Abstract
The cancer preventive action of (−)-epigallocatechin gallate (EGCG), found in green tea, is strongly supported by epidemiology and laboratory research data. However, the mechanism by which EGCG inhibits carcinogenesis and cell transformation is not clear. In this study, we report that EGCG suppressed epidermal growth factor (EGF)-induced cell transformation in JB6 cells. We also found that EGCG inhibited EGF-induced Fyn kinase activity and phosphorylation in vitro and in vivo. Fyn was implicated in the process because EGF-induced JB6 cell transformation was inhibited by small interfering RNA (siRNA)-Fyn-JB6 cells. With an in vitro protein-binding assay, we found that EGCG directly bound with the GST-Fyn-SH2 domain but not the GST-Fyn-SH3 domain. The Kd value for EGCG binding to the Fyn SH2 domain was 0.367 ± 0.122 µM and Bmax was 1.35 ± 0.128 nmol/mg. Compared with control JB6 Cl41 cells, EGF-induced phosphorylation of p38 MAP kinase (p38 MAPK) (Thr180/Tyr182), ATF-2 (Thr71) and signal transducer and activator of transcription 1 (STAT1) (Thr727) was decreased in siRNA-Fyn-JB6 cells. EGCG could inhibit the phosphorylation of p38 MAPK, ATF-2, and STAT1. The DNA binding ability of AP-1, STAT1, and ATF-2 was also decreased in siRNA-Fyn-JB6 cells. Overall, these results demonstrated that EGCG interacted with Fyn and inhibited Fyn kinase activity and thereby regulated EGF-induced cell transformation. Inhibition of Fyn kinase activity is a novel and important mechanism that may be involved in EGCG-induced inhibition of cell transformation.
Keywords: EGCG, Fyn, epidermal growth factor, cell transformation
INTRODUCTION
The cancer preventive effect of the green tea polyphenol, (−)-epigallocatechin gallate (EGCG) is strongly supported by epidemiology, cell culture, animal, and clinical studies. The anticancer properties of EGCG are associated with the inhibition of the cancer-associated enzyme, telomerase [1], repression of cell-cycle progression [2], suppression of the activation of cyclooxygenase-2 (COX-2) activity [3], and inhibition of nuclear factor-kappa B (NF-kappa-B) and activator protein-1 (AP-1) activation [4,5]. We previously found that EGCG inhibited ultraviolet (UV)B- or arsenite-induced AP-1 activity [6,7]. Epidemiology studies have suggested that people who consumed green tea had a lower risk of cancer [8]. Additional evidence indicated that EGCG was a potent compound that could be used to kill cancer cells and has been examined in Phase I and II clinical trials [9,10]. We and other colleagues found that EGCG inhibited EGF- or 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced cell transformation [11,12]. However, the mechanism by which EGCG inhibits cell transformation is not completely clear.
Fyn is a member of the tyrosine kinase family and displays a very wide range of biologic functions. For example, Fyn was reported to regulate the function of the glutamate receptor N-methyl-d-aspartate (NMDA) [13,14] and to determine a cell’s sensitivity to ethanol [14]. Earlier findings showed that neonatal mice lacking Fyn were impeded in their sucking behavior [15] and Fyn mutant mice displayed impaired long-term potentiation, spatial learning, and hippocampal development [16]. Kawakami et al. [17] found that Fyn overexpression in NIH 3T3 cells induced morphologic transformation and anchorage-independent growth. This result strongly indicated that Fyn might have a possible role as an oncogene related to cell transformation. Fyn also was shown to play a vital role in Xiphophorus melanoma by inducing receptor kinase (Xmrk)-stimulated proliferative and anti-apoptotic signaling in pigment cells [18]. An additional study indicated that a complex of Fyn and the receptor tyrosine kinase Xmrk was involved in malignant transformation of pigment cells through activation of phosphatidylinositol-3 (PI-3) kinase [19]. On the other hand, activation of the Src kinase Fyn led to melanocyte dedifferentiation by influencing MAP kinase phosphatase 1 (MKP-1)-regulated MAP kinase signaling [20]. These studies suggested that the tyrosine kinase Fyn played a very important role in cell transformation.
In the present study, we provided strong evidence showing that the tyrosine kinase Fyn is a novel target of EGCG. Our results indicated that EGCG inhibited EGF-induced JB6 Cl41 cell transformation by suppressing EGF-induced Fyn phosphorylation and activity. An in vitro binding assay revealed that EGCG bound to the SH2 domain of Fyn with high affinity. Further, siRNA against Fyn inhibited EGF-induced cell transformation. These results indicated that inhibition of Fyn kinase activity is a novel and important mechanism that provides further clarification of the mechanism of EGCG-mediated inhibition of cell transformation.
MATERIALS AND METHODS
Antibodies and Reagents
Eagle’s minimal essential medium (MEM), fetal bovine serum (FBS), l-glutamine, and gentamicin were purchased from Life Technologies, Inc. (Grand Island, NY). The phospho-Fyn (Thr12) antibody was from Santa Cruz Co. (Santa Cruz, CA) and antibodies for detection of phosphorylation of STAT1 (Thr727) and ATF-2 (Thr71) and nonphosphorylated-Fyn were from Upstate Biotechnology (Lake Placid, NY). Active Fyn was from Upstate (Lake Placid, NY).EGCG (99% purified) was a generous gift from Dr. Chi-Tang Ho (The Rutgers University, Piscataway, NJ). [3H]EGCG (13 Ci/mmol in ethanol containing 8 mg/mL of ascorbic acid) was generously provided by Dr. Yukihiko Hara (Food Research Laboratory, Mitsui Norin Co. Ltd., Fujieda, Shizuoka, Japan). The Modified-Lowry protein assay reagent was from Sigma (St. Louis, MO), the Folin&Ciocalteus Phenol Reagent was from Pierce (Rockford, IL); and the PVDF membrane was purchased from Millipore (Bedford, MA). The LipofectAMINE™ 2000 reagent was from Life Technologies, Inc. The sense and antisense oligonucleotides for siRNA against Fyn were synthesized by Sigma Co. The mU6pro plasmid was kindly provided by Dr. David L. Turner (Mental Health Research Institute, The University of Michigan). The GST-Fyn-SH2 and GST-Fyn-SH3 plasmids were kindly provided by Dr. Isaiah J. Fidler (The University of Texas, M. D. Anderson Cancer Center, Houston, Texas).
Establishing the siRNA-Fyn Stable Expression Cell Line
In order to further study the role of Fyn in the regulation of cell transformation, we successfully knocked down the expression level of Fyn with the siRNA method described previously [21]. In brief, the ligated sense oligonucleotide of Fyn, 5′-TTTGCAGCTCGGAAGGAGATTGG TTCAAGAGACAATCTC CTTCCGAGCTGTTTTT-3′ and the antisense oligonucleotide of Fyn, 5′-CTAGAAAAA CAGCTCGGAAGGAGATTGGTCTCTTGAACCAATCTCCTTCC GAGCTG-3′, were inserted into the mU6pro vector. Annealing, ligation, and colony screening were performed as described previously [22]. The reconstructed plasmid with the mU6pro vector contained the siRNA Fyn sequence and is referred to as the siRNA-Fyn-mU6pro. The plasmid siRNAFyn-mU6pro and pcDNA3.1 were stably co-transfected into JB6 Cl41 cells with the LipofectAMINE™ 2000 reagent. Fyn protein expression level was confirmed by a Western blot assay with an antibody against Fyn.
Cytotoxic Effects of EGCG on JB6 Cells
To determine the effect of EGCG on cell viability, JB6 Cl41 cells were suspended in 100 µL of 5% FBS/MEM and seeded in 96-well plates at 10 000 cells/well. After a 12 h incubation, the media were changed to 0.1% FBS/MEM and cells were then incubated for an additional 24 h. At that time, cells were exposed to EGCG at various doses (0–100 µM) for another 48 h or to 20 µM EGCG for 24, 48, or 72 h. Cells were then subjected to the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay according to the instructions from the manufacturer (CellTiter 96® AQueous One Solution Cell Proliferation assay kit, Promega, Madison, WI). The results were tabulated with the Multiskan MS plate reader (Labsystems, Helsinki, Finland). Untreated cells were used as a positive control.
Anchorage-Independent Transformation Assay
Induction of cell transformation was conducted as described previously [23]. The role of Fyn in EGF-promoted cell transformation was investigated in “mock”-transfected JB6 Cl41 and siRNA-Fyn-JB6 Cl41 cells. In brief, cells (8 × 103/mL) were exposed to mixtures of EGF (5 ng/mL) containing increasing concentrations of EGCG (1–20 µM) in 1 mL of 0.3% basal medium Eagle (BME) agar containing 10% FBS. The cultures were maintained at 37°C in a 5% CO2 incubator for 2 wk and then the cell colonies were automatically counted by a computerized microscope system. Cell transformation is presented as colony number per 8000 seeded cells in soft agar.
In Vitro EGCG Pull-Down Assay
As in our previous study [24], EGCG-Sepharose 4B beads were created by first dissolving EGCG in coupling buffer (0.1MNaHCO3, pH8.3, 0.5MNaCl). Sepharose 4B beads were swollen and washed with 1 mM HCl medium and mixed with the coupling solution containing EGCG and slowly rotated overnight at 4°C. Excess EGCG was washed away with coupling buffer and any remaining active groups were blocked with 0.1 MTris-HCl buffer (pH 8.0) for 2 h. The medium was then washed with 0.1Macetate buffer (pH 4.0) containing 0.5 MNaCl followed by a wash with 0.1 M Tris-HCl (pH 8.0) that also contained 0.5 M NaCl. The EGCG-Sepharose 4B beads were now ready for use in the pull-down assay. Recombinant Fyn (2 µg) or the JB6 Cl41 cellular supernatant fraction (600 µg) was incubated with either EGCG-Sepharose 4B or Sepharose 4B as a negative control (100 µL, 50% slurry) in reaction buffer (50 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP-40, 2 µg/mL of BSA, 0.02mMPMSF, 1 × protease inhibitor cocktail). After incubation with gentle rocking overnight at 4°C, the beads were washed five times with wash buffer (50 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.01% NP-40, 0.02 mM PMSF) and proteins bound to the beads were analyzed by immunoblotting assay with anti-Fyn.
Purification of GST-Fyn-SH2 and GST-Fyn-SH3 Fusion Proteins
GST fusion protein purification was conducted following the company’s (Amersham Biosciences, Piscataway, NJ) instruction manual. In brief, pGEX-Fyn-SH2 and pGEX-Fyn-SH3 recombinant proteins were cultured in LB medium at 37°C with vigorous agitation for 16 h. When the liquid spectrum A600 value reached 0.8, fresh LB medium was used to dilute liquid cultures 20-fold. A final concentration of isopropyl-d-thiogalactoside (IPTG) of 0.1mM was added and these cultures were continuously agitated for another 3 h at 25°C. The bacteria were collected by centrifugation (4000 rpm, 10 min, 4°C) and pellets were washed three times with ice-cold phosphate-buffered saline (PBS). Lysis buffer (400 µL) was added to bacterial pellets and pellets were disrupted by sonication. Suspensions were left on ice for 1 h. The lysate was centrifuged and the supernatant fraction saved. A 50% slurry (40 µL) of glutathione-Sepharose 4B beads was added to each sample and mixed gently for 5 min at room temperature. Beads were washed three times with ice-cold PBS and eluted with buffer (40 µL) and SDS-PAGE was used to separate and analyze the fusion proteins.
Determination of GST-Fyn Binding Affinity With EGCG
As in our previous study [24], we determined the affinity binding between GST-Fyn and EGCG. The expressed GST fusion proteins (1 µg) were incubated with glutathione-Sepharose 4B beads for 1 h at room temperature. The affinity binding assay was carried out overnight at 4°C in a 50 µL mixture containing the reaction buffer described above, 0.5 µCi [3H]EGCG and 1 µg of GST-Fyn-Sepharose 4B or GST-Sepharose 4B. For analyzing concentration-dependent uptake, a range of 76 pM–20 µM concentrations of EGCG was applied. The Kd value was determined by non-linear regression analysis performed with the Prizm 4.0 software program (Graphpad Inc., San Diego, CA).
EGCG Effects on Fyn Kinase Activity In Vitro
Upstate Biotechnology provides a KinaseProfiler™ Selectivity Screening service and the online protocol is found at http://www.upstate.com. /features/ kp_protocols.asp. For this assay, all kinases were pre-diluted in 10× working concentration buffer comprised of 20 mM Tris (pH 7.5), 0.1 mM EGTA, 0.1% β-mercaptoethanol, 1 mg/mL of BSA, 0.01% Brij-35, and 5% glycerol. For the Fyn kinase activity assay, 5 mU Fyn kinase was incubated with 50 mM Tris (pH 7.5) 0.1 mM EGTA, 0.1 mM Na3VO4, 250 µM Cdc2 peptide (KVEKIGEGTYGVVK), 10mM mg acetate, and[γ-32P-ATP]. This reaction was initiated by the addition of the Mg-ATP mix. After incubation for 40 min at room temperature, the reaction was stopped by the addition of 5 µL of a 3% phosphoric acid solution. Then 10 µL of the reaction was spotted onto P30 filtermat paper and washed three times for 5 min in 50 mM phosphoric acid and once in methanol prior to drying and scintillation counting. EGCG (5 µM) was added into the reaction buffer as the experimental group. The total reaction volume was 25 µL. Data from Upstate showed that EGCG (5 µM) inhibited Fyn kinase (5 mU) activity in vitro. In order to confirm the results from Upstate, we performed the in vitro kinase assay again following the instructions provided by Upstate. The reaction contained 6.25 µL of assay buffer (200 mM Tris/HCl, pH 7.5, 0.4 mM EGTA, 0.4 mM sodium orthovanadate), 250 µM Src substrate peptide (KVEKIGEGTYVVYK) corresponding to amino acids 6–20 of p34cdc2 and 10 µL of diluted [γ-32P]ATP solution. Active Fyn (10, 50, or 100 ng) was separately added to different reaction tubes and distilled water was used to bring the final volume to 30 µL. Tubes were incubated at 30°C for 30 min and 25 µL aliquots were transferred onto P81 paper and washed three times with 0.75 % phosphoric acid for 5 min per wash and one time with acetone for 2 min. Radioactive incorporation was determined by scintillation counting. For evaluation of the effect of EGCG, a final concentration of EGCG at 1, 5, 10, or 20 µM was separately incubated with the reaction mixtures described above at 30°C for 30 min. The same experiments were repeated three times.
Immunoprecipitation and Kinase Assays
The immunoprecipitation and kinase assays were carried out as described previously [25]. In brief, JB6 Cl41 cells were cultured to 80% confluence and then starved in 0.1% FBS/MEM for 48 hat 37°C. The media were changed to fresh 0.1% FBS/MEM and the cells were incubated for another 2 h at 37°C. To determine whether EGCG can inhibit EGF-induced Fyn kinase activity, cells were either treated or not treated with different doses of EGCG for 1 h and then incubated with EGF for 30 min and subsequently disrupted with lysis buffer (20 mM Tris/HCl, pH 7.4; 1 mM EDTA; 150 mM NaCl; 1 mM EGTA; 1% Triton X-100; 1 mM β-glycerolphosphate; 1 mg/mL of leupeptin; 1 mM Na3VO4; 1 mg/mL of leupeptin, and 1 mM phenylmethylsulfonyl fluoride) and centrifuged at 14 000 rpm for 10 min in a microcentrifuge. The lysates containing 400 µg protein were used for immunoprecipitation with an antibody against Fyn and incubated overnight at 4°C. Then protein A/G plus-agarose beads were added and the mixture was continuously rotated for another 3 h at 4°C. The beads were washed three times with kinase buffer (200 mM Tris/HCl, pH 7.5, 0.4 mM EGTA, 0.4 mM sodium orthovanadate, 20 mM MOPS, 0.1 mM EDTA, 0.01% BRIJ 35, 0.1 mg/mL of BSA, 0.1% β-mercaptoethanol). The pellets were resuspended in 20 µL of 1 × kinase buffer supplemented with 10 µL of diluted [γ-32P]ATP solution and 2.5 µL of Src substrate peptide (250 µM) and incubated 30 min at 30°C. Samples were denatured at 95°C for 5 min and centrifuged at 4°C, 12 000 rpm for 2 min. Radioactive incorporation was determined by scintillation counting. The same experiments were repeated three times.
Western Blot Assay
To determine whether EGCG has an effect on the phosphorylation of STAT1, ATF-2, and p38 MAPK through Fyn, siRNA-Fyn and mock-transfected JB6 cells were used to detect phosphorylated STAT1, ATF-2, and p38 MAPK with or without EGCG treatment. Briefly, siRNA-Fyn and mock cells were cultured in 10-cm dishes and then incubated in 0.1% FBS/MEM at 37°C in a 5% CO2 incubator for 24 h. Cells were exposed to EGCG (20 µM) for 2 h and then EGF was added (10 ng/mL). The cells were then harvested after 15 min and disrupted in 500 µL of lysis buffer A (50 mM KCl, 0.5% Nonidet P-40, 100 µM dithiothreitol, 25 mM HEPES, pH 7.8, 10 µg/mL of leupeptin, 25 µg/mL of aprotinin, and 1 mM phenylmethylsulfonyl fluoride) 30 min on ice. Protein content was determined by the Modified-Lowry Protein Assay method and 40 µg of each cell lysate were used for polyacrylamide gel electrophoresis (PAGE) and proteins were transferred onto a PVDF membrane. Membranes were incubated with the primary antibody diluted in blocking buffer (Li-COR Biosciences Co., Lincoln, NE) overnight at 4°C. The membranes were then washed with 0.25% Tween-20 in PBS at room temperature for 15 min and then washed with PBS for 5 min. Membranes were incubated with a secondary antibody, which was an infrared dye-linked rabbit or mouse IgG. Protein expression level was analyzed by the Odyssey Scanner (Li-COR Biosciences Co.). Phosphorylation of STAT1, ATF-2, and p38 MAPK were, respectively, detected by Western blot assay with specific antibodies. To ensure equal protein loading and transfer, membranes were immunoblotted with antibodies against the nonphosphorylated proteins.
Determination of AP-1, STAT1, and CREB DNA Binding
Nuclear protein extracts were prepared from cells by a modification of the method of Ye et al. [26]. Briefly, “mock”-transfected and siRNA-Fyn JB6 Cl41 cells were cultured in 10-cm dishes and starved in 0.1% FBS/MEM at 37°C in a 5% CO2 incubator for 24 h. The cells were exposed to different concentrations of EGF for another 12 h and then harvested and disrupted in 500 µL of lysis buffer A (50 mM KCl, 0.5% Nonidet P-40, 100 µM dithiothreitol, 25 mM HEPES, pH 7.8, 10 µg/mL of leupeptin, 25 µg/mL of aprotinin, and 1 mM phenylmethylsulfonyl fluoride). After centrifugation (16 000g at 4°C), the pellets were washed once with 500 µL of Buffer B (Buffer A without Nonidet P-40). The pellets were resuspended in 100 µL of extraction buffer (Buffer B, but with 500 mM KCl and 10% glycerol) and strongly shaken at 4°C for 1 h. After centrifugation (16 000g at 4°C, 10 min), the supernatant solutions were moved into fresh tubes and stored at −70°C until analysis. The DNA-binding reaction was incubated at room temperature for 30 min in a mixture containing 5 µg of nuclear protein, 1 µg of poly (dI • dC), and 15 000 cpm of an α-32P-labeled double-stranded AP-1 oligonucleotide (5′-CGCTTGATGAGTCAGCCGGAA-3′), STAT1 oligonucleotide (5′-CATGTTATGCATATTCCTGTAAGTG-3′), or cAMP regulatory element-binding protein (CREB) oligonucleotide (5′-AGAGATTGCCTGACGTCAGAGAGC TAG-3′). All of these oligonucleotides were purchased from Santa Cruz. The samples were separated on a 5% polyacrylamide gel, and the gels were analyzed with the Storm 840 Phosphor-Imaging system (Amersham Biosciences).
RESULTS
EGCG Inhibits EGF-Induced JB6 Cl41 Cell Transformation in a Dose-Dependent Manner
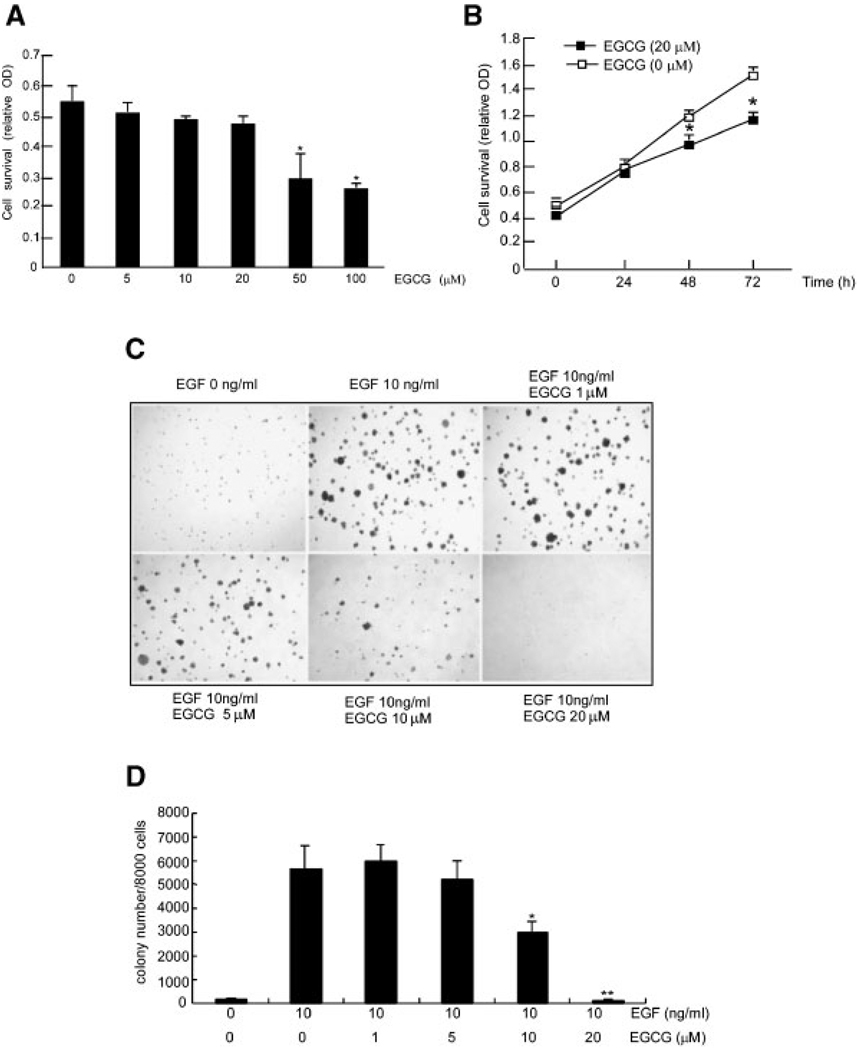
To determine whether EGCG had a cytotoxic effect, we treated JB6 epidermal mouse skin cells (JB6 C141 cells) with EGCG at a range of concentrations (0–100 µM) and assessed viability with the MTS assay. The results showed that EGCG at a concentration of 20 µM or less did not decrease cell viability (Figure 1A). Data also showed that 20 µM EGCG could decrease cell proliferation (Figure 1B). The JB6 C141 cell line is an excellent model to study EGF-[27] or TPA-[28] promoted cell transformation. In this study, EGF was used to induce transformation of JB6 Cl41 cells. Results showed that EGCG treatment significantly decreased EGF-promoted colony number in a dose-dependent manner (Figure 1C and D) with 10 or 20 µM EGCG being most effective. The average colony number from three experiments is shown (Figure 1D).
Figure 1.
EGCG inhibits EGF-induced cell transformation. (A) JB6 Cl41 cells were treated with increasing concentrations of EGCG and viability was assessed with the MTS assay as described in Methods and Materials. (B) For determining the effect of EGCG on proliferation over time, JB6 Cl41 cells were treated with EGCG at 20 µM for different time periods and then proliferation was assessed byMTS assay. For both A and B, data are presented as means ± SD of three independent experiments, each performed in triplicate. The asterisk (*) indicates a significant (*, P<0.05) decrease in viability in EGCG-treated cells relative to untreated control cells. (C) EGCG inhibits JB6 Cl41 anchorage-independent EGF-promoted transformation. Various concentrations of EGCG with or without 10 ng/mL EGF were added into soft agar with JB6 Cl41 cells and colonies were counted automatically after 7 d of incubation at 37°C in a 5% CO2 incubator. Colony formation in JB6 cells without EGF stimulation (1st plate, upper), with EGF (2nd plate, upper), EGF plus 1 µM EGCG (3rd plate, upper), EGF plus 5 µM EGCG (1st plate, lower), EGF plus 10 µM EGCG (2nd plate, lower) or EGF plus 20 µM EGCG (3rd plate lower). (D) Data are represented as the average number of colonies ± SD as determined from three separate experiments ± SD. The asterisk (*) indicates a significant inhibition compared to EGF only (**, P<0.01 and *, P<0.05).
EGCG Inhibits Fyn Kinase Activity in a Dose-Dependent Manner In Vitro and In Vivo
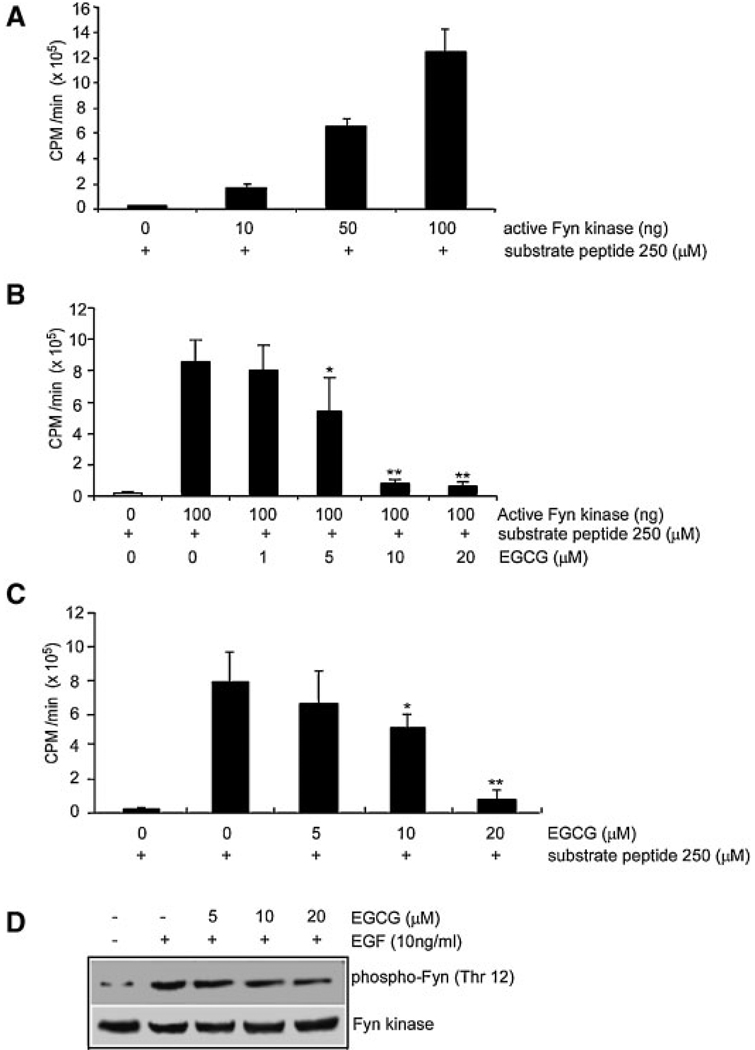
To identify EGCG-targeted kinases, 101 kinases were screened by Upstate Biotechnology with their commercial kinase assay screening system. Their results indicated that EGCG strongly inhibited Fyn kinase activity in vitro (data not shown). We confirmed that the commercially available active Fyn phosphorylated a Src substrate peptide in a dose-dependent manner in vitro (Figure 2A) and that this Fyn kinase activity was inhibited by EGCG in a dose-dependent manner (Figure 2B). Activity was inhibited by about 50% with 5 µM EGCG (Figure 2B, lane 4) and by about 90% with 10 or 20 µM EGCG in vitro (Figure 2B, lanes 5 and 6). In order to determine whether EGCG could inhibit Fyn kinase activity in cells, an immunoprecipitation kinase assay was used. Cells were either treated or not treated with EGCG at various concentrations for 1 h and then treated with EGF. The kinase assay results showed that 10 or 20 µM EGCG significantly inhibited EGF-induced Fyn kinase activity in vivo (Figure 2C, lanes 4 and 5 vs. lane 2).
Figure 2.
EGCG inhibits Fyn kinase activity and phosphorylation of Fyn in a dose-dependent manner. (A) For the in vitro kinase assay, phosphorylation of a Fyn substrate peptide was determined with active Fyn (10, 50, or 100 ng alone or with (B) EGCG (1, 5, 10, 20 µM). (C) For the immunoprecipitation kinase assay, cells were treated or not treated with EGCG (5, 10, or 20 µM) 1 h and then treated with EGF (10 ng/mL) and harvested after 30 min. The average 32P count was determined from three separate experiments. The asterisk (*) indicates a significant EGCG-induced change in kinase activity compared to respective control (**, P<0.01 and *, P<0.05). (D) Cells were treated with various concentrations (0, 5, 10, or 20 µM) of EGCG followed by EGF (10 ng/mL). Phosphorylation of Fyn (Thr12) (upper panel) and total Fyn protein level (lower panel) were then determined by Western blot analysis as described in Materials and Methods with specific antibodies against phosphorylation of Fyn (Thr12) or total Fyn protein.
EGCG Inhibits EGF-Induced Fyn Phosphorylation in a Dose-Dependent Manner
To determine whether EGCG regulates EGF-induced Fyn phosphorylation, a Western blot assay was performed with a specific antibody against phosphorylation of Fyn (Thr12). Results indicated that EGF (10 ng/mL) induced phosphorylation of Fyn (Thr12) and EGCG inhibited EGF-induced phosphorylation of Fyn in a dose-dependent manner (Figure 2D, upper panel) with no effect on total Fyn protein level (Figure 2D, lower panel).
EGCG Specifically Binds With the Fyn SH2 Domain
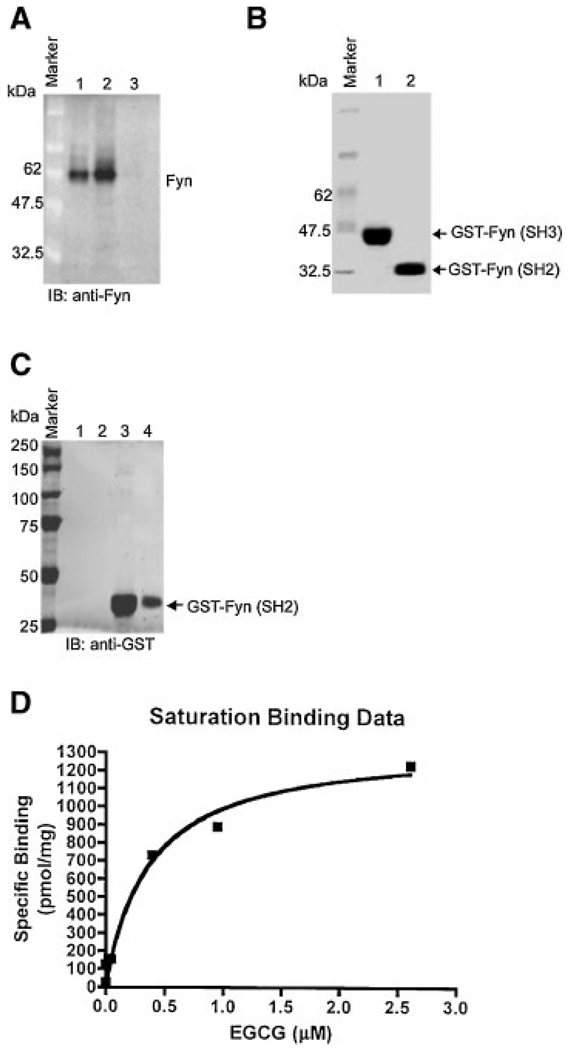
The results above indicated that EGCG is involved in EGF-induced Fyn kinase activity and phosphorylation. To further confirm whether EGCG can directly bind with Fyn in vitro, we used the EGCG pull-down assay. Our results showed that Fyn bound with the EGCG-Sepharose 4B beads (Figure 3A, lane 2) but was not found in the elution buffer (Figure 3A, lane 3). The Fyn protein was loaded as a control (Figure 3A, lane 1). These results indicated that Fyn could bind with EGCG. Next, in order to determine whether EGCG binds with a specific domain of Fyn, we used GST-Fyn (SH3) and GST-Fyn (SH2) fusion proteins (Figure 3B) to perform the EGCG pull-down assay in vitro. We found that the GST-Fyn (SH2) fusion protein bound with the EGCG column (Figure 3C, lanes 3 and 4), but the GST-Fyn (SH3) fusion protein did not (Figure 3C, lanes 1 and 2). We further determined that the SH2 domain of Fyn bound with EGCG with a Kd value of 0.367 ± 0.122 µM and a Bmax of 1.35 ± 0.128 nmol/ mg (Figure 3D). These results indicated that EGCG bound with the Fyn SH2 domain with high affinity.
Figure 3.
EGCG specifically binds with the Fyn SH2 domain and not the SH3 domain. (A) For the in vitro EGCG pull-down assay, lane 1 indicates the Fyn protein detected in the recombinant Fyn sample as a positive control and in lane 2 the cell lysates incubated with EGCG-Sepharose 4B beads. Lanes 3 and 4 show no Fyn protein detected in cell lysates incubated with only Sepharose 4B beads. (B) Purified GST-Fyn (SH2) and GST-Fyn (SH3) are shown. (C) Lanes 1 and 2 show no Fyn protein detected in the mixture of GST-Fyn (SH3) and EGCG-Sepharose 4B beads or elution buffer, respectively. Lanes 3 and 4 show Fyn protein detected in the mixture of GST-Fyn (SH2) with EGCG-Sepharose 4B beads or in the elution solution from this mixture, respectively. (D) GST-Fyn (SH2) binding with EGCG affinity and Kd value (0.367 ± 0.122 µM) are shown.
EGF-Induced Phosphorylation of p38 MAP Kinase (Thr180/Tyr182), ATF-2 (Thr71), and STAT1 (Thr727) is Inhibited in siRNA-Fyn-JB6 Cl41 Cells
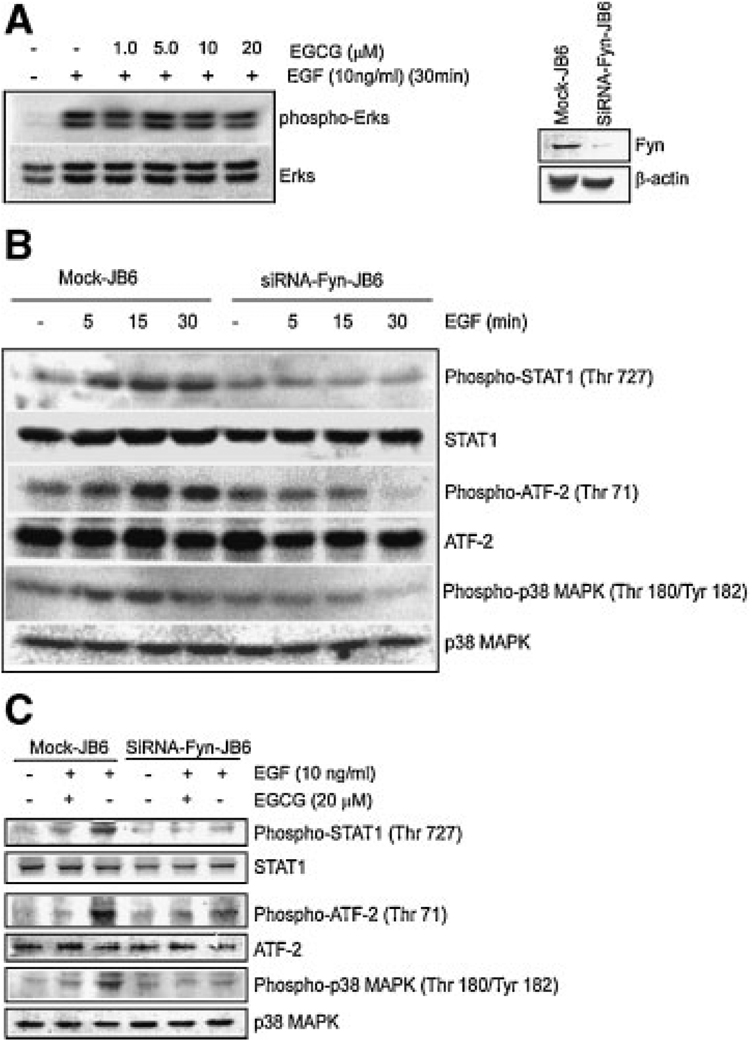
To further explore the mechanism of the EGCG inhibitory effect on EGF-promoted cell transformation, we determined whether EGCG could suppress EGF-induced phosphorylation of ERKs, which are known to be involved in EGF-induced cell transformation. Our results indicated that EGCG had no effect on EGF-induced ERKs phosphorylation (Figure 4A) even though EGCG distinctly inhibited EGF-induced Fyn phosphorylation (Figure 2D). Thus we hypothesized that Fyn might play a more important role than ERKs in the EGCG inhibition of EGF-induced cell transformation and that Fyn may exert its effects through the direct regulation of transformation-related transcription factors or kinases. To test this hypothesis, we successfully knocked down Fyn protein expression with the siRNA method as described [21] (Figure 4A, right panels). Our results showed that phosphorylation of STAT1 (Figure 4B, 1st panel), ATF-2 (Figure 4B, 3rd panel), and p38 MAPK (Figure 4B, 5th panel) was decreased in siRNA-Fyn-JB6 cells compared with mock-transfected JB6 cells. To further confirm whether EGCG regulates these transformation-related proteins, we investigated the effect of EGCG on the phosphorylation level of ATF-2, STAT1, and p38 MAPK in mock-transfected or SiRNA-Fyn cells. The results showed that EGCG could not only inhibit ATF-2 and STAT1 phosphorylation, but also p38 MAPK phosphorylation (Figure 4C, 1st, 3rd, and 5th panels), which is an upstream kinase for both ATF-2 and STAT1. These findings demonstrated that Fyn is involved in the regulation of phosphorylation of transcription factors ATF-2 and STAT1 and their upstream MAP kinase p38, which are all related to cell transformation.
Figure 4.
siRNA-Fyn inhibits phosphorylation of STAT1 (Ser727), ATF-2 (Thr71), and p38MAPK (Thr180/Tyr182). Cells were treated as described in Methods and Materials and phosphorylation of selected proteins was determined by Western blot. (A) Phosphorylation of ERKs (Ser42/Ser 44) was detected with a specific phospho-ERKs (Ser42/44) antibody and total ERKs protein levels were detected by a non-phospho-ERKs antibody (A, left). Fyn expression was confirmed in SiRNA-Fyn JB6 cells and mock cells by Western blot (A, right). (B) Phosphorylation of STAT1 (Ser727), ATF-2 (Thr71), and p38 MAPK (Thr180/Tyr182) were separately detected with specific phospho-STAT1, ATF-2, and p38MAP kinase antibodies. Total protein levels of STAT1, ATF-2, and p38 MAP kinase were detected by a non-phospho-STAT1, ATF-2, or p38 MAP kinase antibody, respectively. (C) SiRNA-Fyn JB6 cells and mock cells were treated with EGCG as described in Methods and Materials. Phosphorylation of STAT1 (Ser727), ATF-2 (Thr71), and p38 MAPK (Thr180/Tyr182) was also detected by Western blot. Respective total protein levels served as controls.
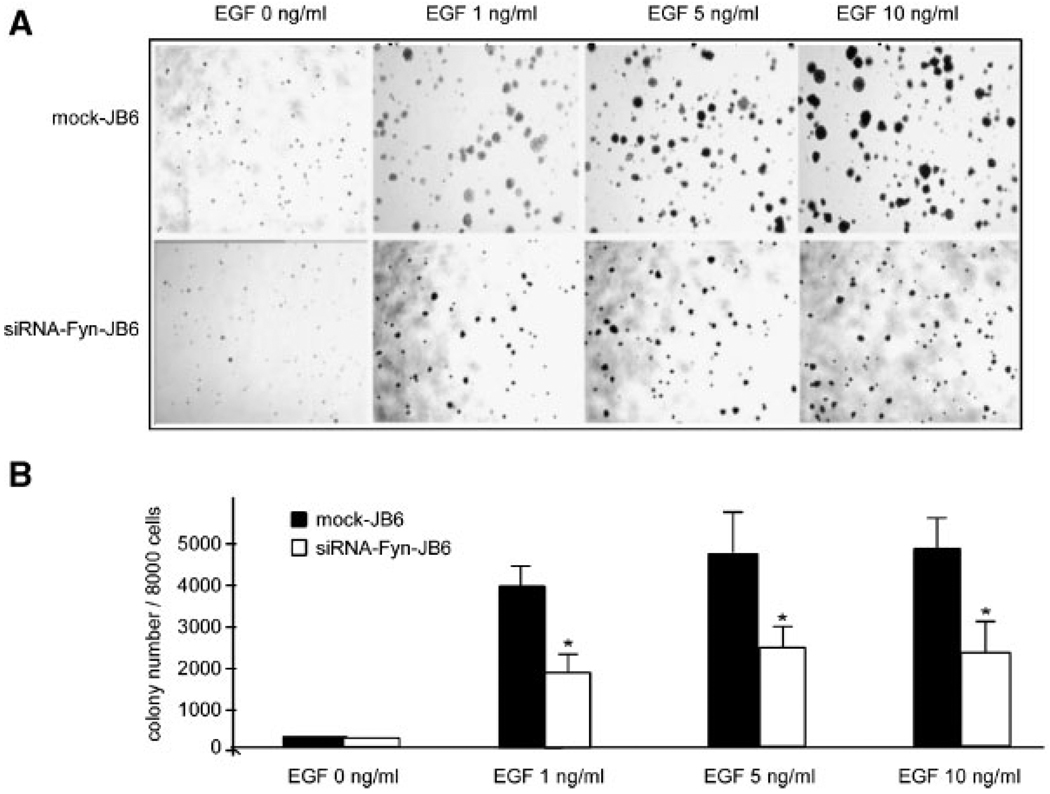
siRNA-Fyn Inhibits EGF-Induced Cell Transformation
To verify whether the inhibitory effect ofEGCGon EGF-promoted JB6 Cl41 cell transformation was regulated by Fyn, we tested the effect of siRNA-Fyn JB6 cells on EGF-induced cell transformation. Our results indicated that colony formation and colony size induced by siRNA-Fyn-JB6 cells was significantly less than that observed in “mock”-transfected JB6 cells (Figure 5A and B). These data demonstrated that decreased Fyn kinase activity suppressed EGF-promoted JB6 cell transformation and thus supports the idea that Fyn is involved in EGF-induced cell transformation.
Figure 5.
siRNA-Fyn inhibits EGF-induced JB6 Cl4l anchorage-independent cell transformation. (A) Colony formation promoted by various concentrations of EGF in mock-transfected JB6 Cl41 cells (upper row) or siRNA-Fyn-JB6 Cl41 cells (lower row) was determined automatically as described in Methods and Materials. (B) Bars indicate the average colony number calculated from three separate experiments ± SD. The asterisk (*) indicates a significant inhibition by siRNA-Fyn-JB6 compared to mock-JB6 cells (*, P<0.05).
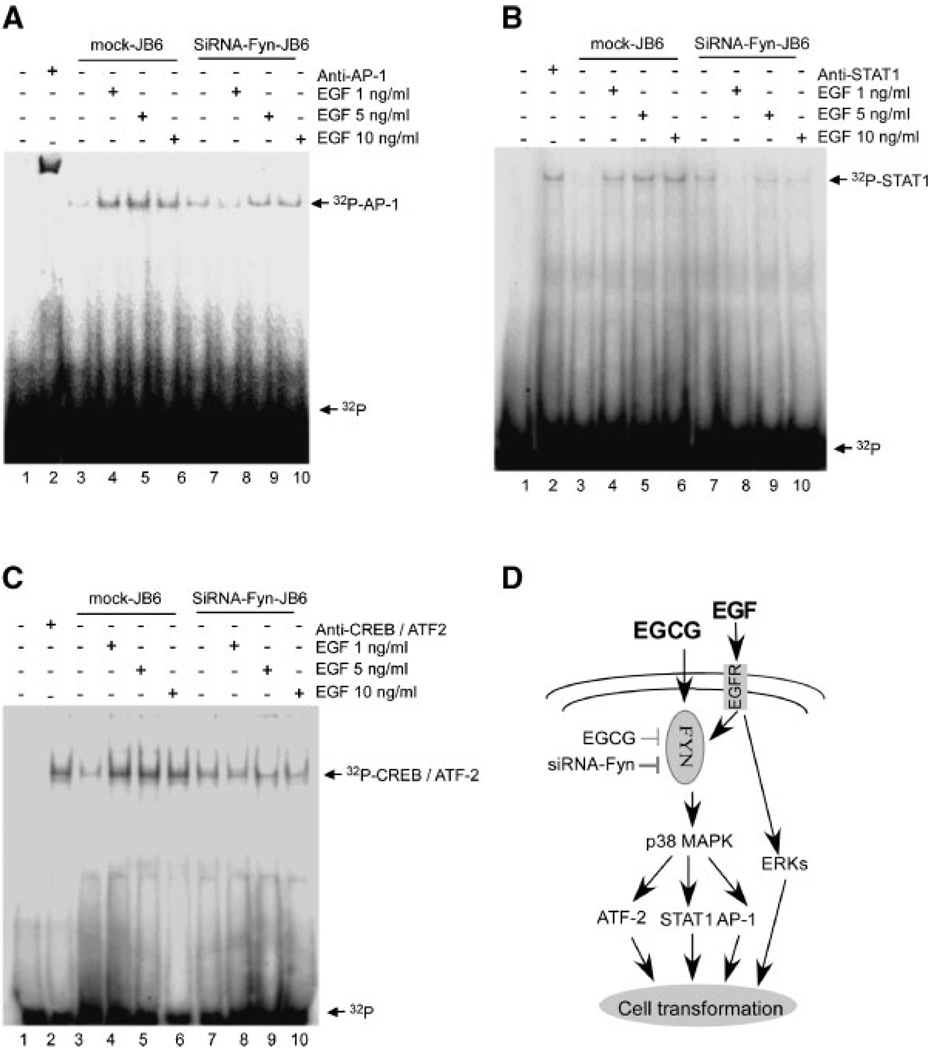
EGF-Induced DNA Binding Ability of AP-1, CREB, and STAT1 is Decreased in siRNA-Fyn-JB6 Cl41 Cells
We also determined the role of Fyn in the regulation of transcription factors AP-1, STAT1, and CREB/ATF-2DNAbinding. Our results indicated that siRNA-Fyn-JB6 cells effectively blocked EGF-induced AP-1 DNA binding (Figure 6A, lanes 8–10), STAT1 DNA binding (Figure 6B, lanes 8–10), and CREB DNA binding (Figure 6C, lanes 8–10) compared with EGF-induced DNA binding of these transcription factors in mock-transfected JB6 Cl41 cells (Figure 6A–C, lanes 4–6). Lane 1 (Figure 6A–C) shows the 32-P labeled probe only.
Figure 6.
siRNA-Fyn inhibits EGF-induced AP-1, STAT1, and CREB/ATF-2 DNA binding. Mock-transfected JB6 Cl41 cells and siRNA-Fyn-JB6 Cl41 cells were treated with EGF and then nuclear proteins were extracted as described in Materials and Methods. siRNA-Fyn cells effectively blocked EGF-induced AP-1 DNA binding (A, lanes 8–10), STAT1 DNA binding (B, lanes 8–10), and CREB/ATF-2 DNA binding (C, lanes 8–10) compared with EGF-induced AP-1 DNA binding in mock-transfected JB6 Cl41 cells (A, lanes 3–5), STAT1 DNA binding (B, lanes 3–5), and CREB/ATF-2 DNA binding (C, lanes 3–5). Lane 1 in each figure shows only 32P as the “cold” probe to confirm specificity and lane 2 indicates that the corresponding antibody was added in the reaction mixture. (D) Proposed signal transduction pathways of EGCG inhibition of JB6 Cl41 cell transformation promoted by EGF. EGCG effectively inhibited EGF-induced Fyn kinase activity and phosphorylation. siRNA-Fyn kinase inhibited EGF-induced phosphorylation of STAT1, ATF-2, and p38 MAP kinase and AP-1, STAT1, and CREB/ATF-2 DNA binding abilities. Thus Fyn plays an important role in EGF-promoted JB6 Cl41 cell transformation through inhibition of its downstream target kinase and transcription factors.
DISCUSSION
EGCG protects against neuronal injury induced by N-methyl-d-aspartate or TRAIL in living mice brain tissue, blocks the formation of neurotoxic reactive oxygen species in neurons, inhibits NF-kappaB activation, abrogates TNF-alpha production in vivo and is regarded as a powerful natural substance for anti-inflammation and neuro-protection [29]. Currently many more researchers are highly interested in the study of EGCG because of its remarkable multifunctional inhibitory effects on carcinogenesis [30–32].
EGCG has been shown to inhibit mouse mammary tumor virus (MMTV)-Her-2/neu-transfected NF639 cell transformation promoted by IL-2 through the inhibition of PI-3 kinase, Akt, and NF-kappaB [33]. In the present study, we found that the inhibition of the tyrosine kinase Fyn was important in EGCG’s suppression of EGF-promoted JB6 cell transformation. Our results also showed that EGCG inhibited Fyn kinase activity in vitro and in vivo and EGCG specifically bound with high affinity to the Fyn Src homology 2 (SH2) domain but not the SH3 domain. Other studies indicated that the SH2 domain interacts with proteins containing phosphorylated tyrosine residues and plays a key role in mediating tyrosine kinase signal transduction [34,35]. Our data strongly suggested that EGCG regulated Fyn kinase activity by binding with the Fyn SH2 domain. We also showed that the colony number and size decreased in siRNA-Fyn-JB6 cells compared with “mock”-transfected JB6 cells. This direct evidence showed that Fyn was involved in EGF-induced JB6 cell transformation.
EGCG is believed to suppress cell transformation through multiple pathways. Studies have indicated that EGCG inhibited growth of the breast cancer cell line NF639 by blocking the Her-2/neu signaling pathway [33] and also suppressed AP-1activity in H-ras-transformed cells [12]. EGCG also has been shown to affect EGF receptor (EGFR) signaling pathways in human head and neck squamous cell carcinomas [36]. Besides the EGFR, EGCG has also been shown to exert its effects on the 67 kDa laminin receptor [37], vascular endothelial growth factor receptor in human colon carcinoma cells [31,38] and on several kinases and phosphatases in human prostate LNCaP cancer cells [39]. Notably, EGCG only appears to exert inhibitory effects on cancer cells, such as H-ras-transformed NIH-pATM fibroblasts, but has little or no effect on normal cells [40].
Accumulating research results show that transcription factors play very important roles in cell transformation. High levels of phosphorylated c-jun, Fra-1, Fra-2, and CREB/ATF-2 proteins correspond with malignant phenotypes in the multistage mouse skin carcinogenesis model [41] and over-expression of ATF-2 is required for growth and progression of mouse skin tumors [42]. STAT1-deficient mice are highly susceptible to chemical carcinogen-induced tumorigenesis [43] and activation of STATs has been found to be associated with viral or oncogene-mediated transformation [44]. STAT1 and STAT5 were shown to be activated in acute lymphocytic leukemia and STAT1, STAT3, and STAT5 are also activated in acute myelocytic leukemia [45]. Nakamura et al. [45] found that human T-cell leukemia virus type 1 (HTLV-1)-transformed T-cells expressed higher amounts of STAT1, STAT3, and STAT5 RNA and proteins than virus-negative T cells. These results suggested that STAT1 and STAT5 play an important role in the cell transformation steps of T-cells promoted by HTLV-1. Our previous study also indicated that STAT1 was involved in EGF-induced JB6 cell transformation [23]. The data presented here showed that EGF-induced phosphorylation of STAT1, ATF-2, and p38 MAP kinase was decreased in siRNA-JB6 cells or with EGCG treatment and that the DNA binding abilities of AP-1, STAT1, and CREB induced by EGF were also decreased in siRNA-Fyn-JB6 cells. Our results further demonstrated that Fyn mediated EGF-induced cell transformation through regulation of its downstream transcription factors AP-1, CREB/ATF-2, and STAT1.
In summary, Fyn participates in cell transformation by inducing p38 MAP kinase, which is an upstream kinase important in activating transcription factors, including ATF-2, STAT1, and AP-1, which are all important in EGF-induced cell transformation (Figure 6D). This hypothesis is supported by a recent study, in which Vittal et al. [46] used DNA microarray analysis to show that EGCG influenced the expression of many genes in human bronchial epithelial 21BES cells. More new EGCG targets are most likely yet to be discovered and EGCG could be utilized as a powerful agent to prevent and treat cancer in the near future.
ACKNOWLEDGMENTS
We thank Andria Hansen for secretarial assistance. Grant support: This work was supported in part by the Hormel Foundation, and NIH grants CA81064, CA88961, CA111536, and CA120388.
REFERENCES
- 1.Naasani I, Oh-Hashi F, Oh-Hara T, et al. Blocking telomerase by dietary polyphenols is a major mechanism for limiting the growth of human cancer cells in vitro and in vivo. Cancer Res. 2003;63:824–830. [PubMed] [Google Scholar]
- 2.Ahmad N, Cheng P, Mukhtar H. Cell cycle dysregulation by green tea polyphenol epigallocatechin-3-gallate. Biochem Biophys Res Commun. 2000;275:328–334. doi: 10.1006/bbrc.2000.3297. [DOI] [PubMed] [Google Scholar]
- 3.Liang YC, Huang YT, Tsai SH, Lin-Shiau SY, Chen CF, Lin JK. Suppression of inducible cyclooxygenase and inducible nitric oxide synthase by apigenin and related flavonoids in mouse macrophages. Carcinogenesis. 1999;20:1945–1952. doi: 10.1093/carcin/20.10.1945. [DOI] [PubMed] [Google Scholar]
- 4.Barthelman M, Bair WB, III, Stickland KK, et al. (−)-Epigallocatechin-3-gallate inhibition of ultraviolet B-induced AP-1 activity. Carcinogenesis. 1998;19:2201–2204. doi: 10.1093/carcin/19.12.2201. [DOI] [PubMed] [Google Scholar]
- 5.Bode AM, Dong Z. Targeting signal transduction pathways by chemopreventive agents. Mutat Res. 2004;555:33–51. doi: 10.1016/j.mrfmmm.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Chen NY, Ma WY, Yang CS, Dong Z. Inhibition of arsenite-induced apoptosis and AP-1 activity by epigallocatechin-3-gallate and theaflavins. J Environ Pathol Toxicol Oncol. 2000;19:287–295. [PubMed] [Google Scholar]
- 7.Nomura M, Ma WY, Huang C, et al. Inhibition of ultraviolet B-induced AP-1 activation by theaflavins from black tea. Mol Carcinog. 2000;28:148–155. [PubMed] [Google Scholar]
- 8.Moyers SB, Kumar NB. Green tea polyphenols and cancer chemoprevention: Multiple mechanisms and endpoints for phase II trials. Nutr Rev. 2004;62:204–211. doi: 10.1111/j.1753-4887.2004.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 9.Jatoi A, Ellison N, Burch PA, et al. A phase II trial of green tea in the treatment of patients with androgen independent metastatic prostate carcinoma. Cancer. 2003;97:1442–1446. doi: 10.1002/cncr.11200. [DOI] [PubMed] [Google Scholar]
- 10.Pisters KM, Newman RA, Coldman B, et al. Phase I trial of oral green tea extract in adult patients with solid tumors. J Clin Oncol. 2001;19:1830–1838. doi: 10.1200/JCO.2001.19.6.1830. [DOI] [PubMed] [Google Scholar]
- 11.Dong Z, Ma W, Huang C, Yang CS. Inhibition of tumor promoter-induced activator protein 1 activation and cell transformation by tea polyphenols, (−)-epigallocatechin gallate, and theaflavins. Cancer Res. 1997;57:4414–4419. [PubMed] [Google Scholar]
- 12.Chung JY, Huang C, Meng X, Dong Z, Yang CS. Inhibition of activator protein 1 activity and cell growth by purified green tea and black tea polyphenols in H-ras-transformed cells: Structure-activity relationship and mechanisms involved. Cancer Res. 1999;59:4610–4617. [PubMed] [Google Scholar]
- 13.Chen M, Hou X, Zhang G. Tyrosine kinase and tyrosine phosphatase participate in regulation of interactions of NMDA receptor subunit 2A with Src and Fyn mediated by PSD-95 after transient brain ischemia. Neurosci Lett. 2003;339:29–32. doi: 10.1016/s0304-3940(02)01439-8. [DOI] [PubMed] [Google Scholar]
- 14.Miyakawa T, Yagi T, Kitazawa H, et al. Fyn-kinase as a determinant of ethanol sensitivity: Relation to NMDA-receptor function. Science. 1997;278:698–701. doi: 10.1126/science.278.5338.698. [DOI] [PubMed] [Google Scholar]
- 15.Yagi T, Aizawa S, Tokunaga T, Shigetani Y, Takeda N, Ikawa Y. A role for Fyn tyrosine kinase in the suckling behaviour of neonatal mice. Nature. 1993;366:742–745. doi: 10.1038/366742a0. [DOI] [PubMed] [Google Scholar]
- 16.Grant SG, O’Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER. Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science. 1992;258:1903–1910. doi: 10.1126/science.1361685. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami T, Kawakami Y, Aaronson SA, Robbins KC. Acquisition of transforming properties by FYN, a normal SRC-related human gene. Proc Natl Acad Sci USA. 1988;85:3870–3874. doi: 10.1073/pnas.85.11.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellbrock C, Gomez A, Schartl M. Melanoma development and pigment cell transformation in xiphophorus. Microsc Res Tech. 2002;58:456–463. doi: 10.1002/jemt.10163. [DOI] [PubMed] [Google Scholar]
- 19.Wellbrock C, Schartl M. Activation of phosphatidylinositol 3-kinase by a complex of p59fyn and the receptor tyrosine kinase Xmrk is involved in malignant transformation of pigment cells. Eur J Biochem. 2000;267:3513–3522. doi: 10.1046/j.1432-1327.2000.01378.x. [DOI] [PubMed] [Google Scholar]
- 20.Wellbrock C, Weisser C, Geissinger E, Troppmair J, Schartl M. Activation of p59(Fyn) leads to melanocyte dedifferentiation by influencing MKP-1-regulated mitogen-activated protein kinase signaling. J Biol Chem. 2002;277:6443–6454. doi: 10.1074/jbc.M110684200. [DOI] [PubMed] [Google Scholar]
- 21.He Z, Cho YY, Ma WY, Choi HS, Bode AM, Dong Z. Regulation of ultraviolet B-induced phosphorylation of histone H3 at serine 10 by Fyn kinase. J Biol Chem. 2005;280:2446–2454. doi: 10.1074/jbc.M402053200. [DOI] [PubMed] [Google Scholar]
- 22.Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Z, Cho YY, Liu G, Ma WY, Bode AM, Dong Z. p38 Mitogen-activated protein kinase regulation of JB6 Cl41 cell transformation promoted by epidermal growth factor. J Biol Chem. 2003;278:26435–26442. doi: 10.1074/jbc.M303859200. [DOI] [PubMed] [Google Scholar]
- 24.Ermakova S, Choi BY, Choi HS, Kang BS, Bode AM, Dong Z. The intermediate filament protein vimentin is a new target for epigallocatechin gallate. J Biol Chem. 2005;280:16882–16890. doi: 10.1074/jbc.M414185200. [DOI] [PubMed] [Google Scholar]
- 25.He Z, Ma WY, Liu G, Zhang Y, Bode AM, Dong Z. Arsenite-induced phosphorylation of histone H3 at serine 10 is mediated by Akt1, extracellular signal-regulated kinase 2, and p90 ribosomal S6 kinase 2 but not mitogen- and stress-activated protein kinase 1. J Biol Chem. 2003;278:10588–10593. doi: 10.1074/jbc.M208581200. [DOI] [PubMed] [Google Scholar]
- 26.Ye J, Zhang X, Dong Z. Characterization of the human granulocyte-macrophage colony-stimulating factor gene promoter: An AP1 complex and an Sp1-related complex transactivate the promoter activity that is suppressed by a YY1 complex. Mol Cell Biol. 1996;16:157–167. doi: 10.1128/mcb.16.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colburn NH, Gindhart TD. Specific binding of transforming growth factor correlates with promotion of anchorage independence in EGF receptorless mouse JB6 cells. Biochem Biophys Res Commun. 1981;102:799–807. doi: 10.1016/s0006-291x(81)80202-1. [DOI] [PubMed] [Google Scholar]
- 28.Colburn NH, Talmadge CB, Gindhart TD. Transfer of sensitivity to tumor promoters by transfection of DNA from sensitive into insensitive mouse JB6 epidermal cells. Mol Cell Biol. 1983;3:1182–1186. doi: 10.1128/mcb.3.7.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aktas O, Prozorovski T, Smorodchenko A, et al. Green tea epigallocatechin-3-gallate mediates T cellular NF-kappa B inhibition and exerts neuroprotection in autoimmune encephalomyelitis. J Immunol. 2004;173:5794–5800. doi: 10.4049/jimmunol.173.9.5794. [DOI] [PubMed] [Google Scholar]
- 30.Paschka AG, Butler R, Young CY. Induction of apoptosis in prostate cancer cell lines by the green tea component, (−)-epigallocatechin-3-gallate. Cancer Lett. 1998;130:1–7. doi: 10.1016/s0304-3835(98)00084-6. [DOI] [PubMed] [Google Scholar]
- 31.Jung YD, Kim MS, Shin BA, et al. EGCG, a major component of green tea, inhibits tumour growth by inhibiting VEGF induction in human colon carcinoma cells. Br J Cancer. 2001;84:844–850. doi: 10.1054/bjoc.2000.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annabi B, Lachambre MP, Bousquet-Gagnon N, Page M, Gingras D, Beliveau R. Green tea polyphenol (−)-epigallocatechin 3-gallate inhibits MMP-2 secretion and MT1-MMP-driven migration in glioblastoma cells. Biochim Biophys Acta. 2002;1542:209–220. doi: 10.1016/s0167-4889(01)00187-2. [DOI] [PubMed] [Google Scholar]
- 33.Pianetti S, Guo S, Kavanagh KT, Sonenshein GE. Green tea polyphenol epigallocatechin-3 gallate inhibits Her-2/neu signaling, proliferation, and transformed phenotype of breast cancer cells. Cancer Res. 2002;62:652–655. [PubMed] [Google Scholar]
- 34.Ladbury JE, Hensmann M, Panayotou G, Campbell ID. Alternative modes of tyrosyl phosphopeptide binding to a Src family SH2 domain: Implications for regulation of tyrosine kinase activity. Biochemistry. 1996;35:11062–11069. doi: 10.1021/bi960543e. [DOI] [PubMed] [Google Scholar]
- 35.Resh MD. Myristylation and palmitylation of Src family members: The fats of the matter. Cell. 1994;76:411–413. doi: 10.1016/0092-8674(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 36.Masuda M, Suzui M, Weinstein IB. Effects of epigallocate-chin-3-gallate on growth, epidermal growth factor receptor signaling pathways, gene expression, and chemosensitivity in human head and neck squamous cell carcinoma cell lines. Clin Cancer Res. 2001;7:4220–4229. [PubMed] [Google Scholar]
- 37.Tachibana H, Koga K, Fujimura Y, Yamada K. A receptor for green tea polyphenol EGCG. Nat Struct Mol Biol. 2004;11:380–381. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- 38.Lee YK, Bone ND, Strege AK, Shanafelt TD, Jelinek DF, Kay NE. VEGF receptor phosphorylation status and apoptosis is modulated by a green tea component, epigallocatechin-3-gallate (EGCG), in B-cell chronic lymphocytic leukemia. Blood. 2004;104:788–794. doi: 10.1182/blood-2003-08-2763. [DOI] [PubMed] [Google Scholar]
- 39.Wang SI, Mukhtar H. Gene expression profile in human prostate LNCaP cancer cells by (−) epigallocatechin-3-gallate. Cancer Lett. 2002;182:43–51. doi: 10.1016/s0304-3835(02)00065-4. [DOI] [PubMed] [Google Scholar]
- 40.Wang YC, Bachrach U. The specific anti-cancer activity of green tea (−)-epigallocatechin-3-gallate (EGCG) Amino Acids. 2002;22:131–143. doi: 10.1007/s007260200002. [DOI] [PubMed] [Google Scholar]
- 41.Zoumpourlis V, Papassava P, Linardopoulos S, Gillespie D, Balmain A, Pintzas A. High levels of phosphorylated c-Jun, Fra-1, Fra-2 and ATF-2 proteins correlate with malignant phenotypes in the multistage mouse skin carcinogenesis model. Oncogene. 2000;19:4011–4021. doi: 10.1038/sj.onc.1203732. [DOI] [PubMed] [Google Scholar]
- 42.Papassava P, Gorgoulis VG, Papaevangeliou D, Vlahopoulos S, van Dam H, Zoumpourlis V. Overexpression of activating transcription factor-2 is required for tumor growth and progression in mouse skin tumors. Cancer Res. 2004;64:8573–8584. doi: 10.1158/0008-5472.CAN-03-0955. [DOI] [PubMed] [Google Scholar]
- 43.Imada K, Leonard WJ. The Jak-STAT pathway. Mol Immunol. 2000;37:1–11. doi: 10.1016/s0161-5890(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 44.Gouilleux-Gruart V, Gouilleux F, Desaint C, et al. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 45.Nakamura N, Fujii M, Tsukahara T, et al. Human T-cell leukemia virus type 1 Tax protein induces the expression of STAT1 and STAT5 genes in T-cells. Oncogene. 1999;18:2667–2675. doi: 10.1038/sj.onc.1202608. [DOI] [PubMed] [Google Scholar]
- 46.Vittal R, Selvanayagam ZE, Sun Y, et al. Gene expression changes induced by green tea polyphenol (−)-epigallocatechin-3-gallate in human bronchial epithelial 21BES cells analyzed by DNA microarray. Mol Cancer Ther. 2004;3:1091–1099. [PubMed] [Google Scholar]