Abstract
Peroxiredoxin 6 (Prdx6) differs from other mammalian peroxiredoxins both in its ability to reduce phospholipid hydroperoxides at neutral pH and in having phospholipase A2 (PLA2) activity that is maximal at acidic pH. We previously showed an active site C47 for peroxidase activity and a catalytic triad S32-H26-D140 necessary for binding of phospholipid and PLA2 activity. This study evaluated binding of reduced and oxidized phospholipid hydroperoxide to Prdx6 at cytosolic pH. Incubation of recombinant Prdx6 with 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine hydroperoxide (PLPCOOH) resulted in peroxidase activity, cys47 oxidation as detected with Prdx6-SO2(3) antibody, and a marked shift in the Prdx6 melting temperature by circular dichroism analysis indicating that PLPCOOH is a specific substrate for Prdx6. Preferential Prdx6 binding to oxidized liposomes was detected by changes in DNS-PE or bis-Pyr fluorescence and by ultrafiltration. Site-specific mutation of S32 or H26 in Prdx6 abolished binding while D140 mutation had no effect. Treatment of A549 cells with peroxides led to lipid peroxidation and translocation of Prdx6 from the cytosol to the cell membrane. Thus, the pH specificity for the two enzymatic activities of Prdx6 can be explained by the differential binding kinetics of the protein; Prdx6 binds to reduced phospholipid at acidic pH but at cytosolic pH binds only phospholipid that is oxidized compatible with a role for Prdx6 in the repair of peroxidized cell membranes.
Keywords: oxidant stress, catalytic triad, MJ33, mercaptosuccinate, protein oxidation, liposomes
INTRODUCTION
Peroxiredoxins are a widely distributed superfamily of peroxidases that use thiols to reduce enzymatically H2O2 and other hydroperoxide substrates [1]. These enzymes have been classified as 2-cys, 1-cys or atypical based on the number of conserved cysteines and consequent mechanism of catalysis[2–6]. Peroxiredoxin 6 is the only mammalian 1-cys peroxiredoxin and unlike the other peroxiredoxins, utilizes glutathione (GSH) [7–9] or ascorbate [10] rather than thioredoxin as the physiological reductant. In addition to H2O2 and short chain aliphatic hydroperoxides, Prdx 6 can reduce phospholipid hydroperoxides such as phosphatidylcholine hydroperoxide (PCOOH1) with a relatively high rate constant similar to that for H2O2 reduction (~106 M−1 s−1) [7, 11]. This property is thought to be of major importance in the ability of Prdx6 to protect cells against oxidant stress. Reduction of the enzyme by GSH requires an intermediary step of protein glutathionylation mediated by π glutathione S-transferase (πGST) [8, 12]. Based on the crystal structure of the protein, the catalytic Cys is situated at the base of a pocket that restricts accessibility of the resolving thiol for interaction with oxidized intermediate and restoration of the reduced state of the protein [13]. Thus, glutathionylation likely induces a conformational change that is conducive to GSH access.
Prdx6 alone among the peroxiredoxins possesses phospholipase A2 (PLA2) activity in addition to its peroxidatic function [5, 9, 11]. The PLA2 activity of Prdx6 has been demonstrated to have an important role in the metabolism of lung surfactant phospholipids [14]. The PLA2 activity is maximal at pH 4 while the peroxidase activity is maximal in the pH 7–8 range [9]. Compatible with the proposed physiological role of these two activities, the presence of Prdx6 has been demonstrated in both the neutral pH (cytosol) and acidic pH (lysosomes; lamellar bodies) compartments of rat lungs [15, 16]. Our previous studies to evaluate PLA2 activity at pH 4 showed binding of Prdx6 to the phospholipid substrate as an important step leading to hydrolysis of the sn-2 phospholipid bond [17]. These studies provided evidence that the phospholipid head group bound at the protein surface, in the vicinity of the PLA2 catalytic triad, while the sn-2 acyl chain is inserted into the protein core allowing alignment of the −OOH group to the catalytic Cys. Thus, specific positioning between the substrate and protein could explain the dual enzymatic function of Prdx6. The present study was designed to evaluate binding of Prdx6 to substrate at cytosolic pH compatible with its peroxidase function. These results can explain the role of cytosolic Prdx6 in protection against oxidant stress.
MATERIALS AND METHODS
Materials
1,2- Bis palmitoyl-sn-glycero-3-phosphocholine (DPPC), 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine (PLPC), egg yolk phosphatidylcholine (PC), phosphatidyl phosphatidylglycerol (PG), phosphatidylserine (PS), and cholesterol (chol) were purchased from Avanti-Polar Lipids (Birmingham, AL). N-(5-dimethylaminonaphthalene-1-sulfonyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (DNS-PE), and 1,2-bis(1-pyrenedecanoyl1)-sn-glycero-3-phosphocholine (bisPyr-PC) were purchased from Molecular Probes (Eugene, OR). MJ33 (1-hexadecyl-3-trifluoroethylglycero-sn-2-phosphomethanol), mercaptosuccinic acid, H2O2, tert butyl hydroperoxide (t-BOOH), catalase and all other chemicals were purchased from Sigma (St. Louis, MO). The polyclonal antibody to Prdx has been described previously [18]. Antibody to oxidized Prdx6 (sulfinic/sulfonic Cys) was obtained from Lab Frontiers (Seoul, Korea). PLPC hydroperoxide (PLPCOOH) was prepared by treatment of PLPC with 15-lipoxygenase obtained from Cayman Chemical (Ann Arbor, MI) and was purified using standard procedures [7, 18]. Anti-caveolin-1 monoclonal antibody was from BD Bioscience (San Jose, CA). A459 cells were obtained by the American Type Culture Collection (ATCC, Manassas, VA).
Expression of recombinant Prdx6
Rat wild type and mutant Prdx6 proteins (S32A, H26A, D140A and DC47S) were expressed in pETBlue-1 Novagen, Madison, WI) as described previously [9, 17]. S32L, S32T and S32V were prepared using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA), following protocols recommended by the supplier. Mutagenic oligonucleotide primer pairs were purchased from Operon Biotechnologies (currently Eurofins Mwg/Operon, Huntsville, AL). The sequences are shown in the Supplementary Data.. S32, H26 and D140 are the putative amino acids constituting the PLA2 catalytic triad and C47 is the catalytic residue for peroxiredoxin activity as described earlier [17]. Rat Prdx6 contains a single Cys residue and lacks the additional non-catalytic Cys present in the human and mouse proteins [17, 19].
Proteins were purified by column chromatography which gave a homogeneous product as determined by SDS-PAGE and Western blot (not shown). Size-exclusion FPLC (YMC-10 column, 500×8 mm ID, 120 Å pore size from Waters, Milford, MA using 1.0 ml/min isocratic elution with 0.15 M NaCl in 0.1 M phosphate buffer, pH 7.0 [8] demonstrated a single peak for the S32A mutant at 25 kDa while two major peaks were seen for the other proteins compatible with the 25 kDa monomeric and 50 kDa homodimeric forms [8, 17]. Treatment with NBD-Cl followed by spectrophotometric detection of the NBD-Prdx6 adduct at 347 nm, using an extinction coefficient of 13,400 M−1 cm−1 [20], indicated that ~70% of cys in recombinant wild type protein was oxidized to the sulfenic acid state (data not shown). Prdx6 was phosphorylated in vitro by incubation with extracellular regulated kinase 2 (Erk 2) as described previously [21].
Liposome preparation
Unilamellar liposomes were prepared from a mixture of DPPC: egg PC: chol: PG (50:25:15:10,mol/mol) by extrusion under pressure [17]. We have used this composition for liposomes as a mimic for the lung surfactant. For fluorescently labeled liposomes, 2 mol% of egg PC was equivalently replaced with DNS-PE or bisPyr-PC. To prepare an oxidized liposomal phospholipid substrate, PLPC was substituted for egg PC; these liposomes were peroxidized by exposure to an •OH-generating system (10 µM Cu2+ in the presence of 0.20 mM ascorbate) [22] for 45 min at room temperature followed by dialysis for 2 h against PBS (pH 7.4) using a 10 kDa molecular mass cut-off Slide-ALyzer ® dialysis cassette (Pierce, Rockford, IL). Liposomal lipids were extracted [23] and analyzed by HPLC using PLPC and PLPCOOH as standards [18]; ~70% of PLPC in the liposomes was oxidized by the Cu2+/ascorbate treatment. The reduced phospholipid (PLPC) was detected by absorbance at 205 nm and the oxidized phospholipid (PLPCOOH) at 235 nm [18]. Analysis by dynamic light scattering (DLS 90 Plus, Particle Size Analyzer, Brookhaven Instruments Corporation, Holtsville, NY) showed a homogeneous population of liposomes with a diameter of ~100–120 nm for the standard vesicles and minimal effect of oxidation or Prdx6 addition as shown previously for study at pH 4 [17]. Assuming a surface area per phospholipid of ~63Å2 [17] and a liposome radius of ~500 Å, each vesicle theoretically contains ~100,000 phospholipid molecules.
Enzymatic activity
Peroxidase activity was measured by the initial slope of the change in NADPH fluorescence in the presence of GSH and GSH reductase with H2O2, PLPCOOH, or oxidized liposomes as substrate in 40 mM PBS, pH 7.4, 5 mM EDTA, 1 mM NaN3, and πGST equimolar to Prdx6; for assay with PLPCOOH, 0.1% Triton X-100 was added [7]. Protein concentration was measured using the Bradford Protein Assay (BioRad, Hercules, CA). To analyze products of the peroxidase reaction, the lipids were extracted from the reaction mixture containing oxidized liposomes at 2 and 5 min [23], dried under N2, dissolved in propanol-2, and analyzed by reverse phase HPLC as described previously [18]. PLA2 activity was measured from the appearance of 3H-palmitate during a 1 hr incubation with the standard liposomes containing DPPC labeled with 9, 10-3H- palmitate in the sn-2 position. Incubation was in Tris-EGTA buffer, pH 7.4 in the absence of Ca2+. At the end of incubation with either reduced or oxidized liposomes, lipids were extracted, separated by TLC, and analyzed by scintillation counting as described previously [24].
Protein fluorescence
Protein fluorescence was measured with a PTI spectrofluorometer (Photon Technology International, Inc., Lawrenceville, NJ) equipped with a single photon counting system for fluorescence intensity detection, dual fluorescence and absorbance channels, and a temperature controlled sample holder using excitation and emission slits of 1 and 2 nm, respectively. For tryptophanyl fluorescence, 0.5µM protein in 40 mM PBS, pH 7.4, in a 5×5 mm quartz cuvette at 22°C was assayed with excitation at 295 nm to minimize input of phenylalanine and tyrosine residues; final spectra were corrected by subtracting the appropriate controls [17].
Fluorescence detection of Prdx6 binding to liposomes
Prdx6 binding was studied with fluorescent liposomes labeled with N-DNS-PE or bisPyr-PC [17]. The buffers used for these studies were equilibrated with 100% N2 before fluorescence measurements (45 min at 4°C) to minimize the possibility of O2-induced quenching. Studies were performed at 22±0.5°C with constant stirring. Kinetics were determined using the time-based radiometric mode of the fluorometer with recording of one measurement per second for ~1000 sec before and ~3000 sec after addition of protein to liposomes (100 µM total phospholipid in 40 mM PBS, pH 7.4). Fluorescence was monitored at 415/505 nm for N-DNS-PE or 382/470 nm for bisPyr-PC. The binding analysis is based on shielding by bound Prdx6 of the DNS chromophore on the liposome surface from the surrounding buffer resulting in a short wavelength shift of DNS fluorescence (from 505 to 415 nm) or, for bisPyr-PC, an increase of monomer (382 nm) fluorescence with consequent decrease of excimer fluorescence (470 nm) [17]. The experimental data were approximated with the use of standard sigmoid (4 parameters) fit using SigmaPlot 2001 (version 9.0) (SPSS, Chicago, IL). For all experiments, the quality of fit (R2) was >0.95.
The linear portion of the curves for binding vs. Prdx6 concentration was used for calculation of an apparent k1, according to the equation:
where [E] is the initial concentration of enzyme (Prdx6), [S] is the initial concentration of peroxidized phospholipids in the outer leaflet of liposomes, and [ES] is the concentration of the enzyme-phopholipid complex [17]. We assume that under these conditions, k1≫k−1 so that k1=d[ES]/dt/[E][S]. The linear part of the kinetic curve represents the formation of the enzyme-substrate complex and is used to calculate d[ES]/dt; [E] is known and [S] can be calculated from the Stokes radius of Prdx6 and the estimated number of oxidized phospholipid molecules per liposome as described previously[17]. Assuming that Prdx6 is bound to all available peroxidized phospholipids on the lipsome surface, we estimate that 0.32 µM Prdx6 is required for enzyme saturation with substrate. We used ~3.2 µM Prdx6 to study the kinetics of protein binding to liposomes in order to have at least 10x excess of protein; using a range of 2 to 20 fold excess of protein gave similar results (not shown). The dissociation constant (Kd) for binding was calculated as 50% of the saturation value for Prdx6 binding to DNS-PE- or bis-Pyr-PC-labeled liposomes as described previously [17].
Immuno-chemical detection of Prdx6 binding
Prdx6 (0.5 or 5.0 µg in 500 µl, 40 mM PBS, pH 7.4) was incubated without or with liposomes (100 µmol total lipid) at room temperature for 45 min as a control and then loaded on a Centricon YM-100 filtration device (Millipore, Bedford, MA) and centrifuged at 1,000 g for 1 h. The Stokes radius for Prdx6 has been calculated as ~25 and ~45Å for the monomer or homodimer, respectively [17]. Since the filtration system with 100 kDa cut-off will allow passage of the Prdx6 mono and homodimer but not the liposomes (~100–120 nm diameter), the Prdx6 in the filtrate and retentate corresponds to “free” and “bound” protein, respectively. Both filtrates and retentates were loaded onto a 12% Tris-glycine precast gel (Invitrogen, CA), resolved by electrophoresis, and transferred to Immobilon-P membranes for western blot analysis and fluorescence detection as described previously [17].
Circular dichroism (CD)
CD measurements were carried out with AVIV 202 and 62 DS CD spectrometers (AVIV Associates, Lakewood, NJ) using a semi-micro quartz rectangular 1×10×40 mm cuvette [17]. The protein samples (50 µM in 40 mM PBS, pH 7.4 buffer) were maintained at 22°C using a Pelletier element. Spectra were recorded while scanning in the far-UV region (190–260 nm) with bandwidth 1.0 nm, step size 0.25 nm, and integration time 30 sec with 3 repeats. Melting curves were recorded as the CD signal at 220 nm as temperature was varied from 10° to 90° C in increments of 0.5°C. The output of the CD spectrometer was recalculated according to the protein concentration, amino acid content, and cuvette thickness into molecular ellipticity units (degrees•cm2•dmol−1). The melting point corresponds to the temperature at 50% loss of molecular ellipticity.
Immunoblots
SDS-PAGE was performed with a 12% Tris-Glycine, 1mm precast gel using the Nu-PAGE system (Invitrogen, Carlsbad, CA) loading ~0.5 –2 µg of protein per lane. The resolved proteins were electroblotted to Immobilon-P membranes (Millipore, Bradford, MA) using a Trans-Blot® SD Semi-Dry Transfer Cell (Bio-Rad) at 12V for 20 min and probed with a monoclonal or polyclonal anti-Prdx6 antibody [17]. The blot was developed using anti-mouse or anti-rabbit IRDye™800 (green) secondary antibody (Rockland, Gilbertsville, PA) and analyzed with an Odyssey dual-color fluorescent infrared-excited imaging system (LI-COR, Lincoln, NE).
Intact cell experiments
A549 cells at confluence on 100 mm culture dishes (BD Biosciences) were incubated at 37° in Minimal Essential Medium equilibrated with 5% CO2 in air and treated for 6 h with 400 µM H2O2 or 800 µM t-BOOH. In some experiments, 100 U/ml catalase was added concurrently with H2O2. Cells were washed and scraped from the dish, disrupted by sonication, and subjected to sucrose gradient centrifugation by previously published methods to isolate a plasma membrane fraction [25]. The membrane fraction was analyzed by western blot for Prdx6 and the marker protein, caveolin-1. In some experiments, the cells were washed after 6 h to remove H2O2 and incubation with fresh medium was continued for an additional 3 or 6 h. In parallel incubations, cells were sonicated for analysis of thiobarbituric acid reactive substances (TBARS) using standard methods [26].
Statistics
Results are expressed as the mean ± SE. Significance was determined by ANOVA or t test as appropriate using SigmaStat (Jandel Scientific, San Jose, CA), and the level of statistical significance was defined as P<0.05.
RESULTS
Peroxidase activity
The peroxidase activity of Prdx6 with PLPCOOH substrate was evaluated from the initial slope of the decrease in NADPH fluorescence using the coupled assay. Minimal activity was observed with untreated recombinant protein consistent with the observation (see above) that at least 70% of the protein is oxidized. Incubation of Prdx6 with πGST in the presence of GSH resulted in enzymatically active protein as indicated by the oxidation of NADPH following substrate addition (Fig. 1A). Activation of the protein under these conditions is consistent with our previous observations that incubation of Prdx6 with πGST results in its glutathionylation and subsequent reduction, thereby restoring the active state. Activity with PLPCOOH and H2O2 as substrates were similar (Table 1) as reported previously [7].
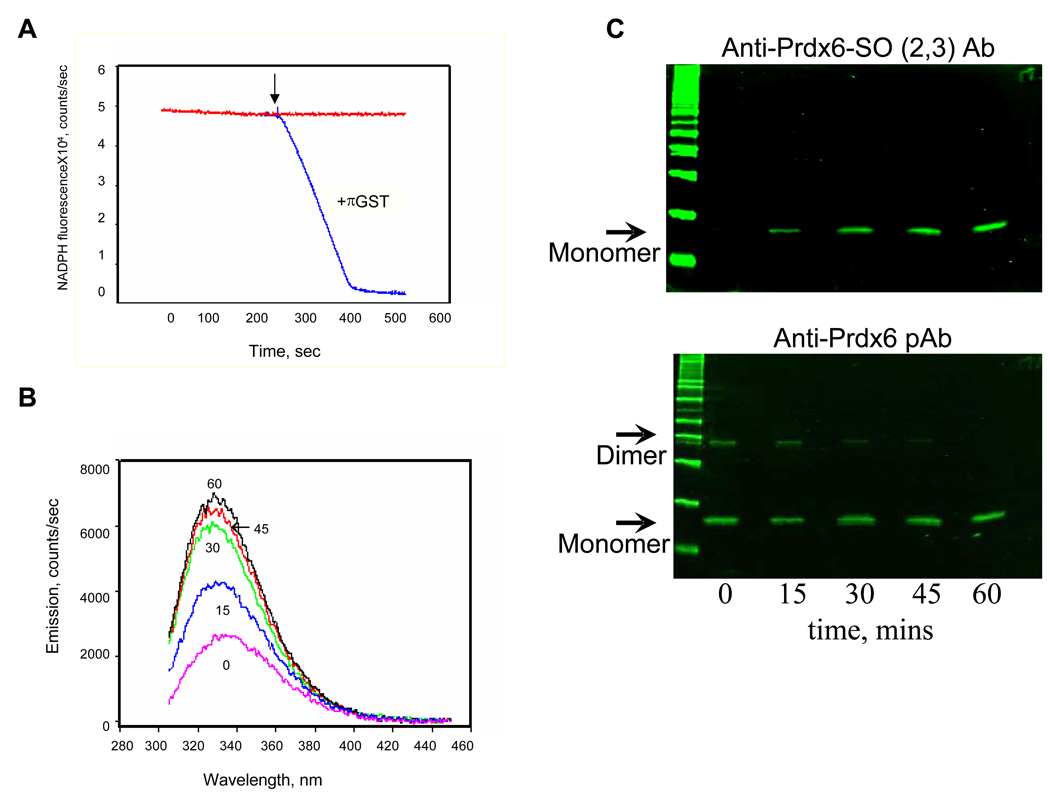
Figure 1. Prdx6 peroxidase activity with phospholipid hydroperoxide (PLPCOOH) substrate.
A. Peroxidase activity measured by the change in NADPH fluorescence in a coupled assay in the presence of GSH and GSH reductase [7]. The reaction was started by the addition of 100 µM PLPCOOH as indicated by the arrow. The reaction was carried out in the presence or absence of GSH S-transferase pi (πGST). B. Prdx6 fluorescence emission spectrum at varying times following addition of PLPCOOH (excitation at 295 nm). Prdx6 that had been reduced by GSH-loaded πGST was incubated with PLPCOOH (100µM); πGST was removed before incubation by GST-trap chromatography. The number associated with each curve indicates the time (mins) following addition of substrate. C. Detection of Prdx6 oxidation state for the experiments described in B. Samples at each time point were analyzed by SDS-PAGE followed by Western blot using a pAb against oxidized Prdx6 (upper panel) or against total Prdx6 (lower panel). The antibody against the oxidized protein detects both the sulfinic (−SO2) and sulfonic (−SO3) states of the protein. All figures are representative results for n=3 separate experiments.
Table 1.
Peroxidase activity and binding constants for wild type and mutant peroxiredoxin 6 proteins
| Peroxidase Activity | Binding to liposomes | ||||
|---|---|---|---|---|---|
| H2O2 µmol/min/mg |
PLPCOOH µmol/min/mg |
Liposomes (oxidized) µmol/min/mg |
k1 107 M−1s−1 |
Kd µM |
|
| Wild type | 6.20 ± 0.31 | 6.00 ± 0.17 | 5.95 ± 0.25 | 0.82 ± 0.2 | 0.23 ± 0.05 |
| C47S | 0 | 0 | 0 | 0.81 ± 0.1 | 0.22 ± 0.1 |
| S32A | 5.72 ± 0.20 | 0 | 0 | ND | ND |
| H26A | 5.60 ± 0.20 | 0.28 ±0.09 | ND | ND | ND |
| D140A | 6.15 ± 0.25 | 5.25 ± 0.31 | 5.76 ± 0.38 | 1.2 ± 0.2 | 0.30 ± 0.09 |
Activity was measured at pH 7.4 in the presence of πGST. H2O2, 200 µM; PLPCOOH, 200µM; oxidized liposomes, 100 µM. PLPCOOH, 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine hydroperoxide. ND, not determined; zero, no signal detected. Results are mean ± SE for n=3.
Oxidation state of Prdx6
Interaction of “activated” Prdx6 with PLPCOOH was studied by measuring tryptophanyl fluorescence of the protein (Figure 1B). There was a progressive increase in fluorescence emission at 332 nm measured at 15 and 30 min after protein addition with a slight further increase between 45 and 60 min to reach a plateau value. Interaction of Prdx6 with the peroxidized substrate should result in oxidation of the catalytic Cys residue in Prdx6; the initial oxidation state in this reaction is a sulfenic acid which is reversible to the sulfhydryl but higher oxidation states are possible. Thus, samples from the incubations were evaluated by western blot using antibodies to Prdx6 and the sulfinic/sulfonic forms of Prdx6 (Figure 1C). This analysis showed absence of the higher oxidized forms in the starting material with a progressive increase in the oxidized species during one hour incubation with PLPCOOH. Adding together the densities of the bands for the dimeric and monomeric forms indicates approximately equal loading for each of the lanes. The dimer band was not detected in the presence of dithiothreitol or other reducing agents (not shown).
Prdx6: lipid interaction by circular dichroism
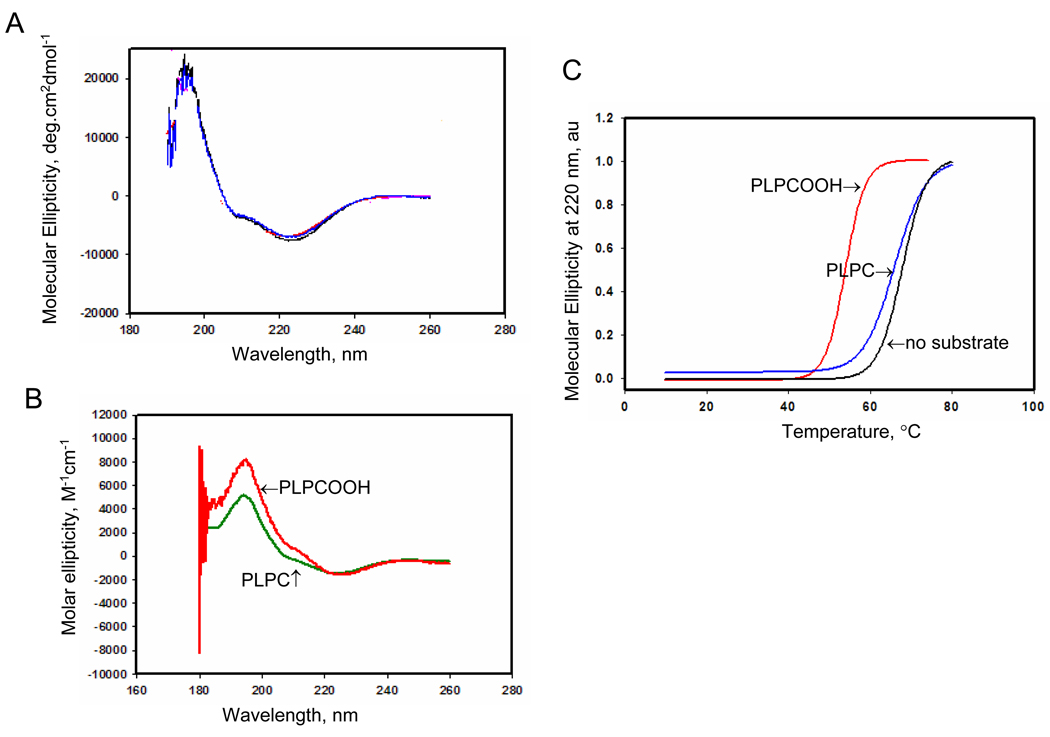
Interaction of Prdx6 with PLPCOOH was further evaluated using circular dichroism (CD). Prdx6 that had not been “activated” with πGST (therefore “inactive”) was used for these assays. The CD spectrum indicating secondary structure of the protein in solution at pH 7.4 (Fig. 2A) was similar to our previous study at pH 4 [17] and also was similar when comparing the wild type and inactive (C47S) mutant proteins. We have reported previously that the CD spectrum of the S32A mutant Prodx6 showed a marked increase in α-helical structure at pH 4 [16] and a similar spectrum was observed at pH 7.4; however, the CD spectra of muntat proteins S32V, S32L, and S32T as well as H26A and D140A were similar to wild type (data not shown). These results suggest that the possible alternation of protein secondary structure due to specific mutations were not responsible for alterations in substrate binding associated with S32 mutation. Incubation of Prdx6 with a 5-fold excess of PLPCOOH resulted in a substantial change of secondary structure (Figure 2B) corresponding to a 15° C decrease in the Prdx6 melting temperature (Figure 2C). Addition of non-oxidized phospholipid (PLPC) had minimal effect on the melting temperature suggesting that the −OOH group resulting from lipid oxidation is important for protein folding upon binding.
Figure 2. Circular dichroism (CD) analysis of Prdx6 in the presence of substrate.
A. CD spectrum of Prdx6 (1.25 mg/ml) in 40 mM phosphate buffered saline, pH 7.4. Spectra are shown for wild type (WT) (blue) and C47S mutant (red) proteins. The spectra are essentially identical. B. CD spectrum for wild type Prdx6 in the presence of reduced (PLPC) or oxidized (PLPCOOH) substrate (200 µM). C. Melting temperature for wild type protein derived from the spectra shown in A and B. Each melting curve was normalized to its maximal CD signal and the melting point was determined as the temperature at 50% loss of molecular ellipiticity (indicated by the arrows). The Prdx6 used in these assays was enzymatically inactive. All results are representative of 3 independent experiments.
Reduction of peroxidized phospholipids
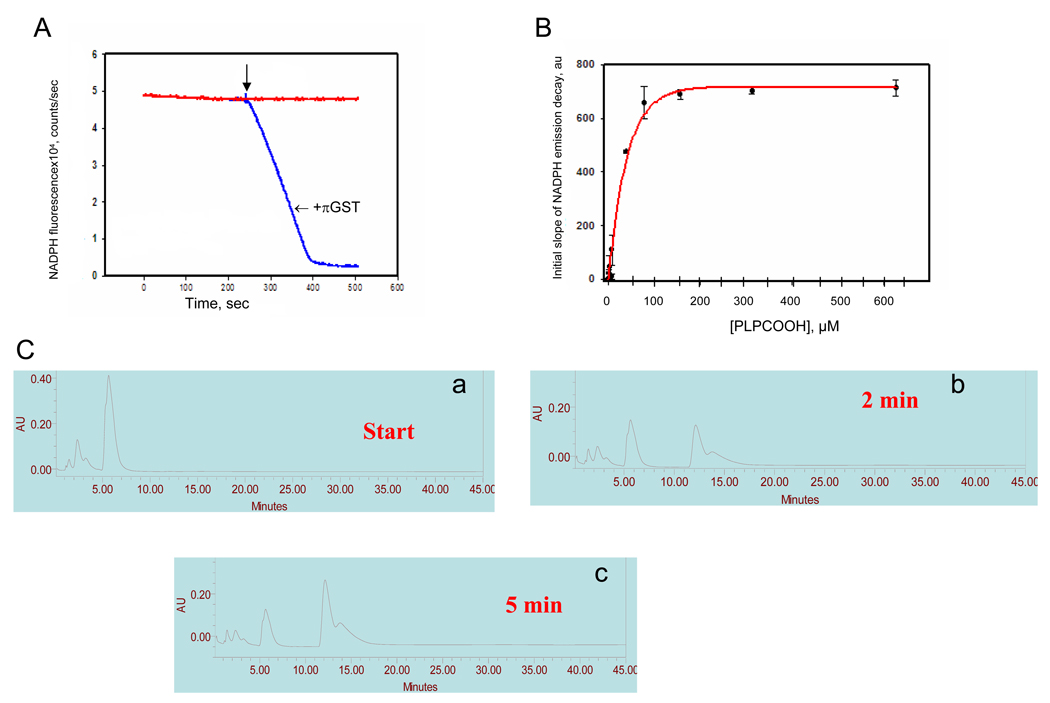
We have postulated that the physiological role for reduction of phospholipid hydroperoxides is related to the reduction of peroxidized phospholipids in biological membranes. To simulate this condition experimentally, we evaluated Prdx6-mediated reduction of peroxidized phospholipid in mixed unilamellar liposomes. Prdx6 that had been activated by reaction with πGST reduced phospholipid hydroperoxide in unilamellar liposomes when assayed with the NADPH/GSH regenerating system (Figure 3A) and the calculated Prdx6 peroxidase activity was similar for oxidized liposomes, PLPCOOH, and H2O2 (Table 1). Peroxidase activity varied with substrate concentration in liposomes and reached a plateau value at approximately 150–200 µM PLPCOOH and 50% of maximal activity at ~30 µM (Fig. 3B). The C47S mutation of Prdx6 abolished peroxidase activity with all three substrates (Table 1) indicating the crucial role of this residue.
Figure 3. Prdx6 peroxidase activity with oxidized liposomes as substrate.
A. Prdx6 peroxidase activity of Prdx6 (1.7 µg/ml) as indicated by oxidation of NADPH in the presence of GSH and GSH reductase. The reaction was started by the addition of oxidized liposomes (100 µM total lipid) as indicated by the arrow. Tracings are shown in the absence or presence of πGST added during the pre-incubation period. B. Plot of the peroxidase activity (initial slope of NADPH fluorescence decay) vs. the concentration of substrate (PLPCOOH) in oxidized liposomes. The substrate concentration was calculated as described in Methods. The initial slope of activity was determined by linear fit for the first 5 seconds after addition of substrate. The results indicate the mean ± SE for n=3 independent experiments. C. Analysis of oxidation products by HPLC, absorbance at 234 nm. a. the elution pattern for the phospholipids extracted from oxidized liposomes before addition of Prdx6; b and c. the lipid elution patterns at 2 and 5 mins after addition of Prdx6 plus GSH-loaded πGST.
To confirm the results of the indirect assay of peroxidase activity, the disappearance of PLPCOOH and appearance of metabolic products was evaluated by HPLC (Fig. 3C). Retention times were ~15 minutes for authentic PLPC and 6 minutes for the oxidized phospholipid (PLPCOOH). At 2 and 5 minutes after the start of incubation with activated Prdx6, there was a progressive decrease in the peak height for PLPCOOH and the appearance of a new major peak at approximately 12 minutes. This indicates the generation of a reaction product, presumably the phospholipid alchohol (PLPCOH).
Binding of Prdx6 to liposomes
Unlike the H2O2 substrate, reduction of peroxidized phospholipid requires proper positioning of the substrate relative to the enzymatic active site. This requires binding of the enzyme to the liposomal surface prior to interfacial catalysis [17, 27]. Binding of “inactive” Prdx6 to peroxidized liposomes was studied by filtration followed by immunodetection of bound and free Prdx6. Prdx6 was incubated with liposomes containing reduced or oxidized phospholipid for 45 minutes and passed through a microfiltration system with 100kDa cut-off, a pore size large enough to allow unbound protein to pass. Thus, the filtrate contains unbound Prdx6 while the retained fraction contains liposomes with bound protein. As a control, liposomes were labeled with DNS-PE and subjected to the filtration protocol; DNS fluorescence was observed only in the retentate (not shown) confirming that liposomes did not pass through the filter. Filtration in the absence of liposomes showed essentially all of the protein recovered in the filtrate (Fig. 4A). The ratio of filtrate to retentate changed only slightly in the presence of the standard non-oxidized liposomes (Fig. 4B). Incubation of Prdx6 with oxidized liposomes resulted in a significant increase in the ratio of protein in the retentate vs. filtrate compatible with binding of Prdx6 to the liposomes (Fig. 4C).
Figure 4. Detection of binding of Prdx6 to oxidized liposomes by filtration analysis.
Prdx6 (80 µg/ml) in 40 mM PBS, pH 7.4, in the absence or presence of liposomes (100 µM total lipid) was processed through a YM-100 microfiltration system with a nominal cutoff of 100 kDa. The filtrate (F) and the retentate (R) were analyzed by SDS-PAGE with Coomassie blue staining (lanes 1–3 in each panel) followed by Western blot for Prdx6 (lanes 4–6 in each panel). Lanes 1 and 4 in each panel are the molecular mass markers; values for the Coomassie-stained gels are indicated at the far left and for the Western blots at the far right. The results are representative of three independent experiments. A. Prdx6 without liposomes. B. Prdx6 with non-oxidized liposomes. C. Prdx6 with oxidized liposomes.
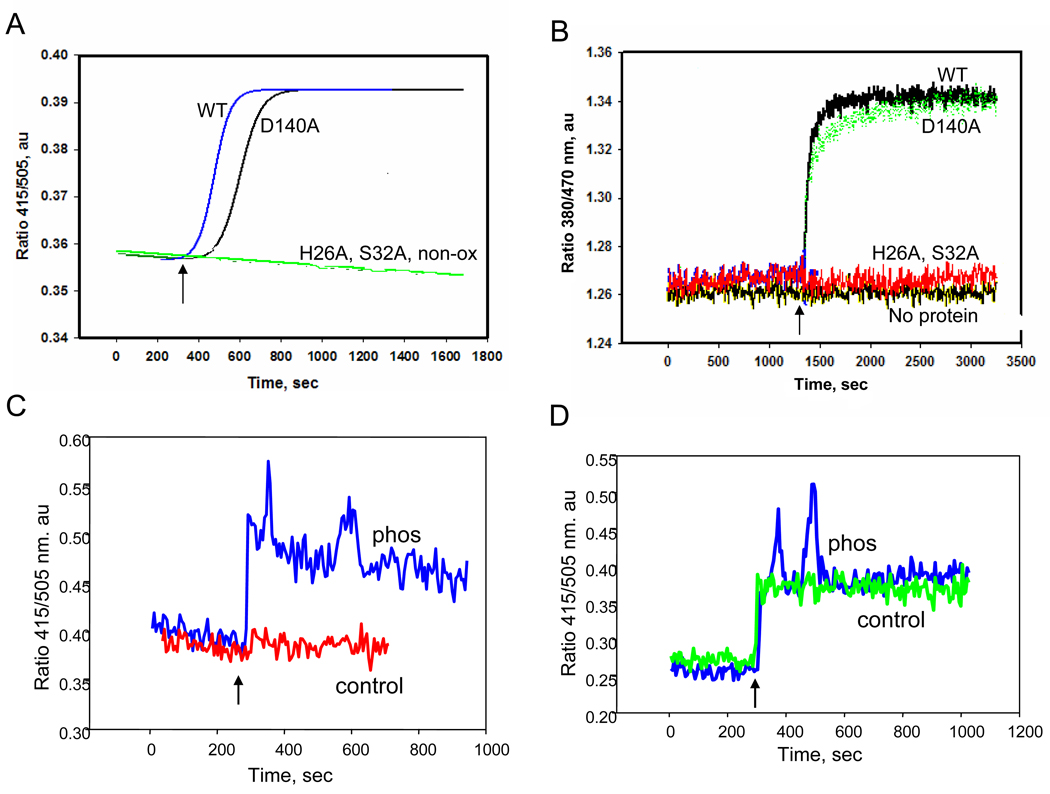
The binding of Prdx6 (“inactive”) to liposomes containing either PLPC or PLPCOOH also was evaluated at pH 7.4 by the fluorescence emission ratio (415 nm/505 nm) for DNS-PE-labeled liposomes or 380 nm/470 nm for bis-Pyr-labeled liposomes. No change in the fluorescence was observed when Prdx6 was added to liposomes containing reduced phospholipids (Fig. 5A, red). However, a distinct increase in fluorescence that reached a plateau in approximately 400 seconds was seen with Prdx6 addition to oxidized liposomes (Figure 5A and B, WT). We have shown recently that phosphorylation of Prdx6 in vitro results in a marked increase in PLA2 activity at pH 7.4, equal to activity at pH 4 [21]. Unlike the non-phosphorylated protein, phosphorylated Prdx6 showed binding at pH 7.4 to reduced liposomes (Fig. 5C). Binding to oxidized liposomes at pH 7.4 was similar for the native and phosphorylated proteins (Fig. 5D).
Figure 5. Fluorescence analysis of Prdx6 to binding liposomes.
Prdx6, phosphorylated Prdx6, and Prdx6 mutants were evaluated for binding to liposomes containing all reduced phospholipids or liposomes that were oxidized by treatment with Cu2+/ascorbate. Prdx6 (3.2 µM) was incubated in 40 mM PBS at pH 7.4 and liposomes (100µM) were added. A. Kinetics of Prdx6 binding to unilamellar liposomes labeled with 2 mol % DNS-PE. The ratio of fluorescence emission at 415/505 nm is shown as a function of time following the addition of lipid (indicated by the arrow). The binding kinetics are shown for addition of oxidized liposomes to wild type (WT) Prdx6 or in the presence of the Prdx6 mutants S32A, D140A or H26A. The binding curve for wild type Prdx6 with non-oxidized liposomes (non-ox) overlapped the H26A and S32A curves. B. Incubation as in A but with bis-Pyr-PC (2 mol%) in oxidized unilamellar liposomes. Fluorescence emission was recorded as the ratio of 380/470 nm for wild type (WT) Prdx6 and S26A, H26A and D140A mutants. ‘No protein’ indicates a control tracing in the absence of Prdx6. C. Fluorescence emission ratio for DNS-PE labeled liposomes (non-oxidized) in the presence of phosphorylated (Phos) or non-phosphorylated (control) wild type Prdx6. D. Same experiment as C but with oxidized liposomes.
Our previous studies of phospholipid binding to Prdx6 were carried out at pH 4 in order to evaluate the role of PLA2 activity of the protein [17]. Under those conditions, mutation of S32 or H26 abolished phospholipid binding while mutation of D140 had no effect. These amino acids constitute the PLA2 catalytic triad. We thus evaluated the effect of these mutations on binding of “inactive” Prdx6 to oxidized liposomes at pH 7.4. The binding of Prdx6 to oxidized liposomes at pH 7.4 was abolished by the S32A and H26A mutations (Fig. 5A,B) as well as by the S32V, S32L, and S32T mutations (data not shown). Mutation of D140 had no effect on binding (Figs. 5A, B). Peroxidase activity with either PLPCOOH or oxidized liposomes as substrate also was abolished by S32A and H26A mutation, while D140 mutation had no effect (Table 1). On the other hand, reduction of H2O2 was unaffected by the S32 and H26 mutations. This supports the importance of binding for the reduction of the phospholipid substrates.
PLA2 activity of Prdx6 was measured with reduced and oxidized liposomes at pH 7.4. Activity with reduced liposomes was essentially nil (0.44 ± 0.07 nmol/min/mg protein) as reported previously for measurement at this pH [24]. However, activity with oxidized liposomes was 80.1 ± 4.8 nmol/min/mg protein, slightly greater than values obtained previously with incubation at pH 4.0 [9]. This result is compatible with binding of the oxidized substrate to Prdx6. Our previous studies have shown that the presence of GSH increases the PLA2 activity of Prdx6 when measured with reduced liposomal substrate [28] but there was no effect of GSH on the activity with oxidized liposomes (data not shown).
The kinetic constants for binding were calculated from data for fluorescence change associated with interactions of DNS-PE liposomes with Prdx6. The calculated rate constant (k1) for wild type protein was approximately 10−7M−1 sec−1 and a dissociation constant (Kd) of 230 nM (Table 1). There was not appreciable effect when phosphatidylserine or phosphatidylethanolamine were substituted for phosphatidylglycerol in the oxidized liposomes (not shown). The binding constants for the D140 mutant protein were similar to the wild type (Table 1).
Effect of inhibitors of enzymatic activity on binding
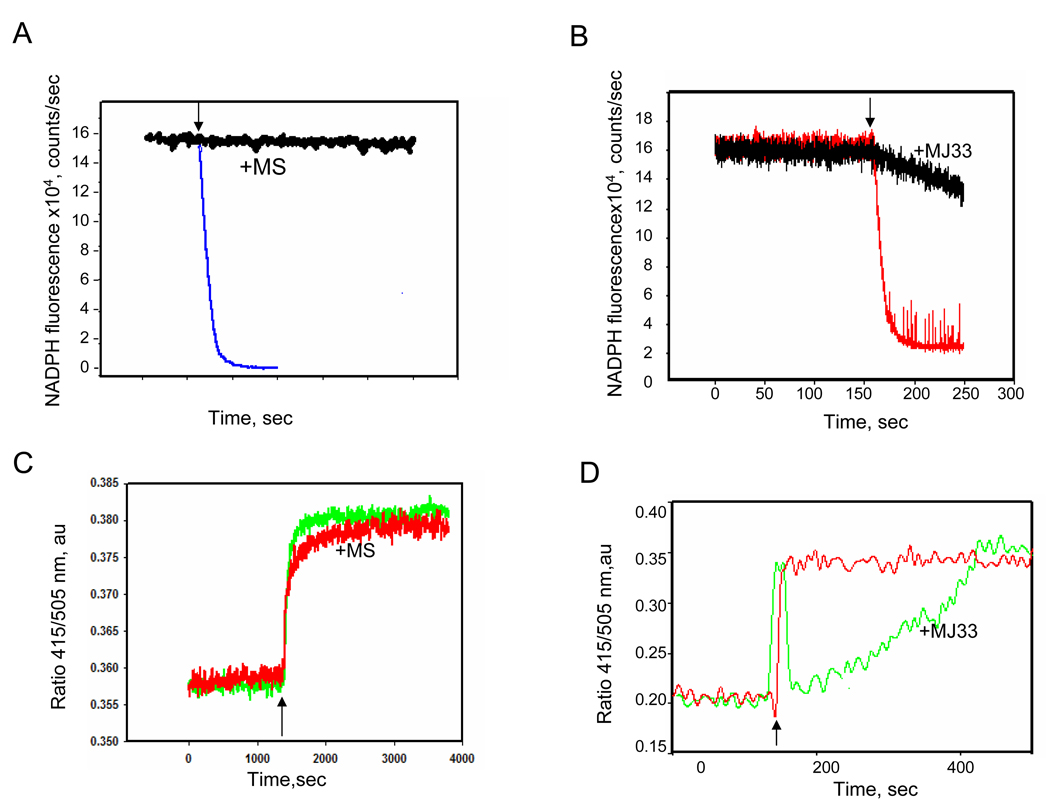
Mercaptosuccinate, a cysteine-active reagent, inhibited the peroxidase activity with oxidized liposomes as substrate (Figure 6A) as previously reported for PLPCOOH [7]. Mercaptosuccinate had no effect on the binding of Prdx6 to peroxidized DNS-PE-labeled liposomes (Figure 6B). The lack of effect is consistent with the proposed site for substrate binding to the surface of Prdx6 which is distant from the catalytic cysteine [17]. We also studied the effect of MJ33, a competitive inhibitor of the PLA2 activity of Prdx6 [9, 19], on binding of the protein to oxidized liposomes. Preincubation of Prdx6 with MJ33 also resulted in a significant decrease in the rate of reduction of oxidized liposomes (Figure 6C). The concentration used for this inhibition, namely a 2-fold molar excess of MJ33, was considerably greater than the effective concentration for inhibition of PLA2 activity. There was no effect of MJ33 on the reduction of H2O2, reported previously [9] and confirmed in the present study (not shown). Preincubation with MJ33 also significantly decreased intrinsic tryptophan fluorescence of Prdx6 (not shown). The change in protein tryptophanyl fluorescence was not observed in the W33F mutant Prdx6 suggesting that this residue (adjacent to Ser32) is associated with MJ33 binding to Prdx6. Finally, MJ33 significantly decreased the rate of binding of Prdx6 to oxidized liposomes as determined by fluorescence of DNS-PE (Figure 6D). These results are compatible with competition between MJ33 and oxidized liposomes for binding to Prdx6 whereas diffusible H2O2 substrate would not bind and would not compete.
Figure 6. The effect of mercaptosuccinate (MS) and MJ33 on Prdx6 peroxidase activity and lipid binding.
A. and B. Peroxidase activity of Prdx6 with oxidized liposomes (100 µM total lipid) in the presence of πGST is indicated by oxidation of NADPH in the coupled assay with GSH/GSH reductase. A. MS (1 µM) was incubated with Prdx6 for 45 min at room temperature prior to assay. B. Prdx6 was incubated in the absence or presence of a 2-fold molar excess of MJ33. C. and D. Measurement of binding of Prdx6 to oxidized liposomes as indicated by DNS-PE fluorescence emission ratio (415/505 nm) using the protocol shown in Fig. 5. C. Binding in the presence or absence of MS (1µM). D. Binding before and after addition of a 2-fold molar excess of MJ33. In all panels, the arrow indicates addition of liposomes.
Prdx6 binding to membrane in intact cells
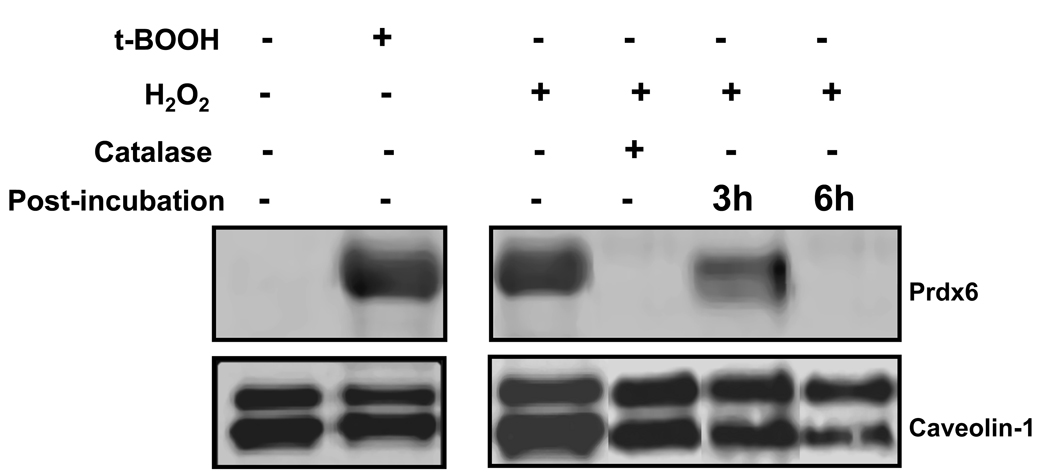
Western blot of the cell membrane fraction from untreated A549 cells showed the absence of Prdx6 (Fig. 7). However, a prominent membrane-associated Prdx6 band was observed following 6 h treatment of cells with either H2O2 or t-BOOH. Addition of catalase at the start of incubation with H2O2 prevented Prdx6 translocation to the membrane fraction. Translocation was partially reversed at 3 h after removal of H2O2 from the incubation medium and was totally reversed at 6 h (Fig. 7 and Table 2). Lipid peroxides in the cell indicated by TBARS measurements (shown below the relevant gel in Fig. 7) were increased significantly with H2O2 treatment; TBARS and had significantly decreased at 3 h and fully returned to baseline at 6 h following oxidant stress (Table 2).
Figure 7. Effect of oxidant stress on Prdx binding to plasma membranes in A549 cells.
A549 cells were treated for 6 h with t-BOOH (800 µM) or H2O2 (400 µM); catalase (100 U/ml) was added in some experiments. In other experiments, cells were washed at the end of the incubation period to remove H2O2 and then incubated for an additional 3 or 6 h in medium free of oxidants. The plasma membrane fraction was isolated and analyzed by western blot under reducing conditions using anti-Prdx6 polyclonal antibody. Caveolin-1 was used as a loading control for the membrane fraction. Blots were analyzed for fluorescence using the Odyssey system.
Table 2.
Plasma membrane-associated Prdx6 and membrane lipid peroxidation in A549 cells treated with tert –butyl hydroperoxide
| Membrane Prdx6 Prdx6/cav-1 |
TBARS pmol/mg protein |
|
|---|---|---|
| Control | 1 | 10.3 ± 0.4 |
| tBOOH | 26.1 ± 0.1 | ND |
| H2O2 | 36.4 ± 1.2 | 36.0 ± 0.8* |
| +3h | 15.4 ± 3.1 | 16.4 ± 0.3* |
| +6h | 1.3 ± 0.2 | 10.7 ± 1.3 |
Results are mean ± SE for the experiments shown in Figure 7. Cells were exposed to tBOOH (800 µM) or H2O2 (400 µM) 6 for 6h and studied immediately or at 3h or 6h following removal of the oxidant. Membrane Prdx6 represents the ratio of fluorescence (arbitrary units) for the Prdx6 and caveolin-1 bands shown in Figure 7. ND, not determined.
P <0.05 vs. control (no exposure).
DISCUSSION
The peroxidation of unsaturated fatty acids in the phospholipids of cellular membranes is a major manifestation of oxidant stress. Lipid peroxidation can dramatically change the biophysical properties of the membrane and result in changes in membrane permeability and cell viability. Two pathways for repair of the peroxidized membrane phospholipids have been proposed. The first involves deacylation of the sn-2 peroxidized fatty acid through activity of phospholipase A2 followed by reacylation of the lyso phospholipid through activity of an acyltransferase [29]. A second mechanism for repair is the direct reduction of the peroxidized fatty acyl moiety to the less toxic fatty acyl alcohol; the latter could be reduced to the native phospholipid. The direct reduction pathway has been estimated as 104 more efficient than deacylation/reacylation [30]. Until recently, the only enzyme known in mammalian systems that was capable of the direct reduction of peroxidized phospholipids at a rate considered physiologically relevant was glutathione peroxidase 4 (GPx4, also called phospholipid hydroperoxide glutathione peroxidase) [31]. Of note, the ubiquitously distributed cystosolic GPx (GPx1) does not have this activity [7]. More recently, Prdx6 has been shown to directly reduce phospholipid hydroperoxides with a rate constant of approximately 106 M−1 s−1 [7]. Although this indicates a rapid reduction of phospholipid hydroperoxides by Prdx6, the rate constant is approximately one order of magnitude lower than that reported for GPx 4 [31]. The relative roles of GPx and Prdx6 under physiological conditions have not been directly compared, but reduction of phospholipid hydroperoxides in homogenates of the lung, an organ especially susceptible to oxidant stress, was decreased by greater than 95% by “knock-out” of Prdx6 [32]. Based on this observation, GPx4 appears to have at best a relatively minor role in PLOOH reduction in the lung. Since Prdx6 null mice, lungs, and lung cells or cells treated with Prdx6 anti-sense RNA show markedly increased sensitivity to oxidant stress compared to the wild type, we conclude that Prdx6 plays an important role in the antioxidant defense of this organ [32–35]. While enzymes that directly reduce PLOOH are limited in number, a spectrum of enzymes including catalase, GSH peroxidases, and peroxiredoxins can participate in removal of H2O2. Further, H2O2 removal is decreased by less than 10% in lungs from Prdx6 null mice [32]. Thus, we propose that the ability to reduce phospholipid hydroperoxides accounts for the major antioxidant-related activity of lung Prdx6.
The present results confirm that phosphatidylcholine hydroperoxide is a substrate for Prdx6. Activity was indicated by the coupled assay which reflects the oxidation of GSH and its subsequent reduction at the expense of NADPH and also by HPLC analysis of products. Activity required pretreatment with πGST in the presence of GSH to reduce the sulfenic acid moiety in the oxidized Prdx6. In the absence of GSH/π GST, interaction of Prdx6 with phospholipid hydroperoxide resulted in progressive oxidation of the protein to sulfinic (or sulfonic) acids as indicated by a progressive time-dependent change in protein tryptophanyl fluorescence and increased immunoreactivity to an antibody specific for the oxidized forms. A band compatible with the Prdx6 dimer is present at zero time and corresponds with reports of dimerization of oxidized protein (sulfenic acid form). Decrease of the Prdx6 dimer intensity with time is compatible with its further oxidation and inability to form the dimer. It is now yet known whether this reaction occurs in vivo and whether hyperoxidation is reversible. Oxidized Prdx6 is not a substrate for sulfiredoxin, the only enzyme identified so far capable of reducing the higher oxides of protein cysteines [36].
We have proposed that peroxidase activity of Prdx6 with PLPCOOH as substrate requires specific binding and positioning between enzyme and substrate on the membrane surface [37]. As evidence for their interaction, incubation of Prdx6 with PLPCOOH led to a marked change in the protein secondary structure as indicated by a shift in the CD spectrum. We postulate that the peroxidized sn-2 acyl chain is inserted into the hydrophobic interior of the protein with the −OOH moiety positioned in the vicinity of the catalytic Cys47. This positioning of the substrate destabilizes the enzyme resulting in altered Prdx6 folding as reflected by a shift in its melting temperature. More physiologically relevant is the binding of Prdx6 to an oxidized lipid membrane. We have used liposomes in the present study as a surrogate for biological membranes and have oxidized the unsaturated phospholipid therein, primarily PLPC, using an •OH-generating system. Binding of Prdx 6 to the surface of oxidized liposomes was indicated by filtration studies showing increased retention of Prdx6. Additional evidence was the increased fluorescence when Prdx6 was added to DNS-PE or bisPyrPC-labeled oxidized liposomes, but not to native liposomes. As we have discussed previously, the shift of the fluorescence maximum for DNS in liposomes from 505 to 415 nm in the presence of Prdx6 reflects a change in polarity of its surroundings attributable to shielding of the DNS chromophore by bound protein [17]. The increase in the monomer fluorescence at the expense of excimer fluorescence of bis-pyrPC is compatible with an increased separation of the pyrene labeled acyl chains. We postulate that the results for filtration analysis and fluorescence of DNS-PE are compatible with binding of Prdx6 to the liposome surface while the result for fluorescence change with bisPyr-PC is compatible with the specific positioning of the phospholipid substrate upon Prdx6 binding.
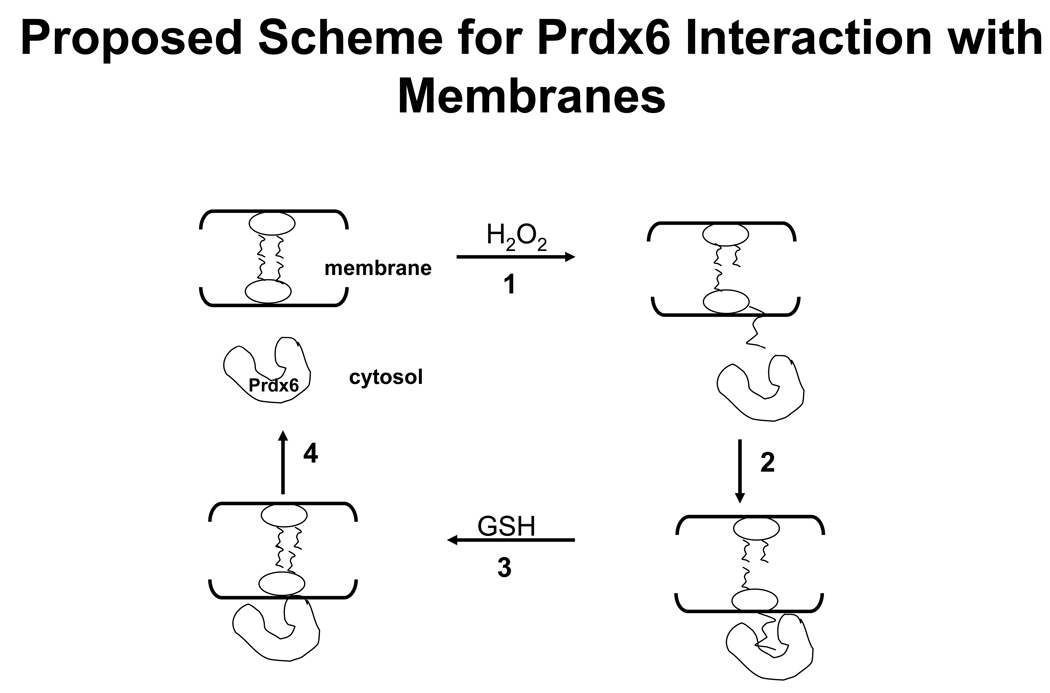
There was relatively little interaction of Prdx6 with non-oxidized phospholipids as indicated by the lack of change in CD spectrum after incubation of Prdx6 with PLPC, significantly less retention of Prdx6 on the filters after incubation with non-oxidized compared to oxidized liposomes, and essentially no change in DNS-PE fluorescence in the presence of non-oxidized liposomes. This lack of binding to non-oxidized phospholipids at pH 7.4 contrasts with our previous results that showed strong binding at pH 4 [17]. We postulate that this pH-dependent differential in binding reflects the charge on the protein and therefore its attraction to negatively-charged liposomes. The protein charge should be significantly less positive at a pH above the pI for Prdx6 (6.3). The pH-dependent activities of Prdx6 reflect the observed binding pattern: PLA2 activity is maximal at pH 4 and is essentially absent at cytosolic pH while peroxidatic activity is found at cytosolic pH [19]. These findings have physiological implications since Prdx6 is present in lung cells in acidic lysosomes and secretory organelles where it can function in degradation of surfactant phospholipids and in the remodeling pathway for synthesis of dipalmitoylphosphatidylcholine [15, 16]. On the other hand, unregulated PLA2 activity would be undesirable for a cytosolic protein. By our scheme, reduction of the −OOH group in PLPCOOH would result in dissociation of the protein: phospholipid complex, since the protein and reduced phospholipid do not bind at pH 7.4, and abrogation of possible PLA2 activity. The relatively high rate constant for the peroxidase activity ensures that this activity takes precedence for the peroxidized phospholipid substrate. Further, the calculated kinetic constants for Prdx6 binding to liposomes were slightly greater than our previous calculated kinetic constants for peroxidase activity [7], suggesting that impaired binding of substrate could be a rate limiting step in reduction of the peroxidized substrate. We showed previously that the binding of Prdx6 at pH 4 was greater for negatively charged as compared to neutral liposomes, as determined by substitution of the uncharged PE for the negatively charged PG or PS in the unilamellar liposomes [17]. This finding is consistent with the known positive charge on Prdx6 at pH 4 (pKa ~6.3). However, at neutral pH, surface charge did not appear to play a role in binding since substitution of PE for the negatively charged phospholipids had no effect on Prdx6 binding. These results suggest that Prdx6 specifically recognizes phospholipid hydroperoxide on the membrane surface and that the surface charge does not play a major role. Studies with intact cells confirmed the biological relevance of the in vitro results. Prdx6 did not co-isolate with the membrane fraction in the control (untreated cells). Peroxidation of cellular lipids, as indicated by TBARS assay, resulted in translocation of Prdx6 indicated by its isolation with the membrane fraction. Both lipid peroxidation and Prdx translocation were reversible. This result is compatible with Prdx6 binding to peroxiredoxin membrane phospholipids, catalyzing their reduction, and dissociation from the now reduced membrane phospholipids (Fig. 8).
Figure 8. Proposed scheme for function of cytosolic Prdx6.
1) Prdx6 is a cytosolic protein in the normal resting cell. 2) With oxidant stress (indicated by H2O2), oxidation of membrane phospholipids leads to an increased Prdx6 accessibility to substrate related to increased hydrophilicity of the phospholipid hydroperoxide. 3) Binding of Prdx6 to the membrane results in glutathione (GSH) – mediated reduction of the oxidized substrate, restoring membrane integrity and 4) leading to release of Prdx6 from binding to the now-reduced phospholipid.
We have previously shown specific binding of Prdx6 to non-oxidized liposomes at pH 4 and demonstrated, using site directed mutagenesis and specific inhibitors, that binding results in interfacial catalysis [17]. We concluded that the surface of the protein in the area of H26 appeared to be crucial for binding of Prdx6 to the liposomal interface while the vicinity of S32 appeared to be crucial for binding of the phospholipid polar head. We have observed previously that the PLA2 activity with peroxidized phospholipid in liposomes at pH 4 was similar to activity with the reduced substrate [15]. This suggests similar positioning of the reduced and peroxidized substrate relative to the protein surface at acidic and neutral pH. The present results provide additional evidence for similar positioning of the peroxidized phospholipid at pH 7 to that observed for the non-oxidized phospholipid at pH 4. First, the effect of site specific mutations was similar for Prdx6 binding to liposomes at pH 4 [17] and pH 7 (this study). Mutation of S32 and H26 abolished binding while mutation of D140 had no effect at either pH. Second, the presence of MJ33, a competitive inhibitor of PLA2 activity at pH 4, bound to Prdx6 at pH 7 and inhibited its peroxidase activity. Third, mutation of the catalytic C47 has no effect on PLA2 activity [9] indicating that this moiety, as expected, does not influence substrate binding. The present findings show that binding also was unaffected by mercaptosuccinate, although peroxidase activity was abolished. Binding of mercaptosuccinate to Prdx6 has been demonstrated to involve positioning of its two carboxylic groups to the R41 and R132 amino acids in the Prdx6 monomer [38]. The observation that mercaptosuccinate does not inhibit binding supports the conclusion that the binding site for the peroxidized phospholipid and the peroxidatic catalytic site are different.
In summary, the present report coupled with our previous publication [17] have provided evidence to explain the two catalytic functions of Prdx6 in terms of their binding kinetics. At pH 7, Prdx6 does not bind to liposomes but does bind if lipids are peroxidized. The result is direct reduction of peroxidized phospholipid and then dissociation of the Prdx6 liposome complex. We cannot exclude some PLA2 activity during this interaction but the rate constant for that activity is significantly less than that observed for peroxidase. At pH 4, Prdx6 binds to non-oxidized phospholipids with consequent PLA2 activity. The present and previous observations can explain the predominantly lysosomal location of the PLA2 activity while peroxidase activity predominates in the cytosolic compartment.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. K.S. Reddy, Suiping Zhou and Walter Englander for assistance with CD measurements, Dr. Sergei Zaitsev for expression of the D140A mutant, Dr. Ibrul Chowdhury for assistance with western blots, Dr. Mehendra Jain for his usual astute advice, and Susan Turbitt for secretarial assistance. Presented in part at the Experimental Biology meetings in Washington, DC, April 2004; San Diego, CA, April 2005; San Francisco, CA, April 2006; and Washington, DC, April 2007. Supported by HL79063 and HL19737.
Footnotes
Abbreviations: Prdx6, peroxiredoxin 6; PCOOH, phosphatidylcholine hydroperoxide; PLPC, 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphocholine; PLPCOOH, hydroperoxide of PLPC; t-BOOH, tert-butyl hydroperoxide; PLA2, phospholipase A2; MJ33, 1-hexadecyl-3-trifluoroethylglycero-sn-2-phosphomethanol; GSH, glutathione; GST, glutathione S-transferase; CD, circular dichroism; TBARS, thiobarbituric acide reactive substances.
REFERENCES
- 1.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. IUBMB Life. 2001;52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
- 2.Poole LB. Subcell. Biochem. 2007;44:61–81. doi: 10.1007/978-1-4020-6051-9_4. [DOI] [PubMed] [Google Scholar]
- 3.Knoops B, Loumaye E, Van Der Eecken V. Subcell. Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 4.Kang SW, Baines IC, Rhee SG. J. Biol. Chem. 1998;273:6303–6311. doi: 10.1074/jbc.273.11.6303. [DOI] [PubMed] [Google Scholar]
- 5.Schremmer B, Manevich Y, Feinstein S, Fisher AB. In: Peroxiredoxin Systems. Flohe L, Harris JR, editors. Springer; 2007. pp. 317–344. [Google Scholar]
- 6.Wood ZA, Schroder E, Harris J Robin, Poole LB. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 7.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. J. Biol. Chem. 1999;274:21326–21334. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 8.Manevich Y, Feinstein SI, Fisher AB. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3780–3785. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JW, Dodia C, Feinstein SI, Jain MK, Fisher AB. J. Biol. Chem. 2000;275:28421–28427. doi: 10.1074/jbc.M005073200. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manevich Y, Fisher AB. Free Radic. Biol. Med. 2005;38:1422–1432. doi: 10.1016/j.freeradbiomed.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Ralat LA, Misquitta SA, Manevich Y, Fisher AB, Colman RF. Arch. Biochem. Biophys. 2008;474:109–118. doi: 10.1016/j.abb.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Nat. Struct. Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- 14.Fisher AB, Dodia C, Feinstein SI, Ho YS. J. Lipid Res. 2005;46:1248–1256. doi: 10.1194/jlr.M400499-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Akiba S, Dodia C, Chen X, Fisher AB. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 1998;120:393–404. doi: 10.1016/s0305-0491(98)10046-9. [DOI] [PubMed] [Google Scholar]
- 16.Fisher AB, Dodia C. J. Lipid Res. 1996;37:1057–1064. [PubMed] [Google Scholar]
- 17.Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. J. Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11599–11604. doi: 10.1073/pnas.182384499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TS, Dodia C, Chen X, Hennigan BB, Jain M, Feinstein SI, Fisher AB. Am. J. Physiol. 1998;274:L750–L761. doi: 10.1152/ajplung.1998.274.5.L750. [DOI] [PubMed] [Google Scholar]
- 20.Poole LB, Ellis HR. Methods Enzymol. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Feinstein S, Manevich Y, Chowdhury I, Pak JH, Kazi A, Dodia C, Speicher DW, Fisher AB. Biochem. J. 2009 doi: 10.1042/BJ20082061. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manevich Y, Held KD, Biaglow JE. Radiat. Res. 1997;148:580–591. [PubMed] [Google Scholar]
- 23.Bligh EG, Dyer WJ. Can J Med Sci. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 24.Kim TS, Sundaresh CS, Feinstein SI, Dodia C, Skach WR, Jain MK, Nagase T, Seki N, Ishikawa K, Nomura N, Fisher AB. J. Biol. Chem. 1997;272:2542–2550. doi: 10.1074/jbc.272.4.2542. [DOI] [PubMed] [Google Scholar]
- 25.Fisher AB, Dodia C, Chander A, Kleinzeller A. Am. J. Physiol. 1992;263:C1250–C1257. doi: 10.1152/ajpcell.1992.263.6.C1250. [DOI] [PubMed] [Google Scholar]
- 26.Fisher AB, Dodia C, Tan ZT, Ayene I, Eckenhoff RG. J. Clin. Invest. 1991;88:674–679. doi: 10.1172/JCI115352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg OG, Gelb MH, Tsai MD, Jain MK. Chem. Rev. 2001;101:2613–2654. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- 28.Wu YZ, Manevich Y, Baldwin JL, Dodia C, Yu K, Feinstein SI, Fisher AB. J. Biol. Chem. 2006;281:7515–7525. doi: 10.1074/jbc.M504525200. [DOI] [PubMed] [Google Scholar]
- 29.van Kuijk FJGM, Sevanian A, Handelman GJ, Dratz EA. Trends Biochem. Sci. 1987;12:31–34. [Google Scholar]
- 30.Zhao L, Wang HP, Zhang HJ, Weydert CJ, Domann FE, Oberley LW, Buettner GR. Arch. Biochem. Biophys. 2003;417:212–218. doi: 10.1016/s0003-9861(03)00342-4. [DOI] [PubMed] [Google Scholar]
- 31.Ursini F, Maiorino M, Gregolin C. Biochim. Biophys. Acta. 1985;839:62–70. doi: 10.1016/0304-4165(85)90182-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Free Radic. Biol. Med. 2004;37:1736–1743. doi: 10.1016/j.freeradbiomed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Feinstein SI, Fisher AB. J. Cell. Biochem. 2008;104:1274–1285. doi: 10.1002/jcb.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Feinstein SI, Manevich Y, Ho YS, Fisher AB. Antioxid Redox Signal. 2006;8:229–237. doi: 10.1089/ars.2006.8.229. [DOI] [PubMed] [Google Scholar]
- 35.Pak JH, Manevich Y, Kim HS, Feinstein SI, Fisher AB. J. Biol. Chem. 2002;277:49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- 36.Chang TS, Jeong W, Woo HA, Lee SM, Park S, Rhee SG. J. Biol. Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 37.Manevich Y, Reddy KS, Shuvaeva T, Feinstein S, Fisher A. J. Lipid Res. 2007 doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Hamza A. J. Biomol. Struct. Dyn. 2002;20:7–20. doi: 10.1080/07391102.2002.10506818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.