Self-propulsion of micro- and nano-scale objects can be achieved by harnessing the chemical free energy of the environment through substrate catalysis. For example, we and others have demonstrated that energy arising from catalytic reactions can drive the movement of asymmetric particles on the micron- and sub-micron length scales by self-electrophoresis, self-diffusiophoresis, and bubble propulsion.1 Autonomous motion of symmetric colloidal particles with enzymatic (catalytic) sites in the presence of a substrate has been proposed,2 but has not yet been demonstrated experimentally. In addition, catalysis-enhanced diffusion at the single molecule scale has not been reported. Due to their great diversity, use of enzymes as catalytic motors would vastly expand the available methods to power nano/micromotors. In this study, we provide the first single-molecule-scale measurements of catalysis-enhanced diffusion of the urease enzyme and calculate the force responsible for this enhancement using Brownian dynamics simulations.
Diffusion coefficients were measured by fluorescence correlation spectroscopy (FCS), using time correlated single photon counting (TCSPC) instrumentation developed previously.3 Briefly, fluctuations in fluorescence intensity arising from diffusion of probe molecules were autocorrelated and fit by a multi-component 3D diffusion model to determine the diffusion coefficient of individual species, e.g. free dye and tagged-enzyme.4,5 Fluorescent tagging of the enzyme was confirmed by the FCS autocorrelation curves by noting that fast and slow diffusing components corresponded to free dye and fluorescently-tagged urease, respectively. Diffusion coefficient of the free dye in buffer was 3.09 × 10−6 cm2/s and exhibited no significant change in the presence of urea. Thus, the diffusion time of the free dye was fixed for all subsequent curve-fits. The diffusion coefficient of urease in buffer was 3.18 × 10−7 cm2/s which corresponded to a hydrodynamic radius of 6.9 nm, consistent with the hydrodynamic radius of 7 nm reported by Follmer et al.6 Urease concentrations used in this study ranged from 10 to 100 μg/ml, well below the reported concentrations at which aggregation of urease occurs.6
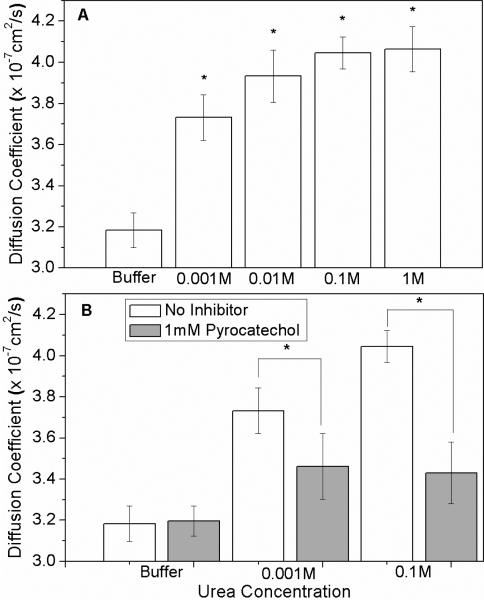
Diffusion coefficients of urease in urea concentrations ranging from 0 to 1 M, increased from 3.18 × 10−7 cm2/s to 4.06 × 10−7 cm2/s, respectively (Fig. 1A), representing a catalysis-induced increase in diffusion coefficient of 28%. This increase in diffusion coefficient of urease saturated at around 0.1 M urea, similar to the concentration at which urea hydrolysis reaction rate saturates. Viscosities of these different urea solutions as measured using a cone-plate viscometer were not significantly different than buffer. Urea concentrations were kept below 1 M to avoid denaturation.7 To test whether enhanced diffusion in the presence of urea was a result of catalysis, urease activity was inhibited by pretreatment of the enzyme with 1 mM pyrocatechol for 2 h.8 In the presence of inhibitor only, the diffusion of urease in buffer was not significantly different from urease without inhibitor suggesting that the inhibitor itself does not alter the hydrodynamic radius of the enzyme. Similarly, when urease was inhibited and exposed to either 0.001 or 0.1 M urea, diffusion coefficients were significantly attenuated compared to matched controls (Fig. 1B).
Figure 1.
(A) Diffusion coefficient of urease increased with increasing substrate concentration. (B) Increase in urease diffusion coefficient was significantly attenuated by the urease inhibitor, pyrocatechol. Error bars represent standard deviation. * indicates a significance value of P < 0.05
We hypothesized that the origin of enhanced diffusion could be the generation of charged reaction products that results in an asymmetric electric field within angstroms of the enzyme as has been observed in asymmetric particles1,9 The diffusion coefficient of NH4+ ions is higher than that of the anions (HCO3− and HPO42−) generated by the hydrolysis of urea in phosphate buffer. As each urea molecule is hydrolyzed, these ions are released at the surface of the enzyme and the faster diffusion of NH4+ creates a local electric field. This field could exert an electrophoretic force in the order of few pN for a short interval of time until the ions diffuse away from the double layer. Since it is technically challenging to measure such short time-scale forces, we estimated the impulsive force due to a single turnover using Brownian dynamics simulations.5
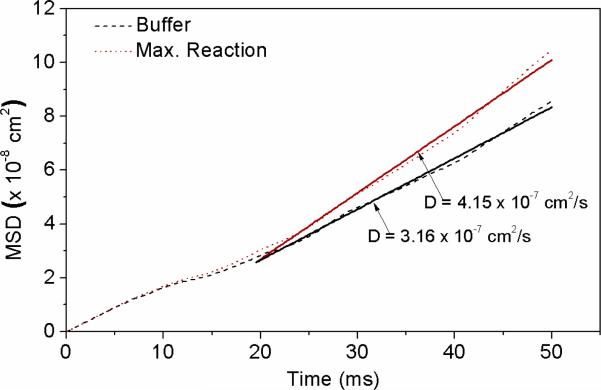
Simulation parameters were as follows. Maximum reaction rate per catalytic site was 2.3 × 104 s−1, as reported by Blakeley et al.10 A 10 ns impulse time per reaction corresponded to the time it would take for the ions to diffuse one Debye length from the urease molecule, dissipating the electrophoretic force. Force per impulse was then adjusted in successive Brownian dynamics simulations until the diffusion coefficient increased from 3.16 × 10−7 cm2/s (no reaction) to 4.15 × 10−7 cm2/s (maximum reaction rate), as observed in experiments. To account for this change, self-electrophoresis would need to produce a force of 12 pN per turnover (Fig. 2.). This is close to the force (20 pN) calculated for an anion (q = 1e) 2 nm from the center of an enzyme (q = 25e) in water (ε = 78). In addition, the electrophoretic force should increase with decreasing ionic strength of the medium, potentially further increasing the diffusion rate.
Figure 2.
Mean square displacement of urease under no reaction (black) and maximum reaction rate assuming a force of 12 pN/turnover (red) obtained from Brownian dynamics simulations. Solid lines are linear fits at longer time scales.
As an alternative explanation, we explored the possibility that local pH changes upon catalysis might enhance diffusion because the hydrolysis of urea should result in an increase in the local pH. This hypothesis is based on a study in which bovine serum albumin diffusion depended strongly on pH, ionic strength, and protein concentration.11 In that study, the diffusion coefficient of the protein increased with increase in pH, with a minimum at the isoelectric point, where the surface charge on the protein is zero.12 Increase in pH is associated with an increase in surface charge of the protein resulting in stronger protein-protein and protein-counterion columbic interactions, which can cause an increase in the protein's diffusion coefficient.
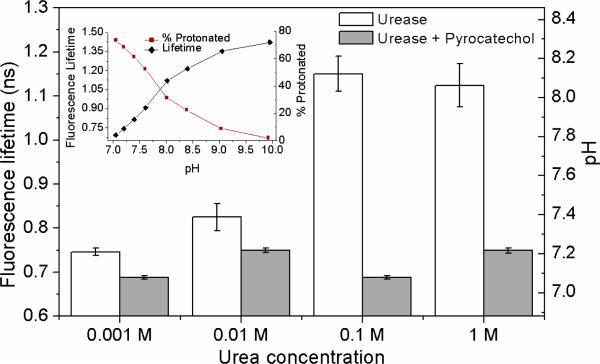
To measure pH in the vicinity of the enzyme, we tagged urease with SNARF-1, the fluorescence lifetime of which changes with pH of the local medium. SNARF-1 based pH measurements reflect the relative fraction of the molecules in the protonated and deprotonated states, each exhibiting different fluorescence lifetimes.13 Fluorescence lifetime curves of SNARF-1 measured in various pH solutions were fit with a double-exponential decay model.4 Decay time constants of 0.44 ns and 1.42 ns, corresponded to the protonated and deprotonated states respectively.14 This result is shown in the inset of Fig. 3, where the relative fraction of protonated SNARF-1 molecules decreases and the amplitude-weighted fluorescence lifetime increases with increase in pH. As shown in Fig. 3, pH in the vicinity of the enzyme increased modestly but significantly at urea concentrations of 0.01 M and above, which would result in less than 5% increase in diffusion, far below the 28% increase measured using FCS.5 No increase in local pH was observed when the enzyme was inhibited suggesting that pH changes were due to catalysis. Thus, although catalysis-induced changes of pH around single enzymes may be of biological and chemical significance, and represents a novel measurement, the observed increase in pH does not explain the large enhancement of diffusion induced by catalysis. In addition, the local rise in the temperature due to urea hydrolysis was estimated to be in μK range, which is too small to explain the observed changes in diffusion coefficient.5 Thus movement through a phoretic mechanism remains the most plausible explanation.
Figure 3.
SNARF-1 fluorescence lifetime was measured as a function of solution pH (inset). Catalysis of urea was accompanied by a significant increase in pH (relative to [urea] = 0.001 M). This increase was significantly attenuated by pyrocatechol for all urea concentrations tested. Error bars represent standard deviation.
In conclusion, we report here that catalysis can alter the diffusive mobility of an enzyme in the presence of a substrate. We anticipate that in the presence of a substrate gradient, the enzyme molecules will exhibit collective directional motion (chemotaxis).15 These observations lay the foundation for development of novel enzyme-driven nanomotors by asymmetric placement of the enzyme on micro- and nano-scale objects. Additionally, many molecular machines in living systems function as Brownian ratchets.16 The movements of these enzyme-based biological machines may be greatly facilitated by catalysis-induced forces resulting from substrate turnover.
Supplementary Material
Acknowledgements
PJB acknowledges financial support from NIH (R01 HL 07754201) and NSF (BES 0238910), and AS from the Penn State Center for Nanoscale Science (NSF-MRSEC, DMR-0820404) and NSF-NIRT CTS-0506967.
Footnotes
Supporting Information Available: Experimental methods, Brownian dynamics simulation methodology, estimates of local temperature and electrolyte friction coefficient. This information is available free of charge via the internet at http://pubs.acs.org.
Contributor Information
Ayusman Sen, Email: asen@psu.edu.
Peter J. Butler, Email: pbutler@psu.edu.
References
- 1.Reviews: Paxton W, Sundararajan S, Mallouk T, Sen A. Angew. Chem. Int. Ed. 2006;45:5420–5429. doi: 10.1002/anie.200600060.Wang J. ACS Nano. 2009;3:4–9. doi: 10.1021/nn800829k.Sanchez S, Pumera M. Chem. Asian J. 2009;4:1402–1430. doi: 10.1002/asia.200900143.
- 2.Golestanian R. Phy. Rev. Lett. 2009;102(14):188305. doi: 10.1103/PhysRevLett.102.188305. [DOI] [PubMed] [Google Scholar]
- 3.Gullapalli RR, Tabouillot T, Mathura R, Dangaria JH, Butler PJ. J. Biomed. Opt. 2007;12:014012. doi: 10.1117/1.2673245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lakowicz JR. Principles of fluorescence spectroscopy. Springer; New York: 2006. [Google Scholar]
- 5.Supporting information.
- 6.Follmer C, Pereira FV, da Silveira NP, Carlini CR. Biophys. Chem. 2004;111:79–87. doi: 10.1016/j.bpc.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 7.a Azari F, Hosseinkhani S, Nemat-Gorgani M. Appl. Biochem. Biotechnol. 2001;94:265–277. doi: 10.1385/abab:94:3:265. [DOI] [PubMed] [Google Scholar]; b Kobayashi S, Yonezu SJ, Kawakita H, Saito K, Sugita K, Tamada M, Sugo T, Lee W. Biotechnol. Progress. 2003;19:396–399. doi: 10.1021/bp020072p. [DOI] [PubMed] [Google Scholar]
- 8.Kot M, Zaborska WJ. Enzyme Inhibition Med. Chem. 2003;18:413–417. doi: 10.1080/1475636031000152268. [DOI] [PubMed] [Google Scholar]
- 9.Paxton WE, Sen A, Mallouk TE. Chem. Europ. J. 2005;11:6462–6470. doi: 10.1002/chem.200500167. [DOI] [PubMed] [Google Scholar]
- 10.Blakeley RL, Webb EC, Zerner B. Biochemistry. 1969;8:1984–1990. doi: 10.1021/bi00833a031. [DOI] [PubMed] [Google Scholar]
- 11.a Gaigalas AK, Hubbard JB, Mccurley M, Woo S. J. Phys. Chem. 1992;96:2355–2359. [Google Scholar]; b Raj T, Flygare WH. Biochemistry. 1974;13:3336–3340. doi: 10.1021/bi00713a024. [DOI] [PubMed] [Google Scholar]
- 12.Doherty P, Benedek GB. J. Chem. Phys. 1974;61:5426–5434. [Google Scholar]
- 13.Brasselet S, Moerner WE. Single Molecules. 2000;1:17–23. [Google Scholar]
- 14.Srivastava A, Krishnamoorthy G. Anal. Biochem. 1997;249:140–146. doi: 10.1006/abio.1997.2164. [DOI] [PubMed] [Google Scholar]
- 15.a Hong Y, Blackman NMK, Kopp ND, Sen A, Velegol D. Phys. Rev. Lett. 2007;99(14):178103. doi: 10.1103/PhysRevLett.99.178103. [DOI] [PubMed] [Google Scholar]; b Sen A, Ibele M, Hong Y, Velegol D. Faraday Discuss. 2009;143:15–27. doi: 10.1039/b900971j. [DOI] [PubMed] [Google Scholar]; c Hong Y, Chaturvedi N, Velegol D, Sen A. Phys. Chem. Chem. Phys. 2010 doi: 10.1039/b917741h. DOI: 10.1039/b917741h. [DOI] [PubMed] [Google Scholar]
- 16.Moran SJ, Flanagan JF, Namy O, Stuart DI, Brierley I, Gilbert RJC. Structure. 2008;16:664–672. doi: 10.1016/j.str.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tomkiewicz D, Nouwen N, Driessen AJM. FEBS Letters. 2007;581:2820–2828. doi: 10.1016/j.febslet.2007.04.015. [DOI] [PubMed] [Google Scholar]; c Ait-Haddou R, Herzog W. Cell Biochemistry Biophysics. 2003;38:191–214. doi: 10.1385/CBB:38:2:191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.