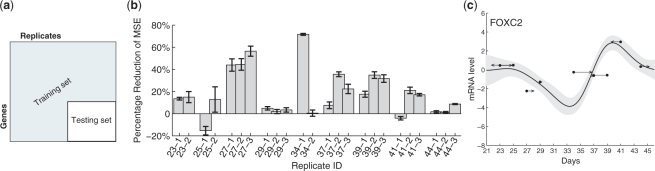
Fig. 6.
(a) Data partitioning for cross-validation. The testing data consists of the intersection of a subset of genes and of replicates; this allows us to predict the profile shape for all genes and time shift for all replicates while withholding unused data for testing. (b) Percent reduction in the average MSE when time shifts are included in the model. Results are grouped by replicate and found using cross-validation; see text. Most replicates have lower MSE (positive reduction) after introducing time shifts. (c) An example curve fit during cross-validation. The curve and time shifts are predicted using only the training data (data not shown); dots show the expression levels of the nine withheld test replicates (one at each time point) at their nominal times, and arrows show the estimated time shifts.