated, embedded and sectioned. Tissue sections were subjected to antigen retrieval using Vector antigen unmasking solution followed by blocking of endogenous peroxidase activity by treating with 1% peroxide for 15 minutes. Sections were then incubated in blocking buffer for 30 minutes followed by incubation with the primary antibody overnight. Sections were washed five times with TT buffer and incubated with a secondary biotin conjugated antibody (Vector Laboratories), washed again and incubated with ABC reagent (Vector Laboratories) followed by detection with DAB substrate (Vector Laboratories). TUNEL analysis was performed according to the manufacture’s suggested protocol (Roche).
Acknowledgements
We would like to thank Anton Berns (The Netherlands Cancer Institute) and John Cleveland (St Jude Children’s Research Hospital) for providing the TDK-TG90 construct. Martine F. Roussel and Charles J. Sherr (SJRH) for providing reagents and helpful suggestions. The transgenic and histology core at the Abramson Family Cancer Research Institute, University of Pennsylvania for technical assistance and Michal Marzec and Fengmin Lu, University of Pennsylvania for helpful suggestions. This work was supported by a grant from the National Institutes of Health (NIH) CA93237(JAD) and CA89194 (MAW).
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Akiyama N, Tsuruta H, Sasaki H, Sakamoto H, Hamaguchi M, Ohmura Y, Seto M, Ueda R, Hirai H, Yazaki Y, et al. Cancer Res. 1994;54:377–379. [PubMed] [Google Scholar]
- Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Embo J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JR, Cleveland JL, Hannink M, Diehl JA. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argatoff LH, Connors JM, Klasa RJ, Horsman DE, Gascoyne RD. Blood. 1997;89:2067–2078. [PubMed] [Google Scholar]
- Bissonnette RP, Echeverri F, Mahboubi A, Green DR. Nature. 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Embo J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzillo GV, Sherr CJ. Mol Cell Biol. 1989;9:3973–3981. doi: 10.1128/mcb.9.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch F, Lopez-Guillermo A, Campo E, Ribera JM, Conde E, Piris MA, Vallespi T, Woessner S, Montserrat E. Cancer. 1998;82:567–575. doi: 10.1002/(sici)1097-0142(19980201)82:3<567::aid-cncr20>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Sherr CJ. Mol Cell Biol. 1997;17:7362–7374. doi: 10.1128/mcb.17.12.7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Mol Cell Biol. 2001a;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Woo D, Roussel MF, Cleveland JL. Mol Cell Biol. 2001b;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanidi A, Harrington EA, Evan GI. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, Albassam M, Zheng X, Leopold WR, Pryer NK, Toogood PL. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- Gladden AB, Diehl JA. J Biol Chem. 2003;278:9754–9760. doi: 10.1074/jbc.M212088200. [DOI] [PubMed] [Google Scholar]
- Gronbaek K, Worm J, Ralfkiaer E, Ahrenkiel V, Hokland P, Guldberg P. Blood. 2002;100:1430–1437. doi: 10.1182/blood-2002-02-0382. [DOI] [PubMed] [Google Scholar]
- Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Hernandez L, Hernandez S, Bea S, Pinyol M, Ferrer A, Bosch F, Nadal A, Fernandez PL, Palacin A, Montserrat E, Campo E. Leukemia. 1999;13:2087–2093. doi: 10.1038/sj.leu.2401599. [DOI] [PubMed] [Google Scholar]
- Hosokawa Y, Joh T, Maeda Y, Arnold A, Seto M. Int J Cancer. 1999;81:616–619. doi: 10.1002/(sici)1097-0215(19990517)81:4<616::aid-ijc18>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Howe D, Lynas C. Haematologica. 2001;86:563–569. [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lai R, McDonnell TJ, O'Connor SL, Medeiros LJ, Oudat R, Keating M, Morgan MB, Curiel TJ, Ford RJ. Leuk Res. 2002;26:849–855. doi: 10.1016/s0145-2126(02)00013-9. [DOI] [PubMed] [Google Scholar]
- Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Embo J. 1994a;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovec H, Sewing A, Lucibello FC, Muller R, Moroy T. Oncogene. 1994b;9:323–326. [PubMed] [Google Scholar]
- Lu F, Gladden AB, Diehl JA. Cancer Res. 2003;63:7056–7061. [PubMed] [Google Scholar]
- Martinez N, Camacho FI, Algara P, Rodriguez A, Dopazo A, Ruiz-Ballesteros E, Martin P, Martinez-Climent JA, Garcia-Conde J, Menarguez J, Solano F, Mollejo M, Piris MA. Cancer Res. 2003;63:8226–8232. [PubMed] [Google Scholar]
- Matsushime H, Quelle DE, Shurtleff SA, Shibuya M, Sherr CJ, Kato JY. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy B, Lundan T, Larramendy ML, Aalto Y, Zhu Y, Niini T, Edgren H, Ferrer A, Vilpo J, Elonen E, Vettenranta K, Franssila K, Knuutila S. Br J Haematol. 2003;120:434–441. doi: 10.1046/j.1365-2141.2003.04121.x. [DOI] [PubMed] [Google Scholar]
- Noelle RJ, Snow EC. Immunol Today. 1990;11:361–368. doi: 10.1016/0167-5699(90)90142-v. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Ashmun RA, Shurtleff SA, Kato JY, Bar-Sagi D, Roussel MF, Sherr CJ. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Seto M, Yamamoto K, Iida S, Akao Y, Utsumi KR, Kubonishi I, Miyoshi I, Ohtsuki T, Yawata Y, Namba M, et al. Oncogene. 1992;7:1401–1416. [PubMed] [Google Scholar]
- Sherr CJ. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- Sofer-Levi Y, Resnitzky D. Oncogene. 1996;13:2431–2437. [PubMed] [Google Scholar]
- Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, Senderowicz AM, Knudsen ES. J Biol Chem. 2003;278:30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Cory S. Oncogene. 1993;8:1–9. [PubMed] [Google Scholar]
- Uchimaru K, Endo K, Fujinuma H, Zukerberg L, Arnold A, Motokura T. Jpn J Cancer Res. 1996;87:459–465. doi: 10.1111/j.1349-7006.1996.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hannon GJ, Beach D. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD, Craig RW. Blood. 2001;97:3902–3909. doi: 10.1182/blood.v97.12.3902. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]