Abstract
Motivation: The rapid development of genotyping technology and extensive cataloguing of single nucleotide polymorphisms (SNPs) across the human genome have made genetic association studies the mainstream for gene mapping of complex human diseases. For many diseases, the most practical approach is the population-based design with unrelated individuals. Although having the advantages of easier sample collection and greater power than family-based designs, unrecognized population stratification in the study samples can lead to both false-positive and false-negative findings and might obscure the true association signals if not appropriately corrected.
Methods: We report PHYLOSTRAT, a new method that corrects for population stratification by combining phylogeny constructed from SNP genotypes and principal coordinates from multi-dimensional scaling (MDS) analysis. This hybrid approach efficiently captures both discrete and admixed population structures.
Results: By extensive simulations, the analysis of a synthetic genome-wide association dataset created using data from the Human Genome Diversity Project, and the analysis of a lactase-height dataset, we show that our method can correct for population stratification more efficiently than several existing population stratification correction methods, including EIGENSTRAT, a hybrid approach based on MDS and clustering, and STRATSCORE , in terms of requiring fewer random SNPs for inference of population structure. By combining the flexibility and hierarchical nature of phylogenetic trees with the advantage of representing admixture using MDS, our hybrid approach can capture the complex population structures in human populations effectively.
Software Availability: Codes can be downloaded from http://people.pcbi.upenn.edu/∼lswang/phylostrat/
Contact: mingyao@upenn.edu; iswang@upenn.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The rapid development of genotyping technology and extensive cataloguing of single nucleotide polymorphisms (SNPs) across the human genome have made genetic association studies the mainstream for gene mapping of complex human diseases. For many diseases, the most practical approach is the population-based design with unrelated individuals. Although having the advantages of easier sample collection and greater power than family-based designs, population-based design is prone to population stratification (Marchini et al., 2004). Population stratification refers to the presence of a systematic difference in allele frequencies between subpopulations in a study due to ancestry difference between study subjects. Unrecognized population stratification can lead to both false-positive and false-negative findings and can obscure the true association signals if not appropriately corrected.
There are three types of population structures that might be observed in genetic association studies: discrete population structure consists of populations that are remotely related (such as Europeans, Africans and Asians) and the population structure is easy to discern as the individuals are clearly separated. Admixed population structure consists of subjects of admixed ancestry (such as African Americans and Hispanic Americans) with different individuals having different degrees of admixture, and that cannot be separated into discrete clusters. Intercontinental gradients can also be considered as admixed, although the degree of admixture is smaller than African Americans and Hispanic Americans. In other scenarios, we may see hierarchical population structure that consists of both discrete and admixed population structures. Hierarchical population structures may be seen in studies that involve multi-ethnic cohorts, which are becoming increasingly common in genetics consortiums (Serre et al., 2008).
Recognizing the issue of population stratification induced by population structures, various methods have been developed to control for population stratification. Two early approaches are genomic control (Devlin and Roeder, 1999) and structured association (Pritchard et al., 2000). The genomic control method corrects for stratification by adjusting association statistics with an overall inflation factor obtained from a set of random markers that are not associated with the phenotypes of interest. However, some markers differ in their allele frequencies across ancestral populations more than others. Thus, the uniform adjustment may be insufficient at markers having strong differentiation across ancestral populations and may be superfluous at markers lacking such differentiation. Structured association uses STRUCTURE program (Pritchard and Rosenberg, 1999) to assign the study subjects to discrete subpopulations and then aggregates evidence of association within each subpopulation. This method is computationally intensive, and assignments of subjects to clusters are sensitive to the number of clusters, which is often ill defined in many real studies.
The current state-of-the-art approach for the correction of population stratification is EIGENSTRAT (Price et al., 2006), which computes principal components for SNPs across the genome to identify population structure. In this approach, a small number of ‘top’ principal components will capture the main axes of genetic variation in the study subjects. Correction for population stratification is carried out by including these top principal components as covariates in a regression framework. Although popular, several studies have demonstrated that EIGENSTRAT will fail to correct for population stratification in certain scenarios (Epstein et al., 2007; Kimmel et al., 2007; Luca et al., 2008).
Recently, Li and Yu (2008) proposed an extension of EIGENSTRAT by incorporating cluster information obtained from multi-dimensional scaling (MDS) analysis as additional covariates in the adjustment. This MDS clustering approach tries to identify both discrete and admixed patterns of genetic variation and correct for their potential confounding effects by adjusting each position of subject along identified axes of genetic variation and the cluster membership simultaneously. This method is a direct extension of EIGENSTRAT when the metric used by EIGENSTRAT is adopted for measuring the genetic correlation between two subjects. Simulation results demonstrate that the MDS clustering method provides a more appropriate correction for population stratification than EIGENSTRAT under simulation settings that they considered (Li and Yu, 2008).
The goal of this article is to utilize phylogenetic trees to correct for population stratification in genetic association studies with unrelated individuals. Widely used in the study of evolution and other fields of biology, a phylogeny represents the evolutionary relationship between species as a tree structure, where each leaf is an observed species, each internal node corresponds to the most recent common ancestors of all species below it and each internal edge represents a bipartition of leaves due to evolutionary divergence. The use of phylogenetics in the study of human genetic diversity has a long history dating back to the sixties: the approach was first proposed by Cavalli-Sforza and Edwards (1967) who developed likelihood estimation methods using allelic frequencies in subpopulations. Distance measures that adjust allelic frequencies are also available (Goldstein et al., 1995; Nei, 1972). The phylogenetic approach has been used extensively in the analysis of the Human Genome Diversity Project (HGDP) (http://www.stanford.edu/group/morrinst/hgdp.html), which genotyped more than 1000 individuals collected at more than 50 geographic locations around the world. The HGDP dataset has well-defined subpopulations and complete population identification. The resulting phylogenies, using microsatellites and SNPs, have been highly consistent and compatible with the widely accepted pattern of human migration (Jakobsson et al., 2008; Li et al., 2008; Rosenberg et al., 2002).
We propose a phylogenetics-based approach for correction of population stratification based on several motivations. First, widely used and thoroughly tested for decades to study evolution, phylogenetics is a natural choice for detecting divergence in human subpopulations. Phylogenetic analysis of the HGDP dataset shows that this approach is robust and sensitive to subtle population structures (Jakobsson et al., 2008; Li et al., 2008). Second, thanks to decades of work by an active research community, many phylogenetic reconstruction algorithms have been developed, and efficient and versatile programs are widely available. Third, the hierarchical characteristic of a phylogeny is easy to interpret and visualize. Finally, phylogenetic trees are more flexible to represent highly complex spatial structures than clusters obtained from clustering-based algorithms (Li and Yu, 2008).
To correct for discrete, admixed and hierarchical population structures, we propose to combine information from phylogeny and MDS together with the phylogenetic tree representing discrete population structure and the principal coordinates of MDS analysis representing admixed population structure. Given the flexible nature and the hierarchical characteristic of phylogenetic trees, we expect this hybrid approach to perform well under complex population structures, such as those from multi-ethnic studies with some of the study samples showing admixed population structure, whereas other study samples showing discrete population structure (Serre et al., 2008).
2 METHODS
2.1 Construction of a phylogenetic tree
The first step in our method is to construct a phylogenetic tree of subjects from the genetic marker data. We opt to use the distance-based approach, which accepts as input a distance matrix, i.e. the pairwise dissimilarities between every pair of individuals based on SNP genotypes and constructs trees entirely from the distance matrix (Saitou and Nei, 1987; Studier and Keppler, 1988). In our analysis, we code the SNP genotypes as 0, 1 and 2, representing the number of minor alleles, and calculate the distance as ‘2—the number of alleles that are identical by state’ between two individuals. We then built the phylogenetic tree using the FastME algorithm (Desper and Gascuel, 2002), a very fast distance-based phylogeny reconstruction algorithm that shows better topological accuracy than the commonly used neighbor joining algorithm (Saitou and Nei, 1987) in simulation studies. Compared with other more computationally intensive methods such as maximum likelihood or maximum parsimony, the distance-based approach is much more tractable, especially when the number of individuals or the number of markers becomes large. Note that the leaves of the constructed phylogenetic tree are individual subjects in the study sample instead of subpopulations.
2.2 Reduced representation for the phylogenetic tree
We reduce the phylogenetic structure into a collection of bipartitions on subjects in order for us to incorporate information from the phylogeny in the association tests as covariates in a regression framework. Each bipartition corresponds to an internal edge in a phylogenetic tree, which divides the data into two groups: a phylogenetic tree with n leaves (subjects) will have up to n−3 internal edges; in turn, the entire phylogenetic tree can be fully recovered by these (n−3) bipartitions. For each bipartition, we can construct a 0-1 vector indicating the bipartition membership by assigning all members from one of the two subsets in a bipartition to have value 0, and all members from the other set to have value 1. We cannot use all of the bipartitions in the regression analysis, because (i) the degrees of freedom in the regression is close to the number of observations (subjects) and significance will never be reached; (ii) most bipartitions, in particular, bipartitions that separate a very small number of subjects from the rest, are not informative for the purpose of population stratification correction; and (iii) the bipartitions are not entirely independent; for example, bipartitions for adjacent edges may only differ by a small number of subjects. We select a subset of representative bipartitions based on the following criteria:
The relative size of either side of a selected bipartition is above some given threshold. We used 2.5% in our analysis.
Filter bipartitions by the correlation threshold so that each remaining bipartition is correlated with at least one selected bipartition. We used 0.7 as the correlation threshold for discrete population structures, and 0.1 for admixed population structures in the simulation studies. For the analysis of the HGDP data and the lactase-height data, we also used 0.7 as the threshold. We discuss the selection of correlation threshold in Section 4.
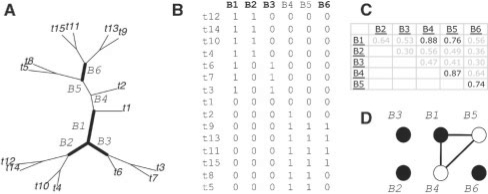
The bipartition selection problem can be formulated as the NP-hard minimum dominating set problem in graph theory as follows (Garey and Johnson, 1979): let G=(V, E) be a graph where each vertex in the vertex set V corresponds to a bipartition passing the size threshold. We add an edge between any two vertices (u, v) to the edge set E if the absolute value of the correlation between bipartitions associated with u and v is above the threshold. The minimum dominating set problem finds a smallest set of vertices V′⊆V such that for every vertex u∈V′−V, there exists a vertex v in V′ such that (u, v)∈E. Our problem can then be solved by finding such a minimum dominating set for G; then every bipartition is correlated with at least one bipartition associated with a vertex in the dominating set. To select bipartitions, we implemented a simple approximation algorithm [where the size of the solution is at most 1 + ln the size of the optimal solution (Garey and Johnson, 1979)] by iteratively (i) selecting a vertex with largest degree and add to the cover, then (ii) removing all adjacent vertices and their incident edges from the graph. The selection step is repeated until all vertices are removed. Figure 1 illustrates the bipartition selection procedure.
Fig. 1.
Illustration of the bipartition selection procedure. (A) A phylogeny with 15 leaves. (B) The matrix of six bipartitions with size threshold of at least three leaves on either side of the bipartition. (C) The pairwise absolute correlation matrix. (D) The corresponding instance to the minimum dominating set problem using correlation threshold 0.7. Black vertices (1, 2, 3, 6) form the solution returned by the greedy algorithm; bipartitions corresponding to the four vertices are also emboldened in the phylogeny (A) and labeled in boldface in the matrix of bipartitions (B). Note that because each vertex in (D) is adjacent to at least one black vertex, this ensures each of the six bipartitions is correlated with at least one of the four chosen bipartitions, so the reduced set of bipartitions still represents the topology of the original phylogeny well.
2.3 MDS analysis
MDS is a statistical technique that aims at displaying the similarity of members of a set of objects. This technique starts with a matrix of similarities or dissimilarities between a set of observations, and embeds the observations as points in a low-dimensional Euclidean space so that Euclidean distances between points in the plot are close to the original dissimilarities. Suppose that T is a (n × n) positive-semidefinite symmetric matrix of similarities among a set of n observations. From the spectral decomposition of T, we have
where τ1≥τ2≥…≥τn are the eigen values of T and b1, b2,…, bn are the corresponding eigen vectors. Alternatively, this may be written as
where cj=τj1/2bj, j=1, 2,…, n. Now consider the n observations as points in n dimensional space with the j-th coordinate for the i-th observation equal to cij, the i-th element of cj. With this geometric interpretation of the n observations, the Euclidean distance between the h-th and i-th observations is
MDS analysis attempts to find the optional q dimensional (q<n) approximation to the n dimensional representation so that the distance is preserved.
2.4 Association test with adjustment of phylogenetic bipartitions and principal coordinates from MDS analysis
The phylogenetic bipartitions can effectively capture the population structure information in a dataset when the structure is discrete or hierarchical. On the other hand, some of the populations, such as admixed populations, may contain continuous patterns of genetic variation. To incorporate both types of population structures, similar to MDS clustering, a hybrid approach proposed by Li and Yu (2008), we adjust for the population structure by the phylogenetic bipartitions and the principal coordinates from MDS analysis. This is done by representing the population structure as a collection of selected bipartitions and principal coordinates from the MDS analysis and introducing them together with the genotypes of the SNP as independent variables, and the binary phenotype as the dependent variable in a regression framework. To test for genetic association for case–control studies, we conduct logistic regression with the following model:
where Y represents disease status (1: affected; 0: unaffected), Ψ represents the selected phylogenetic bipartitions, Γ represents the selected principal coordinates from the MDS analysis, X represents a set of random markers that are used for inference of population structure and g is the genotype score for the testing SNP, with (0, 1, 2) for a multiplicative model, (0, 1, 1) for a dominant model and (0, 0, 1) for a recessive model. To test for association between the SNP and the disease status, either a likelihood ratio test, a score test, or a Wald test can be carried out. For continuous phenotypes, we can conduct linear regression with similar adjustments. We note that principal components analysis (PCA) can be used instead of MDS and the computation for PCA is faster than similarity matrix. We choose to use MDS because the similarity matrix is already calculated when constructing the phylogenetic tree; moreover, the results of MDS and PCA are generally similar.
Note that the selected phylogenetic bipartitions and principal coordinates might be correlated because both of them represent population structure in the data, although with emphasis on different types of variation. To avoid the issue of multi-collinearity, when the correlation between a phylogenetic bipartition and a principal coordinate is greater than 0.7, we will keep only the principal coordinate in the regression model. It is worth noting that multi-collinearity is also a concern for the MDS clustering approach. Similar to what we did for PHYLOSTRAT, when testing the MDS clustering method by Li and Yu (2008), we also used 0.7 as the correlation cut-off point to remove a correlated cluster dummy variable.
2.5 Simulation set up
We conducted extensive simulations under various settings to compare the performance of PHYLOSTRAT with four other methods, including the standard Cochran–Armitage trend test without corrections (Armitage, 1955), the EIGENSTRAT approach (Price et al., 2006), the MDS clustering approach (Li and Yu, 2008) and the STRATSCORE approach (Epstein et al., 2007). We designed three sets of simulations, one for discrete population structures, one for admixed populations and the other based on genotype data from the HGDP samples (Li et al., 2008). In all scenarios, we coded the genotype score for the testing SNP assuming a multiplicative model. In the four methods we tested, either top 10 principal components (EIGENSTRAT and STRATSCORE) or top 10 principal coordinates (MDS clustering and PHYLOSTRAT) were included in the analysis.
Setting 1: Discrete population structures were simulated in a similar method as Price et al. (2006) and Li and Yu (2008). We considered six configurations (Table 1), representing two, three and four subpopulations. In each setting, we generated simulated datasets each consisting of 500 cases and 500 controls, with varying numbers of random SNPs carrying information capable of differentiating among populations. To generate genotypes for the random SNPs, we followed the algorithm of Price et al. (2006). Specifically, for each subpopulation, the allele frequency for each SNP was generated using the Balding–Nichols model (Balding and Nichols, 1995) using FST=0.01. For each SNP, an ancestral population allele frequency p was drawn from the uniform distribution on (0.1, 0.9). The allele frequencies for each subpopulation were drawn from β-distribution with parameters p(1 − FST)/FST and (1 − p)(1 − FST)/FST. This distribution has mean p and variance FSTp(1 − p).
Table 1.
Population stratification configurations for discrete population structure
| Configuration | Group | Population 1 | Population 2 | Population 3 | Population 4 |
|---|---|---|---|---|---|
| C1 | Case | 200 | 300 | ||
| Control | 300 | 200 | |||
| C2 | Case | 250 | 250 | ||
| Control | 0 | 500 | |||
| C3 | Case | 225 | 175 | 100 | |
| control | 175 | 100 | 225 | ||
| C4 | Case | 165 | 335 | 0 | |
| Control | 0 | 165 | 335 | ||
| C5 | Case | 175 | 150 | 100 | 50 |
| Control | 75 | 100 | 150 | 175 | |
| C6 | Case | 125 | 125 | 250 | 0 |
| Control | 0 | 250 | 125 | 125 |
Configurations C1 and C2 contain two subpopulations; configurations C3 and C4 contain three subpopulations; and configurations C5 and C6 contain four subpopulations.
To evaluate the performance of different population stratification correction methods, we considered four categories of testing SNPs: (i) the first category (random SNPs without association with disease) was generated the same way as those SNPs chosen for detecting population structure, i.e. FST=0.01. (ii) For the second category (differentiated SNPs without association with disease), we assumed a large allele frequency difference between the subpopulations. More specifically, for C1 and C2, we chose allele frequencies of 0.2 for population 1 and 0.8 for population 2; for C3 and C4, the allele frequencies are 0.8, 0.8 and 0.2, respectively, for the three subpopulations; and for C5 and C6, the allele frequencies are 0.2, 0.8, 0.2 and 0.8, respectively, for the four subpopulations. (iii) The third category of SNPs (random causal SNPs with association with disease) was for power evaluation in which the allele frequency was generated the same way as those random SNPs, i.e. FST=0.01; we then assumed a multiplicative model with a genotype relative risk of 1.5 for the causal allele to generate genotypes for the causal SNPs conditioned on the disease status. (iv) The fourth category of SNPs (differentiated causal SNPs with association with disease) was generated in a similar fashion as category (iii) except that the marker allele frequency was generated the same way as those from category (ii). Testing SNPs in categories (i) and (ii) allow us to evaluate type I error rates of different methods, whereas testing SNPs in categories (iii) and (iv) allow us to evaluate the power when modest or extreme population stratifications are present.
To evaluate the type I error and power under each population structure, we generated 100 datasets of 500 cases and 500 controls with each dataset consisting of 1000 testing SNPs for each of the above-mentioned four SNP categories. Moreover, for each of the 100 datasets, we also simulated m=100, 300 or 500 random SNPs with FST=0.01 to infer population structure. Type I error and power were estimated based on 100 (datasets) × 1000 (testing SNPs) = 100 000 tests.
Setting 2: Cases and controls from an admixed population were simulated similar to that as described by Price et al. (2006). Disease status for individuals with ancestry proportions a from population 1 and (1 − a) from population 2 were simulated using disease risk proportional to ra, where r is the ancestry risk and a is uniformly distributed on (0, 1). To insure an average value of 0.5 across possible values of a, the probability of being affected was set to 0.5 log(r)ra/(r − 1). The risk model with a genotype relative risk of 1.5 for the disease allele was implemented the same way as discrete populations, with allele frequency ap1 + (1 − a)p2. Similar to the simulations for discrete populations, we also considered two categories of SNPs to evaluate the type I errors and two categories of SNPs to evaluate the power.
Setting 3: A case–control dataset of hierarchical population structure was simulated based on individuals genotyped in HGDP (Li et al., 2008), an international project for studying the diversity and unity of the entire human population. A total of 1064 individuals in this project, representing individuals from 51 populations from sub-Saharan Africa, North Africa, Europe, the Middle East, South/Central Asia, East Asia, Oceania and the Americas, were genotyped by the Illumina HumanHap 650K SNP array, which includes 650 000 SNPs. We downloaded the genotype data and sample description information from http://hagsc.org/hgdp/files.html.
After merging genotype data and sample description and data cleaning, 995 individuals remained for analysis. To obtain case–control data with population structure, we created three artificial subpopulations: (i) subpopulation 1 consists of 107 individuals from Africa, (ii) subpopulation 2 consists of 170 individuals from Middle East and 153 individuals from Europe; and (iii) subpopulation 3 consists of 107 individuals from America, 192 individuals from Central South Asia, 232 individuals from East Asia and 34 individuals from Oceania. For each of the three subpopulations, we selected cases and controls based on the numbers specified in Table 2. To infer population structure, we randomly selected 10 000 autosomal SNPs that have no missing genotypes and are in linkage equilibrium with each other (at r2 < 0.05). We then tested for association with the case–control status using 515 710 autosomal SNPs that satisfy the following quality control criteria: (i) minor allele frequency >1% in both cases and controls, (ii) Hardy–Weinberg equilibrium test P-value >1 × 10−7 and (iii) fraction of missingness <5%. For each method, we estimated the type I error rate at various levels (α = 0.01, 0.005, 0.001 and 0.0005 level) and calculated the genomic control inflation factor (Devlin and Roeder, 1999).
Table 2.
Population stratification configuration in the synthetic HGDP case–control dataset
| Population | Continent | Case (n=452) | Control (n=543) |
|---|---|---|---|
| 1 | Africa | Central African | |
| Republic (n = 29), | Namibia (n = 5) | ||
| Congo (n = 15) | Nigeria (n = 23) | ||
| Kenya (n = 12) | Senegal (n = 23) | ||
| 2 | Middle | Algeria-Mzab (n = 29), | France (n = 48) |
| East/Europe | Israel-Carmel (n = 45), | Italy (n = 35) | |
| Israel-Central (n = 49) | Italy-Bergamo (n = 12) | ||
| Israel-Negev (n = 47) | Orkney Islands (n = 16) | ||
| Russia (n = 25) | |||
| Russia-Caucasus (n = 17) | |||
| 3 | Central South | China (n = 10) | Cambodia (n = 11) |
| Asia/Oceania/ | Pakistan (n = 182) | China (n = 169) | |
| East Asia/ | Bougainville (n = 18) | Japan (n = 27) | |
| America | New Guinea (n = 16) | Siberia (n = 25) | |
| Brazil (n = 45) | |||
| Colombia (n = 13) | |||
| Mexico (n = 49) |
2.6 Application to lactase-height data
A classic way to test the performance of a population stratification correction method is to examine whether the method is able to remove the effect of population stratification between the lactase (LCT) gene and height (Campbell et al., 2005). As we do not have access to the original data reported by Campbell et al. (2005), we created a lactase-height dataset using data from PennCAC, an ongoing candidate-gene study on coronary artery classification that we are working on. PennCAC includes 1361 Caucasians with phenotypes on height. These individuals were genotyped using the ITMAT/Broad/CARe (IBC) 50K SNP array (Keating et al., 2008), which includes 1755 autosomal ancestry informative markers (AIMs) and 21 LCT SNPs. The originally reported LCT SNP, rs4988235, is not included in the IBC array, but two other LCT SNPs, rs3769005 and rs7579771, have strong LD with rs4988235 (r2 = 0.72 in HapMap CEU samples). We, therefore, tested association between height and these two SNPs. For all population stratification correction methods that we considered for comparison, we inferred population structure using 100, 200, 250, 300, 500, 700, 1000 and 1755 autosomal AIMs.
3 RESULTS
To evaluate our proposed method, we carried out extensive simulations, including discrete population structure, admixed population structure and a synthetic case–control GWAS dataset generated from the HGDP data, which represents hierarchical population structure. We assessed whether the proposed method can appropriately correct for population stratification by estimating type I error rate, and also assessed its power in detecting disease association. We compared PHYLOSTRAT with four other methods, including (i) the conventional Cochran–Armitage trend test (Armitage, 1955), which does not control for population stratification, (ii) the EIGENSTRAT approach (Price et al., 2006), (iii) the MDS clustering approach (Li and Yu, 2008); and (iv) the STRATSCORE approach (Epstein et al., 2007). For STRATSCORE, as suggested by the authors (Dr Michael Epstein, personal communication), we adjusted the continuous stratification scores (obtained from principal components) rather than the quartiles of the stratification scores because this modified version of STRATSCORE generally leads to smaller type I errors than the original method under the simulation settings we considered. We did not compare with the genomic control approach because Price et al. (2006) and Li and Yu (2008) have demonstrated its unsatisfactory performance. For type I error and power estimation, significance was evaluated at the 1% level.
3.1 Setting 1: discrete population structure
Tables 3–4 display the results for discrete population structures with two and four subpopulations, respectively. Results for three subpopulations are similar (Supplementary Table 1). In all situations we considered, PHYLOSTRAT has type I error rates that are close to the nominal level. The MDS clustering approach performs well when there are two discrete subpopulations; but when the number of discrete subpopulations is three or four and the population stratification is extreme, its type I error rates can be greater than the nominal level, especially when the number of random SNPs is small. For example, when there are four discrete subpopulations and when the degree of population stratification is extreme (i.e. configuration C6 in Table 4), the type I error rate of the MDS clustering approach can be as high as 10.19% with 100 random SNPs, 2.06% with 300 random SNPs, even with 500 random SNPs, the type I error rate is still slightly inflated. Similar results are observed for EIGENSTRAT, which also yields inflated type I errors. In contrast, for this extreme situation, the type I error rate of PHYLOSTRAT is close to the nominal level even with only 100 random SNPs, much less than the number of random SNPs required by other methods to achieve the same level of type I error rate. This implies that PHYLOSTRAT uses the ancestry information contained in the random SNPs more efficiently. We observed that STRATSCORE can yield either inflated or conservative type I errors. As suggested by the authors (Dr Glen Satten, personal communication), we also implemented a modified version of STRATSCORE with 20 strata. This modified version yields appropriate type I errors when there are two underlying subpopulations; however, when the number of subpopulations is greater than 2, e.g. configurations C4 and C6, the type I errors are still inflated even with 500 random SNPs. It is possible that more strata are needed to appropriately control the type I errors when the number of subpopulations is more than two and the degree of population stratification is extreme.
Table 3.
Empirical type I error rates (%) and power (%) under two discrete populations at 1% significance level
| Configuration | M | Trend | ES | PS | MC | SS | |
|---|---|---|---|---|---|---|---|
| Non-causal SNPs: random (category 1) | C1 | 100 | 45.61 | 1.04 | 1.08 | 1.01 | 0.85 |
| 300 | 45.41 | 0.98 | 1.06 | 1.13 | 0.93 | ||
| 500 | 45.53 | 1.03 | 1.04 | 0.95 | 0.8 | ||
| C2 | 100 | 76.82 | 1.72 | 1.09 | 1.01 | 2.48 | |
| 300 | 77.01 | 1.11 | 1.11 | 1.06 | 1.23 | ||
| 500 | 77.17 | 1 | 1.08 | 1.08 | 1.29 | ||
| Non-causal SNPs: highly differentiated (category 2) | C1 | 100 | 99.84 | 1.18 | 1.1 | 1.05 | 0.59 |
| 300 | 99.87 | 1.02 | 1.08 | 1.03 | 0.5 | ||
| 500 | 99.87 | 1 | 1 | 0.95 | 0.49 | ||
| C2 | 100 | 100 | 3.43 | 1.08 | 1.06 | 4.85 | |
| 300 | 100 | 1.29 | 1.09 | 1.02 | 1.43 | ||
| 500 | 100 | 1.08 | 1.05 | 0.96 | 1.23 | ||
| Causal SNPs: random (category 3) | C1 | 100 | 68.16 | 87.11 | 87.48 | 87.61 | 84.83 |
| 300 | 67.8 | 87.59 | 87.72 | 87.77 | 85.46 | ||
| 500 | 67.9 | 87.59 | 87.44 | 87.72 | 85.21 | ||
| C2 | 100 | 79.62 | 69.09 | 70.19 | 70.17 | 69.62 | |
| 300 | 79.67 | 69.44 | 70.32 | 70.15 | 70.05 | ||
| 500 | 79.6 | 69.25 | 70.09 | 70.47 | 70.01 | ||
| Causal SNPs: highly differentiated (category 4) | C1 | 100 | 23.56 | 75.27 | 82.3 | 82.56 | 67.44 |
| 300 | 23.6 | 80.81 | 82.18 | 82.53 | 72.98 | ||
| 500 | 23.91 | 81 | 81.99 | 82.05 | 73.72 | ||
| C2 | 100 | 100 | 22.15 | 55.17 | 55.15 | 21.29 | |
| 300 | 100 | 39.9 | 55.08 | 55.06 | 40.08 | ||
| 500 | 100 | 44.49 | 55.14 | 55.14 | 45.06 |
m is the number of random SNPs used for inference of population structure. ES: EIGENSTRAT; PS: PYLOSTRAT; MC: MDS clustering; SS: STRATSCORE.
Table 4.
Empirical type I error rates (%) and power (%) under four discrete populations at 1% significance level
| Configuration | m | Trend | ES | PS | MC | SS | |
|---|---|---|---|---|---|---|---|
| Non-causal SNPs: random (category 1) | C5 | 100 | 46.22 | 1.3 | 1.09 | 1.22 | 0.53 |
| 300 | 46.6 | 1.04 | 1.06 | 1.04 | 0.41 | ||
| 500 | 46.17 | 1.02 | 1.05 | 1.02 | 0.22 | ||
| C6 | 100 | 64.21 | 3.31 | 1.19 | 3.52 | 2.19 | |
| 300 | 64.09 | 1.18 | 1.06 | 1.42 | 0.87 | ||
| 500 | 64.11 | 1.11 | 1.05 | 1.15 | 0.75 | ||
| Non-causal SNPs: highly differentiated (category 2) | C5 | 100 | 34.56 | 1.14 | 1.07 | 1.16 | 0.97 |
| 300 | 34.41 | 0.96 | 1.04 | 1.03 | 0.93 | ||
| 500 | 34.39 | 1.02 | 0.98 | 1.04 | 0.92 | ||
| C6 | 100 | 100 | 13.45 | 1.21 | 10.19 | 6.67 | |
| 300 | 100 | 2.2 | 1.09 | 2.06 | 0.68 | ||
| 500 | 100 | 1.55 | 1.11 | 1.45 | 0.39 | ||
| Causal SNPs: random (category 3) | C5 | 100 | 64.42 | 86.82 | 86.42 | 85.67 | 74.42 |
| 300 | 64.24 | 86.06 | 86.33 | 86.28 | 74.73 | ||
| 500 | 64.02 | 86.34 | 86.46 | 86.49 | 74.91 | ||
| C6 | 100 | 70.54 | 70.14 | 71.86 | 71.11 | 62.16 | |
| 300 | 70.33 | 71.58 | 71.98 | 72.06 | 63.86 | ||
| 500 | 70.18 | 71.4 | 71.9 | 72.26 | 64.2 | ||
| Causal SNPs: highly differentiated (category 4) | C5 | 100 | 0.04 | 73.09 | 77.64 | 77.77 | 35.05 |
| 300 | 0.05 | 76.32 | 77.91 | 79.37 | 37.4 | ||
| 500 | 0.05 | 78.7 | 78.6 | 79.88 | 38.59 | ||
| C6 | 100 | 100 | 11.52 | 40.03 | 25.55 | 4.61 | |
| 300 | 100 | 39.06 | 51.27 | 39.49 | 20.77 | ||
| 500 | 100 | 46.78 | 52.45 | 52.18 | 28.08 |
m is the number of random SNPs used for inference of population structure. ES: EIGENSTRAT; PS: PHYLOSTRAT; MC: MDS clustering; SS: STRATSCORE.
For causal SNPs in category (iii), PHYLOSTRAT, MDS clustering and EIGENSTRAT yield similar power, but the power for STRATSCORE is generally lower than the other three methods. For causal SNPs in category (iv), PHYLOSTRAT is more powerful than the other three methods under several settings. For example, for configuration C6 in Table 4, with 300 random SNPs, the power for PHYLOSTRAT is 51.27%, whereas the powers for EIGENSTRAT, MDS clustering and STRATSCORE are 39.06, 39.49 and 20.77%, respectively. This is because EIGENSTRAT and MDS clustering cannot completely remove the confounding effect due to population stratification, which obscures the true association signal, whereas STRATSCORE has conservative type I error rate. For causal SNPs in category (iv), we observed noticeable power change for each method as the number of random SNPs, m, increases, and such power change is also due to difference in ability to remove the confounding effect of population stratification.
3.2 Setting 2: admixed population structure
Table 5 shows the results for admixed populations. We observed similar patterns for PHYLOSTRAT, MDS clustering, EIGENSTRAT and STRATSCORE. All these methods yield type I error rates that are close to the nominal level with STRATSCORE being slightly conservative. The power for detecting causal SNPs is similar for all methods.
Table 5.
Empirical type I error rates (%) and power (%) under admixed populations at 1% significance level
| r | m | Trend | ES | PS | MC | SS | |
|---|---|---|---|---|---|---|---|
| Non-causal SNPs: random (category 1) | 2 | 100 | 21.62 | 1.11 | 1.14 | 1.18 | 1.04 |
| 300 | 21.45 | 1.03 | 1.03 | 1.07 | 1.01 | ||
| 500 | 21.68 | 1.03 | 1.04 | 1.05 | 0.92 | ||
| 3 | 100 | 41.14 | 1.44 | 1.44 | 1.43 | 1.31 | |
| 300 | 41.23 | 1.15 | 1.14 | 1.15 | 0.99 | ||
| 500 | 41.06 | 0.97 | 0.99 | 0.98 | 0.91 | ||
| Non-causal SNPs: highly differentiated (category 2) | 2 | 100 | 62.79 | 1.43 | 1.42 | 1.46 | 1.2 |
| 300 | 62.71 | 1.09 | 1.11 | 1.12 | 0.83 | ||
| 500 | 62.92 | 1.06 | 1.05 | 1.02 | 0.83 | ||
| 3 | 100 | 97.99 | 2.01 | 2.04 | 2.07 | 1.89 | |
| 300 | 98.91 | 1.21 | 1.19 | 1.18 | 1.05 | ||
| 500 | 98.16 | 0.99 | 1.01 | 1 | 0.94 | ||
| Causal SNPs: random (category 3) | 2 | 100 | 74.02 | 90.72 | 90.62 | 90.75 | 90.36 |
| 300 | 74.46 | 91.25 | 91.19 | 91.31 | 90.77 | ||
| 500 | 74.37 | 91.22 | 91.14 | 91.24 | 90.77 | ||
| 3 | 100 | 68.21 | 88.62 | 88.45 | 88.61 | 88.27 | |
| 300 | 68.41 | 89.49 | 89.49 | 89.57 | 89.15 | ||
| 500 | 67.92 | 89.09 | 89.01 | 89.18 | 89.03 | ||
| Causal SNPs: highly differentiated (category 4) | 2 | 100 | 4.97 | 87.96 | 87.54 | 88.1 | 86.8 |
| 300 | 4.45 | 92.36 | 92.23 | 92.43 | 91.19 | ||
| 500 | 5.32 | 92.74 | 92.64 | 92.7 | 91.77 | ||
| 3 | 100 | 2.78 | 80.88 | 80.44 | 80.89 | 79.67 | |
| 300 | 3.06 | 88.83 | 88.56 | 88.82 | 87.89 | ||
| 500 | 2.83 | 90.63 | 90.47 | 90.63 | 89.6 |
m is the number of random SNPs used for inference of population structure. r is the ancestry risk between the two ancestral populations. ES: EIGENSTRAT; PS: PHYLOSTRAT; MC: MDS-clustering; SS: STRATSCORE.
3.3 Setting 3: HGDP data
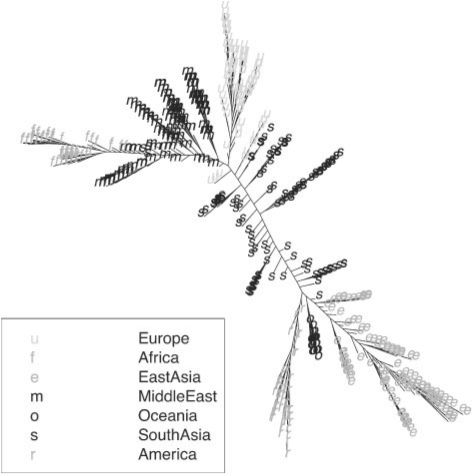
We applied our method to a dataset of 955 individuals from the HGDP data genotyped on the Illumina HumanHap 650 SNP array (Li et al., 2008). To simulate a dataset with population structure, we artificially created three populations (Table 2). The phylogenetic tree built from 10 000 randomly selected autosomal SNPs is shown in Figure 2. With the same 10 000 random SNPs, we also conducted MDS analysis and plotted the first two principal coordinates (Supplementary Fig. 1). As shown in both figures, this synthetic GWAS dataset contains both discrete and admixed population structures. The genomic control inflation factor for unadjusted trend test with 515 710 autosomal SNPs is 17.3, indicating strong population stratification in the data. Such a complex population structure poses a challenge to genetic association analysis, but also offers an opportunity to evaluate various methods.
Fig. 2.
HGDP phylogenetic tree based on 10 000 randomly selected autosomal SNPs.
We analyzed the 515 710 autosomal SNPs using EIGENSTRAT, MDS clustering, STRATSCORE and PHYLOSTRAT, and then estimated type I error rate for each method based on the 515 710 tests as none of the SNPs are associated with case–control status by design. At α = 0.01 significance level, the type I error rates of EIGENSTRAT, MDS clustering, STRATSCORE and PHYLOSTRAT are 0.0428, 0.0290, 0.0373 and 0.0232, respectively; when α = 0.001, the type I error rates of the four methods are 0.0087, 0.0047, 0.0067 and 0.0016, respectively; when α = 0.0005, the corresponding type I error rates are 0.0054, 0.0026, 0.0039 and 0.00048, respectively. We also estimated the genomic control inflation factor for each of the four methods: 1.66 for EIGENSTRAT, 1.45 for MDS clustering, 1.59 for STRATSCORE and 1.35 for PHYLOSTRAT. We also investigated the performance of these three methods with larger number of random SNPs (m = 30 000, 50 000 and 70 000), and obtained similar results. As the continent information is known for each individual, one might consider controlling population stratification by adjusting continent; however, the genomic control inflation factor for this simple approach is 2.67, much higher than the other three methods, suggesting that simply adjusting for continent is not sufficient. We also explored a hybrid approach by adjusting MDS principal coordinates and continent; the genomic control inflation factor for this approach is 1.45, similar to that of MDS clustering. Although none of the methods we considered completely corrected for population stratification for this complex synthetic GWAS dataset, PHYLOSTRAT did show more encouraging result than the other methods.
3.4 Analysis of lactase-height data
Using the naïve Armitage trend test, we observed significant associations between the LCT SNPs and height after adjusting sex (rs3769005: P-value=1.8 × 10−6; rs7579771: P-value=2.1 × 10−6), suggesting the presence of population stratification. We then analyzed this dataset with various population stratification correction methods with different numbers of AIMs (Table 6). We adjusted sex in all analyses.
Table 6.
P-values for analysis of the lactase-height dataset
| No. of AIMs | LCT SNP | ES | PS | MC | SS |
|---|---|---|---|---|---|
| 100 | rs3769005 | 0.0007 | 0.0006 | 0.0006 | 0.0001 |
| rs7579771 | 0.0008 | 0.0007 | 0.0008 | 0.0002 | |
| 200 | rs3769005 | 0.0007 | 0.0016 | 0.0009 | 0.0002 |
| rs7579771 | 0.0009 | 0.0019 | 0.0011 | 0.0002 | |
| 250 | rs3769005 | 0.0358 | 0.0506 | 0.045 | 0.0445 |
| rs7579771 | 0.0523 | 0.0676 | 0.0656 | 0.0506 | |
| 300 | rs3769005 | 0.1331 | 0.2612 | 0.1732 | 0.063 |
| rs7579771 | 0.1637 | 0.329 | 0.217 | 0.0707 | |
| 500 | rs3769005 | 0.0957 | 0.1726 | 0.2141 | 0.0798 |
| rs7579771 | 0.0843 | 0.1932 | 0.2487 | 0.0874 | |
| 700 | rs3769005 | 0.2278 | 0.3994 | 0.2437 | 0.0649 |
| rs7579771 | 0.2485 | 0.4021 | 0.2647 | 0.0699 | |
| 1000 | rs3769005 | 0.5407 | 0.3835 | 0.6098 | 0.1101 |
| rs7579771 | 0.5845 | 0.4075 | 0.647 | 0.1229 | |
| 1755 | rs3769005 | 0.3952 | 0.3739 | 0.3085 | 0.4807 |
| rs7579771 | 0.4261 | 0.4032 | 0.3279 | 0.4516 |
ES: EIGENSTRAT; PS: PHYLOSTRAT; MC: MDS clustering; SS: STRATSCORE.
Our results indicate that PHYLOSTRAT performs favorably against the other methods. For example, with 250 AIMs, among the four methods we considered for comparison, only PHYLOSTRAT yields P-values >0.05 for both SNPs; whereas the other three methods require more AIMs to remove the effect of population stratification. Our results are consistent with Price et al. (2006) and Epstein et al. (2007), who also observed that EIGENSTRAT cannot completely resolve the confounding issue between LCT and height when only a small number of AIMs were used in the analysis.
4 DISCUSSION
We have developed a new method to correct for population stratification in genetic association analysis by combining information obtained from phylogenetic trees and MDS analysis. Our method represents relations between individual's genetic background using a set of phylogenetic bipartitions and principal coordinates from MDS analysis, and incorporates them as covariates in a regression framework to adjust for the confounding effect due to hidden population stratification.
As shown in our simulations, this hybrid approach effectively captures both discrete and admixed population structures. It yields a more appropriate correction for population stratification than EIGENSTRAT (Price et al., 2006), MDS clustering (Li and Yu, 2008) and STRATSCORE (Epstein et al., 2007) under discrete population structures; its performance is similar to these three approaches under admixed population structures. To evaluate the performance of our method when the population structure is hierarchical, we applied our method to the HGDP dataset, which contains real genetic variation patterns. Although none of the methods could completely remove the confounding effect of population stratification, PHYLOSTRAT performs favorably against the other methods and yields type I error rates that are closer to the nominal level. To test the performance of our method in real genetic association studies, we applied our method to a lactase-height dataset and found that PHYLOSTRAT is able to correct for population stratification with only 250 AIMs, smaller than the number of AIMs required by the other methods. Our results suggest that phylogenetics is a robust and useful tool for inferring complex population structures, and appropriate utilization of information captured by phylogenetics trees can help correct for population stratification in genetic association analysis, especially when the number of random SNPs is small.
We note that as the number of random SNPs used for inference of population structure increases (e.g. when m = 50 000), the type I errors of PHYLOSTRAT, EIGENSTRAT and MDS clustering are all close to the nominal level under simulation settings we considered, but the type I errors of STRATSCORE are conservative with the patterns similar to those seen in Tables 3–4 and Supplementary Table 1. These results suggest that when only a small number of random SNPs are available, one might consider using PHYLOSTRAT to control for population stratification, but when the number of random SNPs is large, EIGENSTRAT would be a preferable approach as it is computationally faster. It is worth noting that the bipartitions obtained from the phylogenetics tree can be used together with principal components as basis functions to build the stratification scores (Dr Glen Satten, personal communication), and such a modified version may improve the performance of STRATSCORE.
Our method shares similarity with another hybrid approach MDS clustering (Li and Yu, 2008) in that both methods use the principal coordinates from the MDS analysis to capture admixed population structure. To capture discrete population structure, the MDS clustering method assigns each individual a group membership based on clustering of the MDS principal coordinates, where the number of clusters is determined by the gap statistic (Tibshirani et al., 2001). However, the problem of estimating the number of clusters can be difficult, because in many situations there is no clear definition of a ‘cluster’. Moreover, for data that are not clearly separated into groups, e.g. when there are overlapping classes, determining the number of distinct clusters can be highly subjective. In contrast, the use of phylogenies in our method circumvents this problem because our method does not require such parameter to be predefined. It is worth noting that clustering is a degenerate or a simplified version of phylogenetic tree. Unlike clustering, which can only handle simple population structures, phylogenetic trees are suitable for handling more complex, hierarchical population structures as demonstrated in our analysis of the HGDP data.
Compared with existing methods for population stratification correction, our method has two distinct advantages. First, the phylogenetic approach allows better interpretation and visualization of the population structure in the study sample, especially when the data contain a hierarchical structure. Many tools have been developed for molecular phylogenetic studies since the fifties, and it will be an important research direction to find how these methods can be applied for the population stratification problem. Second, as shown in our simulations and the analysis of the lactase-height data, our method requires fewer markers to infer population structure than other existing methods; therefore our method is ideal for candidate gene studies or replication study of GWAS, in which a large number of random SNPs may not be available. Although we considered a small number of random SNPs in our simulations, our method can be applied to GWAS as demonstrated by the analysis of the HGDP dataset.
Our method has to make a decision on how a set of bipartitions is selected without losing too much information for subsequent association analysis. If the population has a small number of subpopulations, most bipartitions will be similar to one of the bipartitions selected by our bipartition reduction algorithm. Having too many bipartitions retained in the stratification correction procedure will introduce many partially correlated covariates in logistic regression, and can have an adverse effect on the numerical regression procedure; on the contrary, over-reduction of bipartitions may lead to loss of important information on the population structure. Both scenarios can hurt the performance of our algorithm. In our analysis, we tried various thresholds for bipartition selection. We found that using 2.5% as the threshold for the number of leaves and 0.7 as the threshold for the correlation coefficient generally leads to stable results for discrete population structures, and the corresponding thresholds are 2.5% and 0.1 for admixed population structures. In general, we recommend the users to try multiple sets of thresholds and select the threshold that gives the smallest genomic control inflation factor. We recognize that there are other methods such as bootstrapping that can reduce bipartitions in the phylogeny (Felsenstein, 1985); however, we expect these methods are very sensitive to any leaves (individuals) in the tree that cannot be properly placed, which we expect to happen very often in human genetic data. What methods best reduce correlated bipartitions and how the bipartition reduction step affects the stratification correction is an important future research direction.
For PHYLOSTRAT, the running time is dominated by the pairwise distance computation as FastME is a very fast algorithm with sub-quadratic asymptotic running time in practice. Because pairwise distance computation is more time-consuming than PCA-based approaches, the running time of PHYLOSTRAT is longer than EIGENSTRAT and STRATSCORE. For example, for a dataset consisting of 500 cases and 500 controls with 500 random SNPs and 1000 testing SNPs, using an Intel Xeon CPU (2.66 GHz, 8 GB RAM, linux), it took 9 s for EIGENTRAT, 14 s for STRATSCORE, 19 s for PHYLOSTRAT and 40 s for MDS clustering. However, given that high-performance and low-cost computational capabilities are easily accessible, the required computing time for pairwise distance matrix calculation in PHYLOSTRAT would be a small overhead. One possible approach to reduce the running time is to limit the number of markers by LD pruning. Our experience with the analysis of the HGDP data and several other GWAS datasets suggests that this approach generally works well in GWAS.
In summary, we have proposed a new method that combines information from phylogenetic tree and MDS together. Given the flexible nature and the hierarchical characteristic, this hybrid approach is expected to perform well under complex population structures. We expect our method will provide a useful tool in the analysis of both candidate gene and GWAS studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Michael Epstein, Glen Satten, Maja Bucan, Junhyong Kim and the late Richard Spielman for discussions. We also thank three anonymous reviewers for their helpful comments that greatly improved the manuscript.
Funding: Penn Genomics Frontier Institute (an internal grant to M.L. and L.-S.W.). National Institutes of Health (grant R01HG004517 to M.L.).
Conflict of Interest: none declared.
REFERENCES
- Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- Balding DJ, Nichols RA. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica. 1995;96:3–12. doi: 10.1007/BF01441146. [DOI] [PubMed] [Google Scholar]
- Campbell CD, et al. Demonstrating stratification in an European American population. Nat. Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards A.WF. Phylogenetic analysis: models and estimation procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J. Comput. Biol. 2002;19:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Epstein MP, et al. A simple and improved correction for population stratification in case-control studies. Am. J. Hum. Genet. 2007;80:921–930. doi: 10.1086/516842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Garey MR, Johnson DS. Computers and Intractability: A Guide to the Theory of NP-Completeness. New York, NY, USA: WH Freeman; 1979. p. 190. [Google Scholar]
- Goldstein DB, et al. An evaluation of genetic distances for use with microsatellite loci. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1002. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Keating BJ, et al. Concept, design and implementation of a cardiovascular gene-centric 50K SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel G, et al. A randomization test for controlling population stratification in whole-genome association studies. Am. J. Hum. Genet. 2007;81:895–905. doi: 10.1086/521372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yu K. Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet. Epid. 2008;32:215–226. doi: 10.1002/gepi.20296. [DOI] [PubMed] [Google Scholar]
- Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Luca D, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, et al. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Am. Naturalist. 1972;106:283–292. [Google Scholar]
- Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, et al. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Serre D, et al. Correction of population stratification in large multi-ethnic association studies. PLoS ONE. 2008;1:e1382. doi: 10.1371/journal.pone.0001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier JA, Keppler KL. A note on the neighbor-joining algorithm of Saitou and Nei. Mol. Biol. Evol. 1988;5:729–731. doi: 10.1093/oxfordjournals.molbev.a040527. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, et al. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. B. 2001;63:411–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.