Abstract
Motivation: Short sequence motifs are an important class of models in molecular biology, used most commonly for describing transcription factor binding site specificity patterns. High-throughput methods have been recently developed for detecting regulatory factor binding sites in vivo and in vitro and consequently high-quality binding site motif data are becoming available for increasing number of organisms and regulatory factors. Development of intuitive tools for the study of sequence motifs is therefore important.
iMotifs is a graphical motif analysis environment that allows visualization of annotated sequence motifs and scored motif hits in sequences. It also offers motif inference with the sensitive NestedMICA algorithm, as well as overrepresentation and pairwise motif matching capabilities. All of the analysis functionality is provided without the need to convert between file formats or learn different command line interfaces.
The application includes a bundled and graphically integrated version of the NestedMICA motif inference suite that has no outside dependencies. Problems associated with local deployment of software are therefore avoided.
Availability: iMotifs is licensed with the GNU Lesser General Public License v2.0 (LGPL 2.0). The software and its source is available at http://wiki.github.com/mz2/imotifs and can be run on Mac OS X Leopard (Intel/PowerPC). We also provide a cross-platform (Linux, OS X, Windows) LGPL 2.0 licensed library libxms for the Perl, Ruby, R and Objective-C programming languages for input and output of XMS formatted annotated sequence motif set files.
Contact: matias.piipari@gmail.com; imotifs@googlegroups.com
1 INTRODUCTION
Until recent years, studying sequence specificity of transcription factors systematically has been limited to a relatively small number of organisms and transcription factors. High-throughput protein–DNA interaction assays such as protein binding microarrays (Berger et al., 2006), bacterial one-hybrid screens (Meng et al., 2005), large ChIP-chip studies and advances in motif inference algorithms and tools has, however, caused an expansion of motif databases such as UNI-PROBE (Newburger and Bulyk, 2009), TRANSFAC (Matys et al., 2006) and JASPAR (Bryne, 2008).
Sequence motif analysis tools can be hard to deploy and use locally. Many commonly used software packages have therefore been made available as web applications (Mahony and Benos, 2007; Thomas-Chollier et al., 2008). Public servers can, however, be limited in the CPU time given to users which can rule out their use for large-scale studies. Data exchange and usability can also be a challenge. Therefore, we have created an OS X-based desktop software package for sequence motif analysis that is easy to install and update. Compared with previously published desktop-based cis-regulatory sequence analysis tools such as TOUCAN (Aerts et al., 2003) or Sockeye (Montgomery et al., 2004), iMotifs is more focused on visualization and computation of sequence motifs, although it also supports visualizing scored motif matches in sequences.
1.1 Features
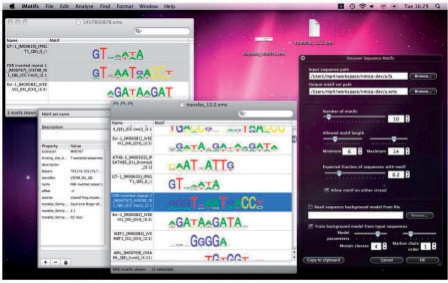
iMotifs is designed for visualization and analysis of cis-regulatory motifs and sequences. It can be used to retrieve sequences (e.g. for a coregulated group of genes), infer cis-regulatory motifs from them and score sequences with motif models, visualize them and their scored matches and compare them against other motifs (Fig. 1 shows the core functionality). A tutorial is included on the web site for common tasks (see Availability). Motifs can be manipulated and moved between sets by dragging and dropping, and filtered using keyword searches. Summary statistics such as entropy, column count or distance from closest pair can also be shown alongside. Free form key-value pair metadata such as database identifiers, species or notes can be viewed and edited. PDF export and printing is available. Import and export of TRANSFAC formatted motif files is also possible.
Fig. 1.
iMotifs can present motif sets and alignments. It integrates with the OS X desktop's previewing functionality and includes a number of analysis tools including an integrated NestedMICA motif inference tool.
iMotifs can be used to retrieve sequences from the Ensembl database (Hubbard et al., 2009). The retrieved sequences can be aligned either to transcription start sites (putative promoter sequence) or ends (e.g. for micro-RNA seed finding), and they can be filtered by gene identifiers. The retrieval tool can fetch specific sequence regions using GFF formatted annotation files, and includes specific support for ranking and retrieving regions of interest based on ChIP-seq ‘peaks’: MACS (Zhang et al., 2008), FindPeaks (Fejes et al., 2008) and SWEMBL formats are supported. Sequences are optionally processed to mask repeats and translated sequence.
iMotifs supports the quick previewing and thumbnailing service native to OS X (QuickLook). Previewing is especially useful for browsing sequence motif sets stored remotely (e.g. on a remote cluster) as no manual transfer or file opening is needed. An automated software update mechanism is included.
Many common motif analysis tasks are supported. These include finding closest matching and reciprocally matching motif pairs between two motif sets with the distance metric and algorithm described in Down et al. (2007). Motif multiple alignments can be visualized and computed with a greedy gapless motif multiple alignment algorithm. Motif inference experiments can be run with the integrated NestedMICA (Down and Hubbard, 2005) tool simply by dragging FASTA formatted sequence files to iMotifs. Downstream analyses such as motif scanning, overrepresentation analysis and motif hit score cutoff assignment as described in (Down et al., 2007) are also possible. Analysis tasks are run in parallel without blocking the user interacting with the application.
1.2 Interoperability
Although iMotifs itself works only on computers running Mac OS X, the analysis tools developed for and included in iMotifs are cross-plaform (Java based) and depend only on libraries included with the package. Most analysis functions are implemented by stand-alone command-line programs. This makes it possible to rapidly integrate unmodified tools into iMotifs. The included analysis tools can also be run on any UNIX system without iMotifs.
We feel that the use of a standard format for exchanging sequence motif data is beneficial for the research community, given the literally hundreds of motif inference tools and databases that are available [reviewed in Das and Dai (2007)]. To encourage the take up of a standard file format for motifs, we provide a programming interface for the input and output of the annotated motif file format XMS for the Perl, Ruby, R and Objective-C languages. The Perl and R libraries can also be used to visualize sequence logos.
2 CONCLUSION
We have created an integrated desktop application for short sequence motif analysis. It incorporates visualization, inference, alignment and comparison tools. The application widens the user base of sequence motif analysis tools and can improve the productivity of researchers working with sequence motif data. We aim to integrate with more sequence motif analysis tools and web services and to develop further the already included basic protein motif visualization and inference support.
We also encourage the introduction of a standard format for exchange of sequence motif data by providing conversion utilities and an API for input and output of XMS motif set files for a number of common bioinformatics programming languages.
ACKNOWLEDGEMENTS
We would like to thank Leopold Parts, Marija Buljan and Stephen Pettitt for critical reading of the manuscript.
Funding: Wellcome Trust (077198/Z/05/Z).
Conflict of Interest: none declared.
REFERENCES
- Aerts S, et al. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC. Jaspar, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das MK, Dai H-K. A survey of dna motif finding algorithms. BMC Bioinformatics. 2007;8(Suppl. 7):S21. doi: 10.1186/1471-2105-8-S7-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, et al. Large-scale discovery of promoter motifs in drosophila melanogaster. PLoS Comput. Biol. 2007;3:e7. doi: 10.1371/journal.pcbi.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Hubbard T.JP. Nestedmica: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 2005;33:1445–1453. doi: 10.1093/nar/gki282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes AP, et al. Findpeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T.JP, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Benos PV. Stamp: a web tool for exploring dna-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. Transfac and its module transcompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, et al. A bacterial one-hybrid system for determining the dna-binding specificity of transcription factors. Nat. Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, et al. Sockeye: a 3d environment for comparative genomics. Genome Res. 2004;14:956–962. doi: 10.1101/gr.1890304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. Uniprobe: an online database of protein binding microarray data on protein-dna interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, et al. Rsat: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Model-based analysis of chip-seq (macs) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]