Abstract
Motivation: High-dimensional data are frequently generated in genome-wide association studies (GWAS) and other studies. It is important to identify features such as single nucleotide polymorphisms (SNPs) in GWAS that are associated with a disease. Random forests represent a very useful approach for this purpose, using a variable importance score. This importance score has several shortcomings. We propose an alternative importance measure to overcome those shortcomings.
Results: We characterized the effect of multiple SNPs under various models using our proposed importance measure in random forests, which uses maximal conditional chi-square (MCC) as a measure of association between a SNP and the trait conditional on other SNPs. Based on this importance measure, we employed a permutation test to estimate empirical P-values of SNPs. Our method was compared to a univariate test and the permutation test using the Gini and permutation importance. In simulation, the proposed method performed consistently superior to the other methods in identifying of risk SNPs. In a GWAS of age-related macular degeneration, the proposed method confirmed two significant SNPs (at the genome-wide adjusted level of 0.05). Further analysis showed that these two SNPs conformed with a heterogeneity model. Compared with the existing importance measures, the MCC importance measure is more sensitive to complex effects of risk SNPs by utilizing conditional information on different SNPs. The permutation test with the MCC importance measure provides an efficient way to identify candidate SNPs in GWAS and facilitates the understanding of the etiology between genetic variants and complex diseases.
Contact: heping.zhang@yale.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Successes of genome-wide association studies (GWAS) have demonstrated that single nucleotide polymorphisms (SNPs) can be used to identify genetic variants underlying complex diseases, such as age-related macular degeneration (AMD) and coronary artery disease (CAD) (Edwards et al., 2005; Haines et al., 2005; Helgadottir et al., 2007; Klein et al., 2005; McPherson et al., 2007; Samani et al., 2007). GWAS has emerged as the most effective study design for identifying candidate genes in the scenario of ‘common disease, common variant’ (CD–CV). With the advance of genotyping technology, the number of available SNPs in one assay has soared from hundreds of thousand to more than 1 million in the past few years, leading to an outburst of high-dimensional data.
On the contrary, statistical methods available for GWAS remain limited. Single marker univariate tests such as the chi-square test or likelihood ratio test are commonly used. While simple, the univariate tests have produced many successes in identifying genetic variants for complex diseases (Klein et al., 2005; Li et al., 2006), which some may regard as ‘low hanging fruits’. In reality, biological systems are more complex than single variants acting independently, and most likely, multiple genes may work together in a complex system. For example, in the case of CD–CV, multiple loci and environment factors may be involved, and the individual effects may not be large enough to be detectable with thousands of study subjects. Some approaches have been proposed for GWAS to consider the effects of multiple SNPs (Li et al., 2006; Zhang et al., 2008). But, there are practical limitations in those approaches. For example, SNPs interactions can be examined only under very limited configurations. Moreover, genetic variants may manifest different effects such as multiplicative and heterogeneity effects (Meng et al., 2009; Risch, 1990a, b), further complicating the analysis.
To meet the growing computational demand of the analytic methods, machine-learning approaches have attracted more attention in detecting significant SNPs in GWAS. For example, classification trees and forest-based methods (Breiman, 2001; Breiman et al., 1984; Zhang and Ye, 2008; Zhang et al., 2003) are powerful tools for identifying complex relationships between a trait and a large number of predictors, and these methods also have been found useful in the analysis of gene expression data (Bureau et al., 2005; Chen et al., 2007; Diaz-Uriarte and Alvarez de Andres, 2006; Ye et al., 2005; Zhang and Bonney, 2000). A random forest consists of many classification trees, and at each node of the trees, a small subset of randomly selected predictors, instead all predictors, are considered to split on the node. Within a random forest, the effect of a predictor is measured by either the permutation importance or Gini importance (Breiman, 2001). The Gini importance of a specific predictor directly sums the improvement of weighted Gini index when this variable is used for splitting a node among all trees in the forest (Friedman, 2001). The permutation importance of a predictor calculates the increase of the out-of-bag errors as a result of permuting the values of the predictor (Breiman, 2001), because the permutation destroys any potential predictive power of the predictor on the trait. Previous studies (Jiang et al., 2009; Wang et al., 2009) have demonstrated that the random forest-based approach is feasible and efficient for GWAS, although two major issues hamper further applications of random forests. Firstly, although large importance scores are often indicative of SNPs associated with the trait, it is usually not clear how large is large, and the importance score is not coupled with the rigorous statistical significance of the investigated SNPs. Secondly, both the permutation and Gini importance scores measure the ‘average’ effect of a predictor in a random forest, and can be easily altered by the presence of the other SNPs.
To overcome the problems stated above, we introduce an alternative importance score using maximal conditional chi-square (MCC) statistic to assess the conditional significance of SNPs in GWAS. For example, with two SNPs (A and B), if SNP B confounds the effect of SNP A on the trait, failure to control the effects of SNP B may lead to inefficient tests for SNP A. Like the Mantel–Haenszel test, a test by stratifying by SNP B is an effective approach to adjusting for the confounding effect of SNP B.
We make use of the hierarchical tree structure to assess the effect of SNP A through stratification of the SNPs that precede SNP A in splitting the nodes in a tree. A conditional chi-square statistic for SNP A can be calculated whenever it is used to split a node of any tree in a random forest. The maximal value among all these chi-squares calculated for SNP A is obtained after the construction of the random forest. This maximum indicates the relationship between the trait and the SNP given its preceding SNPs in the random forest, and can serve as an importance measure. The reason we select the largest chi-square statistic, instead of the average as used in the Gini and permutation importance, is that disease-associated SNPs are usually very rare in the data, and therefore, most of the conditional chi-square statistics come from the SNPs that are not associated with the trait. Thus averaging all chi-square values limits the sensitivity of the permutation test.
Based on the MCC importance score, we developed a permutation procedure to estimate the significance of each SNP. Simulated data sets based on various multiplicative and heterogeneity genetic models were generated to evaluate the performance of the proposed approach. We compared the results among Gini, permutation, and MCC importance scores. Finally, the proposed method was applied to a real data set for GWAS.
2 METHODS
2.1 Definition of MCC importance
Let Lij be the list of SNPs that precede SNP i in its split of the j-th node of a tree. Let Xij be the chi-square statistic resulting from the split of the j-th node from SNP i . Define set Si as:
| (1) |
where M is the number of SNPs and ni is the number of nodes split by SNP i in the forest.
The MCC importance of SNP i is then defined as:
| (2) |
Let mi be the node at which MCCi is reached. If it is not unique, we select the first one by starting from the root node and left to right. Let
| (3) |
We should note that when the node size is small, the chi-square statistic is not reliable. Thus we used the corrected chi-square statistic. In addition, we imposed a reasonable minimum size (5) on a node for splitting as typically done.
2.2 Random forest construction
Computing importance scores through permutation for the whole genome is theoretically possible but practically unrealistic. To overcome this computational issue, SNPs are screened using a single-marker analysis before being used in the construction of a random forest. Obviously, we need to choose a threshold for the screening. Because the concern is computational, the threshold can be relatively flexible based on the number of SNPs and computing capacity. Whenever feasible, we should try to be inclusive. For this report, we chose the threshold corresponding to the false discovery rate (FDR) below 0.75, which is a high threshold with a low chance of missing any important SNPs while enough to reduce the computational burden due to the much fewer number of SNPs considered. In each random forest, 1000 trees were generated, and the number of SNPs to be considered for splitting a node was set to be the square root of the total number of the post screening SNPs (Breiman, 2002).
2.3 Permutation test
Theoretical understanding of tree and forest based methods is known to be very difficult if not impossible. As a result, permutation procedures are commonly adopted to assess the significance level of a test (Chen et al., 2007; Rodenburg et al., 2008; Wang et al., 2009). In this study, we also applied permutation to estimate empirical P-values for MCC, Gini and permutation importance scores in random forests. After a random forest was built from the original data, the trait values were permuted randomly. Then, a new random forest was built and the importance values were recalculated for the permuted data set. The maximum importance value over all the SNPs in every permutation was recorded and thereby an empirical distribution of the maximum importance was estimated. We used this estimate to further assess the significance of each SNP in the data. To balance the heavy computational burden and the size of the probability to be estimated, we performed 1000 replications that seemed large enough to estimate the empirical P-value at significance levels of 0.01 and 0.05 in simulation studies. Others have taken similar strategies in related simulations (McDonough et al., 2009; Sohn et al., 2009). In a real analysis, more permutations could be carried out if necessary.
2.4 Genetic models for simulation
To reflect complex diseases under the CD–CV assumption, we adopted the genetic models studied by Lunetta et al. (2004) and Meng et al. (2009), which incorporate both genetic heterogeneity and multiplicative interactions in terms of penetrance factors defined by Risch (1990a and b). For example, in a simple two-locus model, denote the genotypes of the two SNPs (namely A and B) by Ai, i=0, 1, 2, and Bj, j=0, 1, 2, respectively, where i and j denotes the number of risk alleles, and p=(p0, p1, p2) and q=(q0, q1, q2) are penetrance factors for SNPs A and B, respectively. Then, for a multiplicative model, the penetrance of genotype AiBj is wij=piqj; for a heterogeneity model, wij=1−(1−pi)(1−qj)=pi+qj−piqj (Meng et al., 2009; Risch, 1990a, b).
For clarity, the penetrance factors for 0, 1 and 2 risk alleles are set the same at each risk SNP in each simulation model, namely q=(q0, q1, q2). Furthermore, in a combined heterogeneity and multiplicative model, define a multi-locus genotype as G=(g11, g12,…, gHM), where H denotes the number of heterogeneous model, M denotes the number of multiplicatively acting loci in each model, and g is denoting the number of risk alleles at each locus. Then the penetrance for genotype G is calculated as:
| (4) |
According to the notation in Lunetta et al. (2004) and Meng et al. (2009), each model is abbreviated as ‘HhMm’, where h is the number of heterogeneous groups and m is the number of multiplicatively acting loci in each group. These groups are sometimes referred to as genetic networks or systems. For example, the genetic model
H4M4 means that the model contains four heterogeneous groups each with four multiplicative risk SNPs (Table 1). As in real data, noise SNPs in linkage equilibrium with allele frequencies distributed uniformly (0.01, 0.99) were generated. For each simulated data set, 200 cases and 200 controls were generated with a total of either 100 or 1000 SNPs. See Table 1 for more details.
Table 1.
Genetic models used to simulate GWAS data
| Model | Risk alleles |
Penetrance factor |
|||||
|---|---|---|---|---|---|---|---|
| Total number | Heterogeneity alleles | Multiplicative alleles | Allele frequency | 0 | 1 | 2 | |
| H4M2 | 8 | 6 | 2 | 0.160 | 3.8E−4 | 0.5 | 1 |
| H4M4 | 16 | 12 | 4 | 0.282 | 1.2E−8 | 0.79 | 1 |
3 RESULTS
3.1 Performance evaluation
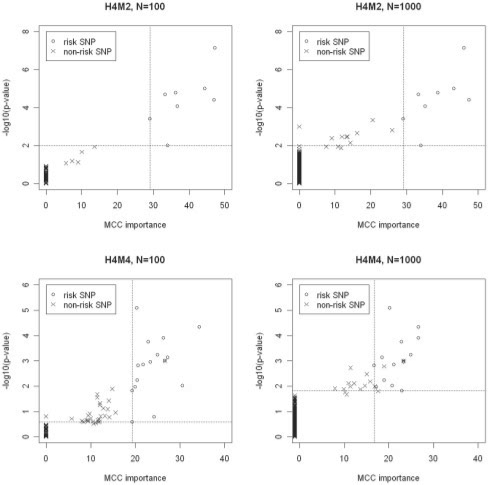
As a starting point, we analyzed the data generated by models H4M2 and H4M4 to show whether the MCC importance is a preferable statistic in association studies. Scatter plots of raw MCC importance scores and P-values from univariate tests (uncorrected for multiple comparisons) were calculated for these genetic models, as shown in Figure 1. The agreement between uncorrected single marker P-values and MCC importance scores for risk SNPs is apparent in Figure 1. While for most of the noise SNPs with single marker P-value ranging from 0.01 to 1, the corresponding MCC importance scores are 0, indicating that the MCC importance is robust to the abundant, non-informative SNPs in the data. At the same time, with the increase of complexity in the genetic models and decrease of the signal-to-noise ratio in data, the overlap in MCC between risk and noise SNPs is expected. As indicated by the vertical and horizontal lines, if we try to distinguish risk SNPs from noise SNPs, the MCC importance is far more effective than the single marker P-values (see Fig. 1). Thus, our results suggest that the MCC importance has gained discriminant power by incorporating conditional information in multiple SNPs.
Fig. 1.
Scatter plots of raw MCC importance scores and P-values from single-SNP tests in models H4M2 and H4M4.
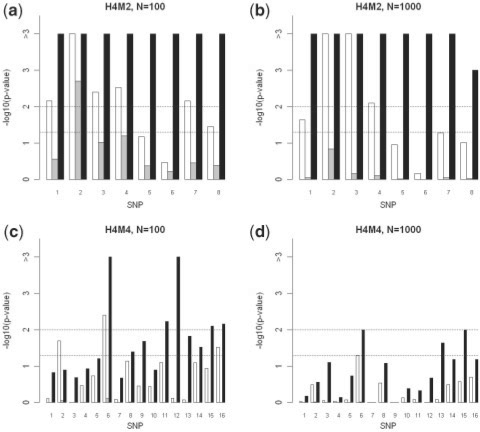
Next, we compared the empirical P-values obtained from the permutation test using different kinds of importance measures in random forests. Figure 2 illustrates the estimated genome-wide significance levels for all risk SNPs in the simulation data sets. For genetic model H4M2, both the MCC and permutation importance scores performed well in terms of identifying risk factors under the genome-wide 0.05 significance level (indicated by the horizontal line), although the MCC revealed much stronger evidence. The Gini importance, however, failed to identify seven out of eight risk SNPs with a genome-wide significance level of 0.05 when a total of 100 SNPs are tested. When a total of 1000 SNPs are included, none of the risk SNPs were identified. For the data sets generated from model H4M4 with more interactions, the MCC importance score consistently produced the best results. Moreover, we replicated the simulation 50 times and calculated the average of sensitivity and specificity under two thresholds, genome-wide significance levels of 0.05 and 0.01, based on the empirical P-values from the permutation test using different importance measures and univariate test (after Bonferroni correction for multiple comparisons) (shown in Table 2). All four methods investigated in this study have 100% specificity throughout all tests. This is a critical feature for GWAS as there are usually more than 100 k SNPs in one assay and most of them are non-informative. For the sensitivity, the permutation test using the Gini importance has the highest missing rate in all tests, even worse than the simple univariate test. The permutation test using the MCC importance is significantly (P < 10−4) superior to the other three tests, and for instance, increases the sensitivity by a range of 6–40% relative to the univariate test. These results underscore the usefulness of the MCC importance in identifying SNPs for complex diseases. We also performed a limited number of simulations using larger sample sizes. The results are shown in Supplementary Figure S1 and Supplementary Table S1, and confirm that the MCC importance is consistently better than other methods.
Fig. 2.
Empirical P-values of risk SNPs in models H4M2 and H4M4. White, grey and black bars represent the permutation importance, Gini importance and MCC importance scores, respectively. Two dash lines represent the genome-wide significance level of 0.05 and 0.01, respectively.
Table 2.
Comparison of prediction performance of different methods with different significance levels using models H4M2 and H4M4
| H4M2 |
H4M4 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
N=100 |
N=1000 |
N=100 |
N=1000 |
||||||
| sn (%) | sp (%) | sn (%) | sp (%) | sn (%) | sp (%) | sn (%) | sp (%) | ||
| Significant level 0.01 | MCC RF | 95.2*** | 100 | 68.0*** | 100 | 38.9*** | 100 | 9.4* | 100 |
| Permutation RF | 69.5 | 100 | 28.5 | 100 | 12.2 | 100 | 2.2 | 100 | |
| Gini RF | 27.2 | 100 | 13.0 | 100 | 3.0 | 100 | 2.0 | 100 | |
| Univariate | 55.0 | 100 | 27.5 | 100 | 7.6 | 100 | 2.8 | 100 | |
| Significant level 0.05 | MCC RF | 98.0** | 100 | 78.0*** | 100 | 53.1*** | 100 | 15.8** | 100 |
| Permutation RF | 85.0 | 100 | 47.2 | 100 | 21.9 | 100 | 5.2 | 100 | |
| Gini RF | 36.2 | 100 | 21.0 | 100 | 8.8 | 100 | 4.8 | 100 | |
| Univariate | 67.0 | 100 | 43.8 | 100 | 15.9 | 100 | 6.5 | 100 | |
sn: sensitivity; sp: specificity; N: number of SNPs.
Significance of paired t-test between MCC RF and the best among other three methods: *: <1E−2, **: <1E−5, ***: <1E−10.
3.2 Effect of linkage disequilibrium (LD)
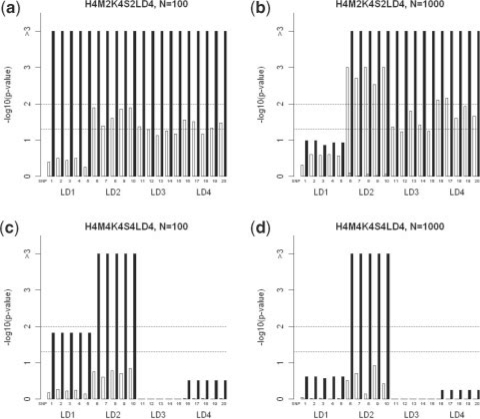
We should note that LD may reduce the importance scores of risk SNPs in random forests due to the strong correlation among them. Also, it is possible that some of the risk SNPs are not genotyped. To assess the impact of LD and ungenotyped risk SNPs on the performance of the MCC importance, we followed the approach in Lunetta et al. (2004) and Meng et al. (2009). Let K represent the number of genotyped risk SNPs, S the number of genotyped SNPs within each multiplicative model, and LD the number of SNPs in LD with a risk SNP. The SNPs in LD with a risk SNP but without functional effect on the trait were treated equally as the risk SNP, since they identify the correct region of the genome associated with the trait. We selected four risk SNPs in models H4M2 (SNPs 1–4 of the first two heterogeneity groups in Fig. 2a and b) and H4M4 (SNPs 5–8 of the second heterogeneity group in Fig. 2c and d), and added four extra SNPs in LD with each genotyped risk SNP. To indicate the genetic models, for example, H4M2K4S2LD4 means that two heterogeneous groups are genotyped out of all four groups, and four additional SNPs are in LD for each risk SNP. Following Lunetta et al. (2004) and Meng et al. (2009), all LD levels were simulated using r2=1. The empirical genome-wide P-values are shown in Figure 3. The performance of the permutation test using the MCC importance is largely unaffected by LD. In model H4M2K4S2LD4 with 100 SNPs (Fig. 3a), all four risk SNPs and the SNPs in LD with them show similar significance levels as they did in the unmodified model H4M2 (Fig. 3a). Also the significant risk SNP in the selected heterogeneity group (SNP 6) of H4M4 with 100 SNPs still can be distinguished in the more complicated new model (Figs 2c and 3c). There is one risk SNP with reduced significance in each model. For example, the first risk SNP in H4M2K4S2LD4 with 1000 SNPs (Fig. 2b) has genome-wide P-value <0.01; however, the corresponding LD region 1 in the modified model is no longer significant (Fig. 3b), probably due to missing information of other risk SNPs. At the same time, the permutation test using Gini or permutation importance become less powerful in both models.
Fig. 3.
Empirical P-values of risk SNPs and SNPs in LD with them in models H4M2K2S2LD4 and H4M4K4S2LD4. White, grey and black bars represent the permutation importance, Gini importance, and MCC importance scores, respectively. Two dash lines represents the genome-wide significance level of 0.05 and 0.01, respectively.
We further compared performance of different methods on these modified models based on 50 replications (see in Table 3), and the results show that the permutation test using the MCC importance achieves the best performance and it is significantly (P<10−5) better than the next runner-up in all scenarios. For example, compared to the other methods, at least 25% improvement in sensitivity can be observed by using the MCC importance in models H4M4K4S4LD4 with 1000 SNPs at genome-wide significance level of 0.01. Without exceptions, the permutation tests using the other importance measures perform worse than the univariate test. We also performed a limited number of simulations using larger sample sizes. The results are shown in Supplementary Figure S2 and Supplementary Table S2.
Table 3.
Comparison of prediction performance of different methods with different significance levels using models H4M2K2S2LD4 and H4M4K4S4LD4
| H4M2K2S2LD4 |
H4M4K4S4LD4 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
N=100 |
N=1000 |
N=100 |
N=1000 |
||||||
| sn (%) | sp (%) | sn (%) | sp (%) | sn (%) | sp (%) | sn (%) | sp (%) | ||
| Significant level 0.01 | MCC RF | 76.6** | 100 | 54.2** | 100 | 45.0*** | 100 | 11.0** | 100 |
| Permutation RF | 18.6 | 100 | 12.8 | 100 | 2.3 | 100 | 1.2 | 100 | |
| Gini RF | 0 | 100 | 4.4 | 100 | 0 | 100 | 0.3 | 100 | |
| Univariate | 56.0 | 100 | 29.5 | 100 | 7.0 | 100 | 1.0 | 100 | |
| Significant level 0.05 | MCC RF | 89.6** | 100 | 66.5** | 100 | 58.6*** | 100 | 17.3*** | 100 |
| Permutation RF | 45.9 | 100 | 26.9 | 100 | 10.9 | 100 | 2.9 | 100 | |
| Gini RF | 0 | 100 | 10.2 | 100 | 0 | 100 | 1.2 | 100 | |
| Univariate | 72.0 | 100 | 42.0 | 100 | 18.5 | 100 | 4.0 | 100 | |
sn: sensitivity; sp: specificity; N: number of SNPs.
Significance of paired t-test between MCC RF and the best among other three methods: *: <1E−2, **: <1E−5, ***: <1E−10.
3.3 Inference on SNP interactions
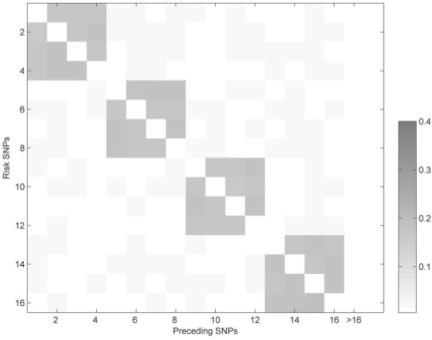
We have demonstrated the use of the MCC importance in random forests. Another important question remains: can we identify risk SNPs and their interactions beyond chance from the list of preceding SNPs during the course of computing the MCC importance? To answer this question, we investigated the relationship between a risk SNP and its preceding SNPs. First let us define Pa to be the probability that the specific SNP appears in LMCC. Pa was then estimated from a random forest with 1000 replications from the H4M4 model (see Fig. 4). The 16 × 16 grids with grayscale represent Pa for all 16 risk SNPs in the data, the column ‘>16’ represents the average Pa for all noise SNPs. We can easily identify four 4 × 4 blocks in Figure 4, which represent four heterogeneity groups.
Fig. 4.
Probability of SNPs in LMCC of risk SNPs in model H4M4. 1–16: risk SNPs, > 16: noise SNPs.
Within these blocks, Pa scores in the diagonal are zero, because a SNP can not be in LMCC of itself. The averaged Pa of multiplicative interacting SNPs is 0.18, ∼10 times of the averaged probability of the risk SNPs outside the corresponding group. At the same time, the results in Figure 4 also indicate that noise SNPs have a much lower chance to be included in LMCC, and the averaged Pa for noise SNPs is as low as 2.6 × 10−3. Moreover, also let us define Pn to be the probability that for a specific SNP, none of risk SNPs is included in LMCC. It is very unlikely for LMCC of the risk SNPs to include only noise SNPs: the averaged Pn of all risk SNPs is 1.4 × 10−2, suggesting that the MCC importance for the risk SNPs is indeed dependent on the other risk SNPs, beyond chance. These results suggest that using the MCC importance random forests makes it feasible to identify multiple risk SNPs.
3.4 Application in GWAS
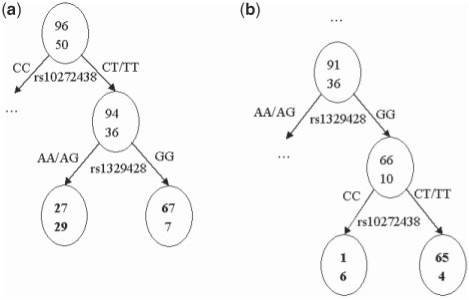
We applied the proposed method to a GWAS of AMD (Klein et al., 2005). AMD is the most common cause of vision loss in the elderly. Many researchers have studied the genetic mechanism of this complex disease (Daiger, 2005; Marx, 2006). This dataset contains 116 212 SNPs in 96 cases and 50 controls, and we removed SNPs that had more than 5% missing ratio or <5% minor allele frequency. Two significant SNPs: rs1329428 and rs10272438 were successfully identified under genome-wide significance level of 0.05, which have been previously reported by different studies of AMD (Chen et al., 2007; Klein et al., 2005; Ng et al., 2008). We then investigated the LMCC of these two SNPs and showed detailed information in Figure 5. It turns out that they cooperate in a scenario that both SNPs have the strongest association with the disease when they are dependant on each other. For example, in the 94 patients with AMD and 36 healthy people which have at least one ‘T’ allele in rs10272438, genotype ‘GG’ in rs1329428 was identified in 67 patients while only seven in the control group (Fig. 5a), resulting in a maximal chi-square score 28.52. For rs10272438, a maximal Chi-square score 28.87 was achieved with the same SNPs and genotype splitting criteria (Fig. 5b). Furthermore, we performed a logistic regression using both rs10272438 and rs1329428, and the coefficient of the interaction term is −0.1234 with an insignificant P-value of 0.84, suggesting that these two SNPs belong to a heterogeneity genetic model involved in AMD. It is noteworthy that Figure 5 is presented to illustrate the tree structures in the random forest and how the selected SNPs may interact. However, in the context of random forest, we are not aimed in comparing the performance of one tree against another in the forest.
Fig. 5.
Illustration of LMCC for two significant SNPs identified: rs10272438 and rs1329428.
4 DISCUSSION
Random forests, as a powerful machine learning method, has been successfully applied in many classification problems, especially with a large number of predictors. The permutation and Gini importance scores are commonly computed for random forests to evaluate the overall contribution of a predictor in classification. The reason for the reduced power they exhibited in identification of risk SNPs in GWAS is probably that the risk SNPs are extremely sparse in the data and they usually cooperate as a complex system associated with the phenotype; therefore averaging over all scores may significantly reduce its sensitivity in the permutation test. Moreover, there are other practical issues with the permutation importance. For example, highly correlated SNPs due to link age disequilibrium act as surrogates to each other, causing an underestimation of the permutation importance when they appear in one tree. During the calculation of permutation importance, the permutation of one SNP will break its intrinsic relationship with other SNPs which leads to inaccurate estimation of permutation importance. Some approaches (Amaratunga et al., 2008; Jiang et al., 2009; Meng et al., 2009) have been proposed to address these issues, but they are too computationally intensive for ultra high throughput data. Therefore, a powerful and yet simple statistic is very important to detect subtle effects between casual SNPs.
In this article, we proposed and studied the maximal chi-square statistic as a new importance measurement in random forests and its application in the permutation test for GWAS. We first evaluated the performance of the MCC importance in detecting risk SNPs using empirical P-values under null hypothesis and discovered that there is no association between SNPs and a trait. We also compared this method with the permutation tests using different importance scores in random forests and single marker analysis. We further modified the genetic models in simulation by including LD, reflecting real data in GWAS. The results indicated that the permutation test using the MCC importance was consistently the best. Moreover, we showed that it is possible to make inference on risk SNPs from the preceding list of SNPs while deriving the MCC importance. Finally we applied this method to a GWAS data for AMD. Two AMD-related SNPs: rs10272438 and rs1329428 were successfully identified with the genome-wide significance level 0.05. Our analysis suggested that these two SNPs belong to a heterogeneity model involved in etiology.
Funding: This research was supported in part by grant R01DA016750 from the National Institutes of Health.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Amaratunga D, et al. Enriched random forests. Bioinformatics. 2008;24:2010–2014. doi: 10.1093/bioinformatics/btn356. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Machine Learn. 2001;45:5–32. [Google Scholar]
- Breiman L. Manual On Setting Up, Using, And Understanding Random Forests V3.1. 2002 http://oz.berkeley.edu/users/breiman/Using_random_forests_V3.1.pdf. [Google Scholar]
- Breiman L, et al. Classification and Regression Trees. New York: Chapman and Hall; 1984. [Google Scholar]
- Bureau A, et al. Identifying SNPs predictive of phenotype using random forests. Genet. Epidemiol. 2005;28:171–182. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. A forest-based approach to identifying gene and gene gene interactions. Proc. Natl Acad. Sci. USA. 2007;104:19199–19203. doi: 10.1073/pnas.0709868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger SP. Genetics. Was the Human Genome Project worth the effort? Science. 2005;308:362–364. doi: 10.1126/science.1111655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Friedman JH. Greedy function approximation: a gradient boosting machine. Ann. Stat. 2001;29:1189–1232. [Google Scholar]
- Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Jiang R, et al. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinformatics. 2009;10(Suppl. 1):S65. doi: 10.1186/1471-2105-10-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat. Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, et al. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx J. Gene offers insight into macular degeneration. Science. 2006;314:405. doi: 10.1126/science.314.5798.405a. [DOI] [PubMed] [Google Scholar]
- McDonough CW, et al. The influence of carnosinase gene polymorphisms on diabetic nephropathy risk in African-Americans. Hum. Genet. 2009;126:265–275. doi: 10.1007/s00439-009-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YA, et al. Performance of random forest when SNPs are in linkage disequilibrium. BMC Bioinformatics. 2009;10:78. doi: 10.1186/1471-2105-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TK, et al. Multiple gene polymorphisms in the complement factor h gene are associated with exudative age-related macular degeneration in Chinese. Invest. Ophthalmol. Vis. Sci. 2008;49:3312–3317. doi: 10.1167/iovs.07-1517. [DOI] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am. J. Hum. Genet. 1990a;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am. J. Hum. Genet. 1990b;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- Rodenburg W, et al. A framework to identify physiological responses in microarray-based gene expression studies: selection and interpretation of biologically relevant genes. Physiol. Genomics. 2008;33:78–90. doi: 10.1152/physiolgenomics.00167.2007. [DOI] [PubMed] [Google Scholar]
- Samani NJ, et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn I, et al. A permutation-based multiple testing method for time-course microarray experiments. BMC Bioinformatics. 2009;10:336. doi: 10.1186/1471-2105-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, et al. Detecting significant SNPs in rheumatoid arthritis study with random forests. BMC Proc. 2009;3:S69. doi: 10.1186/1753-6561-3-s7-s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, et al. A genome-wide tree- and forest-based association analysis of comorbidity of alcoholism and smoking. BMC Genet. 2005;6(Suppl. 1):S135. doi: 10.1186/1471-2156-6-S1-S135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bonney G. Use of classification trees for association studies. Genet. Epidemiol. 2000;19:323–332. doi: 10.1002/1098-2272(200012)19:4<323::AID-GEPI4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ye Y. A tree-based method for modeling a multivariate ordinal response. Stat. Interface. 2008;1:169–178. doi: 10.4310/sii.2008.v1.n1.a14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Cell and tumor classification using gene expression data: construction of forests. Proc. Natl Acad. Sci. USA. 2003;100:4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. The NEI/NCBI dbGAP database: genotypes and haplotypes that may specifically predispose to risk of neovascular age-related macular degeneration. BMC Med. Genet. 2008;9:51. doi: 10.1186/1471-2350-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.